

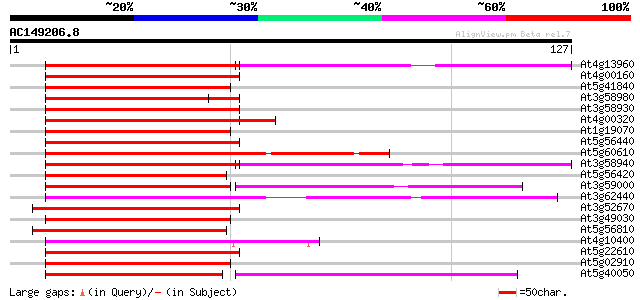
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149206.8 - phase: 0
(127 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g13960 hypothetical protein 61 1e-10
At4g00160 unknown protein 58 1e-09
At5g41840 putative protein 57 2e-09
At3g58980 unknown protein 57 2e-09
At3g58930 putative protein 57 3e-09
At4g00320 57 3e-09
At1g19070 hypothetical protein 57 3e-09
At5g56440 putative protein 56 4e-09
At5g60610 unknown protein 55 7e-09
At3g58940 putative protein 55 7e-09
At5g56420 unknown protein 55 1e-08
At3g59000 unknown protein 55 1e-08
At3g62440 putative protein 54 2e-08
At3g52670 putative protein 54 2e-08
At3g49030 putative protein 54 2e-08
At5g56810 putative protein 54 3e-08
At4g10400 putative protein 54 3e-08
At5g22610 unknown protein 53 5e-08
At5g02910 unknown protein 53 5e-08
At5g40050 putative protein 52 6e-08
>At4g13960 hypothetical protein
Length = 434
Score = 61.2 bits (147), Expect = 1e-10
Identities = 26/44 (59%), Positives = 35/44 (79%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPNAD 52
D +S LPDE++YHILS+L K+ ALTS+LSKRW+ L+ +PN D
Sbjct: 2 DHVSSLPDEVLYHILSFLTTKEAALTSILSKRWRNLFTFVPNLD 45
Score = 38.9 bits (89), Expect = 7e-04
Identities = 25/76 (32%), Positives = 41/76 (53%), Gaps = 5/76 (6%)
Query: 52 DRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVLNKILETNHIHYDDDSDSDMEEEE 111
D +S+LP EVL IL L TK+ +T+ILS RWR + + + D D + +E
Sbjct: 2 DHVSSLPDEVLYHILSFLTTKEAALTSILSKRWRNLFTFV-----PNLDIDDSVFLHPQE 56
Query: 112 EEEEEEEEEEEQQEYV 127
+E+ E ++ ++V
Sbjct: 57 GKEDRYEIQKSFMKFV 72
>At4g00160 unknown protein
Length = 453
Score = 58.2 bits (139), Expect = 1e-09
Identities = 23/44 (52%), Positives = 34/44 (77%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPNAD 52
DRIS+LPD ++ ILS+LP K + TS+ SK+W+PLW+ +PN +
Sbjct: 16 DRISELPDALLIKILSFLPTKIVVATSVFSKQWRPLWKLVPNLE 59
Score = 36.2 bits (82), Expect = 0.005
Identities = 24/66 (36%), Positives = 32/66 (48%), Gaps = 2/66 (3%)
Query: 46 RAMPNADRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVLNKI--LETNHIHYDDDS 103
R + DRIS LP +L IL LPTK T++ S +WR + + LE + YDD
Sbjct: 10 RVVMGKDRISELPDALLIKILSFLPTKIVVATSVFSKQWRPLWKLVPNLEFDSEDYDDKE 69
Query: 104 DSDMEE 109
E
Sbjct: 70 QYTFSE 75
>At5g41840 putative protein
Length = 540
Score = 57.4 bits (137), Expect = 2e-09
Identities = 26/42 (61%), Positives = 33/42 (77%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPN 50
DRIS LPD +I HILS+LP K+ A T++L+KRWKPL +PN
Sbjct: 14 DRISGLPDALICHILSFLPTKEAASTTVLAKRWKPLLAFVPN 55
Score = 34.7 bits (78), Expect = 0.013
Identities = 19/52 (36%), Positives = 32/52 (61%), Gaps = 3/52 (5%)
Query: 50 NADRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVLNKILETNHIHYDD 101
+ DRIS LP ++ IL LPTK+ T +L+ RW+ +L + ++++DD
Sbjct: 12 SGDRISGLPDALICHILSFLPTKEAASTTVLAKRWKPLLAFV---PNLNFDD 60
>At3g58980 unknown protein
Length = 977
Score = 57.4 bits (137), Expect = 2e-09
Identities = 25/44 (56%), Positives = 33/44 (74%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPNAD 52
D +S+LP+E+ HILS+LP K ALTS+LSK W LW+ +PN D
Sbjct: 608 DLVSNLPEEVRCHILSFLPTKHAALTSVLSKSWLNLWKLVPNLD 651
Score = 48.9 bits (115), Expect = 7e-07
Identities = 22/37 (59%), Positives = 30/37 (80%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLW 45
DRIS+LP+EII HI+S+L K+ A S+LSKRW+ L+
Sbjct: 2 DRISNLPNEIICHIVSFLSAKEAAFASILSKRWRNLF 38
Score = 33.1 bits (74), Expect = 0.039
Identities = 18/50 (36%), Positives = 29/50 (58%), Gaps = 3/50 (6%)
Query: 52 DRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVLNKILETNHIHYDD 101
DRIS LP E++ I+ L K+ +ILS RWR + +++ + +DD
Sbjct: 2 DRISNLPNEIICHIVSFLSAKEAAFASILSKRWRNLFTIVIK---LQFDD 48
Score = 32.7 bits (73), Expect = 0.051
Identities = 26/63 (41%), Positives = 34/63 (53%), Gaps = 4/63 (6%)
Query: 52 DRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVLNKILETNHIHYDDDSDSDMEEEE 111
D +S LP EV IL LPTK +T++LS W L K++ I DDS+ + EEE
Sbjct: 608 DLVSNLPEEVRCHILSFLPTKHAALTSVLSKSWLN-LWKLVPNLDI---DDSEFLLPEEE 663
Query: 112 EEE 114
E
Sbjct: 664 IPE 666
>At3g58930 putative protein
Length = 455
Score = 57.0 bits (136), Expect = 3e-09
Identities = 26/44 (59%), Positives = 33/44 (74%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPNAD 52
DR+S+LPD + HILS+LP K IALTS+LSK W LW+ +P D
Sbjct: 2 DRVSNLPDGVRGHILSFLPAKHIALTSVLSKSWLNLWKLIPILD 45
Score = 30.0 bits (66), Expect = 0.33
Identities = 25/66 (37%), Positives = 33/66 (49%), Gaps = 4/66 (6%)
Query: 52 DRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVLNKILETNHIHYDDDSDSDMEEEE 111
DR+S LP V IL LP K +T++LS W L K++ I DDS+ EE
Sbjct: 2 DRVSNLPDGVRGHILSFLPAKHIALTSVLSKSWLN-LWKLIPILDI---DDSEFLHPEEG 57
Query: 112 EEEEEE 117
+ E E
Sbjct: 58 KAERLE 63
>At4g00320
Length = 773
Score = 56.6 bits (135), Expect = 3e-09
Identities = 28/52 (53%), Positives = 35/52 (66%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPNADRISALPFE 60
D IS LPD +I HILS+LP K A T++L+KRWKPL MPN D + F+
Sbjct: 279 DGISGLPDAMICHILSFLPTKVAASTTVLAKRWKPLLAFMPNLDFDESFRFD 330
Score = 51.6 bits (122), Expect = 1e-07
Identities = 22/44 (50%), Positives = 29/44 (65%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPNAD 52
D+ S LPDE++ +LS+LP K TSLLS RWK LW +P +
Sbjct: 2 DKTSQLPDELLVKVLSFLPTKDAVRTSLLSMRWKSLWMWLPKLE 45
Score = 33.1 bits (74), Expect = 0.039
Identities = 20/50 (40%), Positives = 29/50 (58%), Gaps = 2/50 (4%)
Query: 52 DRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVLNKI--LETNHIHY 99
D+ S LP E+L +L LPTK T++LS RW+ + + LE + HY
Sbjct: 2 DKTSQLPDELLVKVLSFLPTKDAVRTSLLSMRWKSLWMWLPKLEYDFRHY 51
Score = 29.6 bits (65), Expect = 0.43
Identities = 16/37 (43%), Positives = 22/37 (59%)
Query: 52 DRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVL 88
D IS LP ++ IL LPTK T +L+ RW+ +L
Sbjct: 279 DGISGLPDAMICHILSFLPTKVAASTTVLAKRWKPLL 315
>At1g19070 hypothetical protein
Length = 83
Score = 56.6 bits (135), Expect = 3e-09
Identities = 27/42 (64%), Positives = 33/42 (78%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPN 50
DRI+ L DEII HILS+LP K+ ALTS+LSKRW+ L+ PN
Sbjct: 2 DRINGLSDEIICHILSFLPTKESALTSVLSKRWRNLFALTPN 43
Score = 38.5 bits (88), Expect = 0.001
Identities = 22/51 (43%), Positives = 31/51 (60%), Gaps = 3/51 (5%)
Query: 52 DRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVLNKILETNHIHYDDD 102
DRI+ L E++ IL LPTK+ +T++LS RWR N T ++H DD
Sbjct: 2 DRINGLSDEIICHILSFLPTKESALTSVLSKRWR---NLFALTPNLHLVDD 49
>At5g56440 putative protein
Length = 430
Score = 56.2 bits (134), Expect = 4e-09
Identities = 23/44 (52%), Positives = 34/44 (77%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPNAD 52
DRIS LPD++++ ILS++P K + T+LLSKRW+ LW+ +P D
Sbjct: 2 DRISLLPDDVVFKILSFVPTKVVVSTNLLSKRWRYLWKHVPKLD 45
Score = 33.9 bits (76), Expect = 0.023
Identities = 23/57 (40%), Positives = 31/57 (54%), Gaps = 3/57 (5%)
Query: 52 DRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVLNKILETNHIHYDDDSDSDME 108
DRIS LP +V+ IL +PTK T +LS RWR + + + + Y D S D E
Sbjct: 2 DRISLLPDDVVFKILSFVPTKVVVSTNLLSKRWRYLWKHVPK---LDYRDPSLVDTE 55
>At5g60610 unknown protein
Length = 388
Score = 55.5 bits (132), Expect = 7e-09
Identities = 32/78 (41%), Positives = 47/78 (60%), Gaps = 2/78 (2%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPNADRISALPFEVLSCILYN 68
DRIS LPDE++ I+S++P K TS+LSKRW+ LW+ +P + P + IL N
Sbjct: 2 DRISGLPDELLVKIISFVPTKVAVSTSILSKRWESLWKWVPKLECDCTEP-ALRDFILKN 60
Query: 69 LPTKQFFVTAILSWRWRR 86
LP + + + L R+RR
Sbjct: 61 LPLQARIIES-LYLRFRR 77
>At3g58940 putative protein
Length = 618
Score = 55.5 bits (132), Expect = 7e-09
Identities = 25/44 (56%), Positives = 32/44 (71%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPNAD 52
DR+S+LP+E+ HILS+LP K ALTS+LSK W LW+ N D
Sbjct: 2 DRVSNLPEEVRCHILSFLPTKHAALTSVLSKSWLNLWKFETNLD 45
Score = 39.3 bits (90), Expect = 5e-04
Identities = 28/76 (36%), Positives = 38/76 (49%), Gaps = 5/76 (6%)
Query: 52 DRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVLNKILETNHIHYDDDSDSDMEEEE 111
DR+S LP EV IL LPTK +T++LS W + ETN D D + EE
Sbjct: 2 DRVSNLPEEVRCHILSFLPTKHAALTSVLSKSWLNLWK--FETN---LDIDDSDFLHPEE 56
Query: 112 EEEEEEEEEEEQQEYV 127
+ E +E + E+V
Sbjct: 57 GKAERDEIRQSFVEFV 72
>At5g56420 unknown protein
Length = 422
Score = 54.7 bits (130), Expect = 1e-08
Identities = 23/41 (56%), Positives = 31/41 (75%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMP 49
DR+S LPD+ + ILS+LP K + +TSLLSKRW+ LW +P
Sbjct: 6 DRLSQLPDDFLLQILSWLPTKDVLVTSLLSKRWRFLWTLVP 46
Score = 37.4 bits (85), Expect = 0.002
Identities = 20/45 (44%), Positives = 27/45 (59%)
Query: 52 DRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVLNKILETNH 96
DR+S LP + L IL LPTK VT++LS RWR + + N+
Sbjct: 6 DRLSQLPDDFLLQILSWLPTKDVLVTSLLSKRWRFLWTLVPRLNY 50
>At3g59000 unknown protein
Length = 491
Score = 54.7 bits (130), Expect = 1e-08
Identities = 25/42 (59%), Positives = 32/42 (75%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPN 50
DR+ LPDE++ HILS+L K+ ALTSLLSKRW+ L +PN
Sbjct: 2 DRVGSLPDELLSHILSFLTTKEAALTSLLSKRWRYLIAFVPN 43
Score = 38.9 bits (89), Expect = 7e-04
Identities = 25/65 (38%), Positives = 38/65 (58%), Gaps = 3/65 (4%)
Query: 52 DRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVLNKILETNHIHYDDDSDSDMEEEE 111
DR+ +LP E+LS IL L TK+ +T++LS RWR + I ++ +DD EE +
Sbjct: 2 DRVGSLPDELLSHILSFLTTKEAALTSLLSKRWRYL---IAFVPNLAFDDIVFLHPEEGK 58
Query: 112 EEEEE 116
E +E
Sbjct: 59 PERDE 63
>At3g62440 putative protein
Length = 457
Score = 53.9 bits (128), Expect = 2e-08
Identities = 37/116 (31%), Positives = 63/116 (53%), Gaps = 11/116 (9%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPNADRISALPFEVLSCILYN 68
DRIS+LPDEII HI S+L ++ A T++LSKRW L+ +P+ S++ +
Sbjct: 2 DRISNLPDEIICHIGSFLSAREAAFTTVLSKRWHNLFTIVPDLHFDSSVK---------D 52
Query: 69 LPTKQFFVTAILSWRWRRVLNKILETNHIHYDDDSDSDMEEEEEEEEEEEEEEEQQ 124
+ FV +++ +NK+ + +D+D+DS +E+ E E+ E + Q
Sbjct: 53 GESLTDFVDRVMALPASSRVNKL--SLKWWFDEDTDSAQFDEDTEPEDTEPAQFDQ 106
>At3g52670 putative protein
Length = 384
Score = 53.9 bits (128), Expect = 2e-08
Identities = 21/47 (44%), Positives = 35/47 (73%)
Query: 6 TTTDRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPNAD 52
T DR++ LP+++I ILS+LP + + TS+LSK+W+ LW+ +PN +
Sbjct: 7 TKEDRMNQLPEDLILRILSFLPTELVIATSVLSKQWRSLWKLVPNLE 53
Score = 31.2 bits (69), Expect = 0.15
Identities = 16/51 (31%), Positives = 30/51 (58%), Gaps = 3/51 (5%)
Query: 52 DRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVLNKILETNHIHYDDD 102
DR++ LP +++ IL LPT+ T++LS +WR + + ++ +D D
Sbjct: 10 DRMNQLPEDLILRILSFLPTELVIATSVLSKQWRSLWKLV---PNLEFDSD 57
>At3g49030 putative protein
Length = 443
Score = 53.9 bits (128), Expect = 2e-08
Identities = 21/42 (50%), Positives = 33/42 (78%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPN 50
DRIS+LP++++ ILS +P + + TS+LSKRW+ LW+ +PN
Sbjct: 21 DRISELPEDLLLQILSDIPTENVIATSVLSKRWRSLWKMVPN 62
Score = 37.0 bits (84), Expect = 0.003
Identities = 17/34 (50%), Positives = 24/34 (70%)
Query: 52 DRISALPFEVLSCILYNLPTKQFFVTAILSWRWR 85
DRIS LP ++L IL ++PT+ T++LS RWR
Sbjct: 21 DRISELPEDLLLQILSDIPTENVIATSVLSKRWR 54
>At5g56810 putative protein
Length = 435
Score = 53.5 bits (127), Expect = 3e-08
Identities = 22/44 (50%), Positives = 32/44 (72%)
Query: 6 TTTDRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMP 49
T+ DRIS LP+++++ ILS +P TSLLSKRWK +W+ +P
Sbjct: 12 TSPDRISQLPNDLLFRILSLIPVSDAMSTSLLSKRWKSVWKMLP 55
Score = 33.1 bits (74), Expect = 0.039
Identities = 22/56 (39%), Positives = 35/56 (62%), Gaps = 3/56 (5%)
Query: 52 DRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVLNKILETNHIHYDDDSDSDM 107
DRIS LP ++L IL +P T++LS RW+ V K+L T + Y+++S S++
Sbjct: 15 DRISQLPNDLLFRILSLIPVSDAMSTSLLSKRWKSVW-KMLPT--LVYNENSCSNI 67
>At4g10400 putative protein
Length = 326
Score = 53.5 bits (127), Expect = 3e-08
Identities = 31/67 (46%), Positives = 40/67 (59%), Gaps = 5/67 (7%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMP----NADRISALPFEVLSC 64
DRIS LPDE++ ILS++P K TS+LSKRW+ LW + + R S F+ L C
Sbjct: 2 DRISGLPDEVLVKILSFVPTKVAVSTSILSKRWEFLWMWLTKLKFGSKRYSESEFKRLQC 61
Query: 65 IL-YNLP 70
L NLP
Sbjct: 62 FLDRNLP 68
Score = 35.4 bits (80), Expect = 0.008
Identities = 20/33 (60%), Positives = 22/33 (66%)
Query: 52 DRISALPFEVLSCILYNLPTKQFFVTAILSWRW 84
DRIS LP EVL IL +PTK T+ILS RW
Sbjct: 2 DRISGLPDEVLVKILSFVPTKVAVSTSILSKRW 34
>At5g22610 unknown protein
Length = 502
Score = 52.8 bits (125), Expect = 5e-08
Identities = 24/44 (54%), Positives = 30/44 (67%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPNAD 52
D IS LP+ ++ ILSYLP K I TS+LSKRWK +W +P D
Sbjct: 18 DLISKLPEVLLSQILSYLPTKDIVRTSVLSKRWKSVWLLIPGLD 61
Score = 35.0 bits (79), Expect = 0.010
Identities = 19/36 (52%), Positives = 23/36 (63%)
Query: 52 DRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRV 87
D IS LP +LS IL LPTK T++LS RW+ V
Sbjct: 18 DLISKLPEVLLSQILSYLPTKDIVRTSVLSKRWKSV 53
>At5g02910 unknown protein
Length = 458
Score = 52.8 bits (125), Expect = 5e-08
Identities = 24/42 (57%), Positives = 30/42 (71%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPN 50
D IS LPDEI++HILS +P K TSLLSKRW+ +W P+
Sbjct: 11 DFISSLPDEILHHILSSVPTKSAIRTSLLSKRWRYVWSETPS 52
Score = 37.0 bits (84), Expect = 0.003
Identities = 19/36 (52%), Positives = 25/36 (68%)
Query: 52 DRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRV 87
D IS+LP E+L IL ++PTK T++LS RWR V
Sbjct: 11 DFISSLPDEILHHILSSVPTKSAIRTSLLSKRWRYV 46
>At5g40050 putative protein
Length = 415
Score = 52.4 bits (124), Expect = 6e-08
Identities = 24/40 (60%), Positives = 31/40 (77%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAM 48
D IS LPDE++ HILS+LP K+ A TS+LSKRW+ L+ M
Sbjct: 14 DSISPLPDELLSHILSFLPTKRAASTSILSKRWRTLFPLM 53
Score = 44.7 bits (104), Expect = 1e-05
Identities = 29/64 (45%), Positives = 36/64 (55%)
Query: 52 DRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVLNKILETNHIHYDDDSDSDMEEEE 111
D IS LP E+LS IL LPTK+ T+ILS RWR + + Y DD+D E +
Sbjct: 14 DSISPLPDELLSHILSFLPTKRAASTSILSKRWRTLFPLMNHLCASLYLDDTDLLYPERQ 73
Query: 112 EEEE 115
EEE
Sbjct: 74 TEEE 77
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.133 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,545,614
Number of Sequences: 26719
Number of extensions: 171913
Number of successful extensions: 12835
Number of sequences better than 10.0: 900
Number of HSP's better than 10.0 without gapping: 847
Number of HSP's successfully gapped in prelim test: 54
Number of HSP's that attempted gapping in prelim test: 3865
Number of HSP's gapped (non-prelim): 4381
length of query: 127
length of database: 11,318,596
effective HSP length: 87
effective length of query: 40
effective length of database: 8,994,043
effective search space: 359761720
effective search space used: 359761720
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 54 (25.4 bits)
Medicago: description of AC149206.8


