

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148819.7 - phase: 0
(343 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
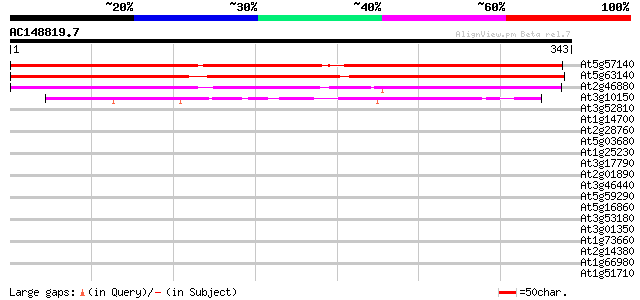
At5g57140 unknown protein 458 e-129
At5g63140 unknown protein 326 1e-89
At2g46880 unknown protein 294 5e-80
At3g10150 hypothetical protein 92 3e-19
At3g52810 purple acid phosphatase-like protein 35 0.078
At1g14700 purple acid phosphatase -like protein 33 0.17
At2g28760 putative nucleotide-sugar dehydratase 32 0.39
At5g03680 GT2 -like protein 32 0.66
At1g25230 unknown protein 32 0.66
At3g17790 acid phosphatase type 5 30 1.5
At2g01890 purple acid phosphatase like protein 30 1.5
At3g46440 dTDP-glucose 4-6-dehydratases-like protein 30 2.5
At5g59290 dTDP-glucose 4-6-dehydratase - like protein 29 3.3
At5g16860 putative protein 29 3.3
At3g53180 nodulin / glutamate-ammonia ligase - like protein 29 3.3
At3g01350 putative peptide transporter 29 3.3
At1g73660 putative protein kinase 29 3.3
At2g14380 putative retroelement pol polyprotein 29 4.3
At1g66980 putative kinase 28 5.6
At1g51710 ubiquitin-specific protease 6 (UBP6) 28 5.6
>At5g57140 unknown protein
Length = 397
Score = 458 bits (1179), Expect = e-129
Identities = 221/339 (65%), Positives = 265/339 (77%), Gaps = 15/339 (4%)
Query: 1 MHFGNG-ITKCRDVLASEFEFCSDLNTTLFLKRVIQDETPDFIAFTGDNIFGPSSHDAAE 59
MHFG G IT+CRDVL SEFE+CSDLNTT FL+R+I+ E PD IAFTGDNIFG S+ DAAE
Sbjct: 66 MHFGMGMITRCRDVLDSEFEYCSDLNTTRFLRRMIESERPDLIAFTGDNIFGSSTTDAAE 125
Query: 60 SMFKAFGPAMESGLPWAAILGNHDQESTLNREELMSLISLMDYSVSQINPSADSLTNSAK 119
S+ +A GPA+E G+PWAA+LGNHD ESTLNR ELM+ +SLMD+SVSQINP + T K
Sbjct: 126 SLLEAIGPAIEYGIPWAAVLGNHDHESTLNRLELMTFLSLMDFSVSQINPLVEDET---K 182
Query: 120 GHKMSKIDGFGNYNLRVYGAPGSMMANSSVLNLFFLDSGDRVVYQGIRTYDWIKDSQLHW 179
G M IDGFGNY +RVYGAPGS++ANS+V +LFF DSGDR + QG RTY WIK+SQL W
Sbjct: 183 GDTMRLIDGFGNYRVRVYGAPGSVLANSTVFDLFFFDSGDREIVQGKRTYGWIKESQLRW 242
Query: 180 LRHVSQEPQAQEQDPLHSTDHVTSPITPPALAFFHIPIPEVRQLFYKQIVGQFQEGVACS 239
L+ S + +Q +H + PPALAFFHIPI EVR+L+Y +GQFQEGVACS
Sbjct: 243 LQDTSIQGHSQR---IH--------VNPPALAFFHIPILEVRELWYTPFIGQFQEGVACS 291
Query: 240 RVNSAVLQTFVSMGDVKAVFIGHDHTNDFCGNLDGIWFCYGGGFGYHGYGKAGWPRRARI 299
V S VLQTFVSMG+VKA F+GHDH NDFCG L G+WFCYGGGFGYH YG+ W RRAR+
Sbjct: 292 IVQSGVLQTFVSMGNVKAAFMGHDHVNDFCGTLKGVWFCYGGGFGYHAYGRPNWHRRARV 351
Query: 300 ILAELQKGKESWTSVQKIMTWKRLDDEKMSKIDEQILWD 338
I A+L KG+++W ++ I TWKRLDDE +SKIDEQ+LW+
Sbjct: 352 IEAKLGKGRDTWEGIKLIKTWKRLDDEYLSKIDEQVLWE 390
>At5g63140 unknown protein
Length = 389
Score = 326 bits (836), Expect = 1e-89
Identities = 166/342 (48%), Positives = 217/342 (62%), Gaps = 18/342 (5%)
Query: 1 MHFGNGI-TKCRDVLASEFEFCSDLNTTLFLKRVIQDETPDFIAFTGDNIFGPSSHDAAE 59
MHF NG T+C++VL S+ CSDLNTT+F+ RVI E PD I FTGDNIFG DA +
Sbjct: 55 MHFANGAKTQCQNVLPSQRAHCSDLNTTIFMSRVIAAEKPDLIVFTGDNIFGFDVKDALK 114
Query: 60 SMFKAFGPAMESGLPWAAILGNHDQESTLNREELMSLISLMDYSVSQINPSADSLTNSAK 119
S+ AF PA+ S +PW AILGNHDQEST R+++M+ I + ++SQ+NP
Sbjct: 115 SINAAFAPAIASKIPWVAILGNHDQESTFTRQQVMNHIVKLPNTLSQVNPP--------- 165
Query: 120 GHKMSKIDGFGNYNLRVYGAPGSMMANSSVLNLFFLDSGDRVVYQGIRTYDWIKDSQLHW 179
IDGFGNYNL+++GA S + N SVLNL+FLDSGD + YDWIK SQ W
Sbjct: 166 -EAAHYIDGFGNYNLQIHGAADSKLQNKSVLNLYFLDSGDYSSVPYMEGYDWIKTSQQFW 224
Query: 180 LRHVSQEPQAQEQDPLHSTDHVTSPITPPALAFFHIPIPEVRQLFYKQIV-GQFQEGVAC 238
S+ + + + + + P LA+FHIP+PE K G QEG +
Sbjct: 225 FDRTSKRLKREYNAKPNPQEGIA-----PGLAYFHIPLPEFLSFDSKNATKGVRQEGTSA 279
Query: 239 SRVNSAVLQTFVSMGDVKAVFIGHDHTNDFCGNLDGIWFCYGGGFGYHGYGKAGWPRRAR 298
+ NS T ++ GDVK+VF+GHDH NDFCG L G+ CYGGGFGYH YGKAGW RRAR
Sbjct: 280 ASTNSGFFTTLIARGDVKSVFVGHDHVNDFCGELKGLNLCYGGGFGYHAYGKAGWERRAR 339
Query: 299 IILAEL-QKGKESWTSVQKIMTWKRLDDEKMSKIDEQILWDH 339
+++ +L +K K W +V+ I TWKRLDD+ +S ID Q+LW++
Sbjct: 340 VVVVDLNKKRKGKWGAVKSIKTWKRLDDKHLSVIDSQVLWNN 381
>At2g46880 unknown protein
Length = 401
Score = 294 bits (752), Expect = 5e-80
Identities = 160/343 (46%), Positives = 202/343 (58%), Gaps = 21/343 (6%)
Query: 1 MHFGNGI-TKCRDVLASEFEFCSDLNTTLFLKRVIQDETPDFIAFTGDNIFGP-SSHDAA 58
MH+G G T+C DV +EF +CSDLNTT FL+R I E PD I F+GDN++G + D A
Sbjct: 54 MHYGFGKETQCSDVSPAEFPYCSDLNTTSFLQRTIASEKPDLIVFSGDNVYGLCETSDVA 113
Query: 59 ESMFKAFGPAMESGLPWAAILGNHDQESTLNREELMSLISLMDYSVSQINPSADSLTNSA 118
+SM AF PA+ESG+PW AILGNHDQES + RE +M I + S+SQ+NP L
Sbjct: 114 KSMDMAFAPAIESGIPWVAILGNHDQESDMTRETMMKYIMKLPNSLSQVNPPDAWLY--- 170
Query: 119 KGHKMSKIDGFGNYNLRVYGAPGSMMANSSVLNLFFLDSGDRVVYQGIR-TYDWIKDSQL 177
+IDGFGNYNL++ G GS + S+LNL+ LD G G YDW+K SQ
Sbjct: 171 ------QIDGFGNYNLQIEGPFGSPLFFKSILNLYLLDGGSYTKLDGFGYKYDWVKTSQQ 224
Query: 178 HWLRHVSQEPQAQEQDPLHSTDHVTSPITPPALAFFHIPIPEVRQLFYK--QIVGQFQEG 235
+W H S+ + + H T P L + HIP+PE LF K ++ G QE
Sbjct: 225 NWYEHTSKWLEME-----HKRWPFPQNSTAPGLVYLHIPMPEFA-LFNKSTEMTGVRQES 278
Query: 236 VACSRVNSAVLQTFVSMGDVKAVFIGHDHTNDFCGNLDGIWFCYGGGFGYHGYGKAGWPR 295
+NS V G+VK VF GHDH NDFC L GI CY GG GYHGYG+ GW R
Sbjct: 279 TCSPPINSGFFTKLVERGEVKGVFSGHDHVNDFCAELHGINLCYAGGAGYHGYGQVGWAR 338
Query: 296 RARIILAELQKGKES-WTSVQKIMTWKRLDDEKMSKIDEQILW 337
R R++ A+L+K W +V I TWKRLDD+ S ID Q+LW
Sbjct: 339 RVRVVEAQLEKTMYGRWGAVDTIKTWKRLDDKNHSLIDTQLLW 381
>At3g10150 hypothetical protein
Length = 367
Score = 92.4 bits (228), Expect = 3e-19
Identities = 86/332 (25%), Positives = 137/332 (40%), Gaps = 63/332 (18%)
Query: 23 DLNTTLFLKRVIQDETPDFIAFTGDNIFGPSSHDAAESMF--KAFGPAMESGLPWAAILG 80
D+N+ + V+ ETPDF+ + GD + + S+F KA P + G+PWA + G
Sbjct: 58 DVNSVNVMSAVLDAETPDFVVYLGDVVTANNIAIQNASLFWDKAISPTRDRGIPWATLFG 117
Query: 81 NHDQESTLNREELMSLISLMDYS--------------------VSQINPSADSLTNSAKG 120
NHD S + + +S + + + S+++L+ S
Sbjct: 118 NHDDASFVWPLDWLSSSGIPPLRCPAASDDDGCTFRGTTRVELIQEEIKSSNALSYSMIS 177
Query: 121 HKMSKIDGFGNYNLRVYGAPGSMMANSSVLNLFFLDSGDRVVYQGIRTYDWIKDSQLHWL 180
K NY L V + S V L+FLDSG G + I ++Q+ W
Sbjct: 178 PK-ELWPSVSNYVLLVESSDHS---KPPVALLYFLDSG------GGSYPEVISNAQVEWF 227
Query: 181 RHVSQEPQAQEQDPLHSTDHVTSPITPPALAFFHIPIPEVRQL-----FYKQIVGQF-QE 234
+ S + + P L F+HIP +++ K VG +E
Sbjct: 228 KTKSNT--------------LNPYLRIPELIFWHIPSKAYKKVAPRLWITKPCVGSINKE 273
Query: 235 GVACSRVNSAVLQTFVSMGDVKAVFIGHDHTNDF-CGNLDGIWFCYGGGFGYHGYGKAGW 293
V + +++ + VKAVF+GH+H D+ C D +W C+ GY GYG W
Sbjct: 274 KVVAQEAENGMMRVLENRSSVKAVFVGHNHGLDWCCPYKDKLWLCFARHTGYGGYG--NW 331
Query: 294 PRRARIILAELQKGKESWTSVQKIMTWKRLDD 325
PR +RI+ E +I TW R++D
Sbjct: 332 PRGSRIL--------EISEMPFRIKTWIRMED 355
>At3g52810 purple acid phosphatase-like protein
Length = 437
Score = 34.7 bits (78), Expect = 0.078
Identities = 32/107 (29%), Positives = 47/107 (43%), Gaps = 14/107 (13%)
Query: 17 EFEFCSDLNTTLFLKRVI-QDETPDFIAFT--GDNIFGPSSHDAAESMFKAFGPAME--- 70
EF DL T + R + Q DF F GD + D + ++ +FG +E
Sbjct: 146 EFAVAGDLGQTDWTVRTLDQIRKRDFDVFLLPGDLSYA----DTHQPLWDSFGRLLETLA 201
Query: 71 SGLPWAAILGNHDQESTLNREELMSLISLMDYSVSQINPSADSLTNS 117
S PW GNH+ ES + IS Y+ + P A+SL++S
Sbjct: 202 STRPWMVTEGNHEIESFPTNDH----ISFKSYNARWLMPHAESLSHS 244
>At1g14700 purple acid phosphatase -like protein
Length = 366
Score = 33.5 bits (75), Expect = 0.17
Identities = 21/66 (31%), Positives = 31/66 (46%), Gaps = 5/66 (7%)
Query: 40 DFIAFTGDNIFG---PSSHDAA-ESMFKAFGPAMESGLPWAAILGNHDQESTLNREELMS 95
DF+ TGDN + S HD + F A PW ++LGNHD + R +L
Sbjct: 107 DFVISTGDNFYDNGLTSLHDPLFQDSFTNIYTAPSLQKPWYSVLGNHDYRGDV-RAQLSP 165
Query: 96 LISLMD 101
++ +D
Sbjct: 166 MLRALD 171
>At2g28760 putative nucleotide-sugar dehydratase
Length = 343
Score = 32.3 bits (72), Expect = 0.39
Identities = 30/126 (23%), Positives = 52/126 (40%), Gaps = 21/126 (16%)
Query: 171 WIKDSQLHWLRHVSQEPQAQEQDPLHSTDHVTSPITPPALAFFHIPIPEVRQLFYKQIVG 230
WI + +RH EP E D ++ H+ P +P + + + P+ ++ ++G
Sbjct: 75 WIGHPRFELIRHDVTEPLFVEVDQIY---HLACPASP--IFYKYNPVKTIK----TNVIG 125
Query: 231 QFQEGVACSRVNSAVLQTFVSMGDVKAVFIGHDHTNDFCGNLD--GIWFCYGGG------ 282
RV + +L T S +V + H T + GN++ G+ CY G
Sbjct: 126 TLNMLGLAKRVGARILLT--STSEVYGDPLVHPQTESYWGNVNPIGVRSCYDEGKRVAET 183
Query: 283 --FGYH 286
F YH
Sbjct: 184 LMFDYH 189
>At5g03680 GT2 -like protein
Length = 591
Score = 31.6 bits (70), Expect = 0.66
Identities = 23/63 (36%), Positives = 30/63 (47%), Gaps = 13/63 (20%)
Query: 268 FCGNLDGIWFCYGGGFGYHGYGKAGWPRRARIILAELQ-----KGKES------WTSVQK 316
F G LDG F G G G G G WPR+ + L E++ K KE+ W V +
Sbjct: 97 FSGFLDGGGF--GSGVGGDGGGTGRWPRQETLTLLEIRSRLDHKFKEANQKGPLWDEVSR 154
Query: 317 IMT 319
IM+
Sbjct: 155 IMS 157
>At1g25230 unknown protein
Length = 339
Score = 31.6 bits (70), Expect = 0.66
Identities = 23/78 (29%), Positives = 37/78 (46%), Gaps = 5/78 (6%)
Query: 28 LFLKRVIQDETPDFIAFTGDNIF--GPSSHD--AAESMFKAFGPAMESGLPWAAILGNHD 83
L + R+ ++ +F+ TGDNI+ G S D A + F + PW +LGNHD
Sbjct: 67 LQMGRIGEEMDINFVVSTGDNIYDNGMKSIDDPAFQLSFSNIYTSPSLQKPWYLVLGNHD 126
Query: 84 QESTLNREELMSLISLMD 101
+ +L ++ MD
Sbjct: 127 YRGDV-EAQLSPILRSMD 143
>At3g17790 acid phosphatase type 5
Length = 338
Score = 30.4 bits (67), Expect = 1.5
Identities = 18/48 (37%), Positives = 22/48 (45%), Gaps = 4/48 (8%)
Query: 40 DFIAFTGDNIFGP---SSHDAA-ESMFKAFGPAMESGLPWAAILGNHD 83
DF+ TGDN + S HD E F A W ++LGNHD
Sbjct: 79 DFVVSTGDNFYDNGLFSEHDPNFEQSFSNIYTAPSLQKQWYSVLGNHD 126
>At2g01890 purple acid phosphatase like protein
Length = 335
Score = 30.4 bits (67), Expect = 1.5
Identities = 19/60 (31%), Positives = 29/60 (47%), Gaps = 4/60 (6%)
Query: 28 LFLKRVIQDETPDFIAFTGDNIFGP---SSHDAA-ESMFKAFGPAMESGLPWAAILGNHD 83
L + ++ +D DF+ TGDN + S +D+ + F A PW +LGNHD
Sbjct: 66 LQMGKIGKDLNIDFLISTGDNFYDDGIISPYDSQFQDSFTNIYTATSLQKPWYNVLGNHD 125
>At3g46440 dTDP-glucose 4-6-dehydratases-like protein
Length = 341
Score = 29.6 bits (65), Expect = 2.5
Identities = 29/126 (23%), Positives = 51/126 (40%), Gaps = 21/126 (16%)
Query: 171 WIKDSQLHWLRHVSQEPQAQEQDPLHSTDHVTSPITPPALAFFHIPIPEVRQLFYKQIVG 230
WI + +RH EP E D ++ H+ P +P + + + P+ ++ ++G
Sbjct: 73 WIGHPRFELIRHDVTEPLLIEVDQIY---HLACPASP--IFYKYNPVKTIK----TNVIG 123
Query: 231 QFQEGVACSRVNSAVLQTFVSMGDVKAVFIGHDHTNDFCGNLD--GIWFCYGGG------ 282
RV + +L T S +V + H + GN++ G+ CY G
Sbjct: 124 TLNMLGLAKRVGARILLT--STSEVYGDPLIHPQPESYWGNVNPIGVRSCYDEGKRVAET 181
Query: 283 --FGYH 286
F YH
Sbjct: 182 LMFDYH 187
>At5g59290 dTDP-glucose 4-6-dehydratase - like protein
Length = 342
Score = 29.3 bits (64), Expect = 3.3
Identities = 29/126 (23%), Positives = 51/126 (40%), Gaps = 21/126 (16%)
Query: 171 WIKDSQLHWLRHVSQEPQAQEQDPLHSTDHVTSPITPPALAFFHIPIPEVRQLFYKQIVG 230
WI + +RH EP E D ++ H+ P +P + + + P+ ++ ++G
Sbjct: 74 WIGHPRFELIRHDVTEPLLIEVDRIY---HLACPASP--IFYKYNPVKTIK----TNVIG 124
Query: 231 QFQEGVACSRVNSAVLQTFVSMGDVKAVFIGHDHTNDFCGNLD--GIWFCYGGG------ 282
RV + +L T S +V + H + GN++ G+ CY G
Sbjct: 125 TLNMLGLAKRVGARILLT--STSEVYGDPLIHPQPESYWGNVNPIGVRSCYDEGKRVAET 182
Query: 283 --FGYH 286
F YH
Sbjct: 183 LMFDYH 188
>At5g16860 putative protein
Length = 850
Score = 29.3 bits (64), Expect = 3.3
Identities = 15/60 (25%), Positives = 28/60 (46%), Gaps = 2/60 (3%)
Query: 56 DAAESMFKAFGPAMESGLPWAAILGNHDQESTLNREELMSLISLMDYSVSQINPSADSLT 115
D A +MF + P + W ++G + Q N+ + L+S M Q P+A +++
Sbjct: 424 DTARAMFDSLSPKERDVVTWTVMIGGYSQHGDANK--ALELLSEMFEEDCQTRPNAFTIS 481
>At3g53180 nodulin / glutamate-ammonia ligase - like protein
Length = 845
Score = 29.3 bits (64), Expect = 3.3
Identities = 19/62 (30%), Positives = 29/62 (46%), Gaps = 3/62 (4%)
Query: 13 VLASEFEFCSDL---NTTLFLKRVIQDETPDFIAFTGDNIFGPSSHDAAESMFKAFGPAM 69
VL EF+ + N LK V+++ +++ F SS DAA +F PA+
Sbjct: 520 VLKDEFDLVMNAGFENEFYLLKNVVREGKEEYMPFDFGPYCATSSFDAASPIFHDIVPAL 579
Query: 70 ES 71
ES
Sbjct: 580 ES 581
>At3g01350 putative peptide transporter
Length = 563
Score = 29.3 bits (64), Expect = 3.3
Identities = 14/54 (25%), Positives = 23/54 (41%)
Query: 72 GLPWAAILGNHDQESTLNREELMSLISLMDYSVSQINPSADSLTNSAKGHKMSK 125
GL W A G+ T++ L S + L+ + +NPS + H + K
Sbjct: 95 GLTWTAFAGSRSATKTISSYFLYSSLCLVSIGLGVLNPSLQAFGADQLDHDLDK 148
>At1g73660 putative protein kinase
Length = 1030
Score = 29.3 bits (64), Expect = 3.3
Identities = 20/80 (25%), Positives = 34/80 (42%), Gaps = 4/80 (5%)
Query: 49 IFGPSSHDAAESMFKAFGPAMESGLPWAAILGNHDQESTLNREELMSLISLMDYSVSQIN 108
+ SS + + G + G+ W A+L N +S L R E M+L D + +
Sbjct: 208 VLNASSAERIPPLLDLQGTPVSDGVTWEAVLVNRSGDSNLLRLEQMAL----DIAAKSRS 263
Query: 109 PSADSLTNSAKGHKMSKIDG 128
S+ NS K++ + G
Sbjct: 264 VSSSGFVNSELVRKLAILVG 283
>At2g14380 putative retroelement pol polyprotein
Length = 764
Score = 28.9 bits (63), Expect = 4.3
Identities = 15/46 (32%), Positives = 24/46 (51%), Gaps = 2/46 (4%)
Query: 6 GITKCRDVLASEFEFCSDLNTTLFLKRVIQDETPDFIAFTGDNIFG 51
G+T CRD S E+ TL+ ++ TP + FT +++FG
Sbjct: 296 GLTACRDSFRSIKEYIKSGAATLWSSPATKEMTP--LTFTSEDLFG 339
>At1g66980 putative kinase
Length = 1109
Score = 28.5 bits (62), Expect = 5.6
Identities = 12/28 (42%), Positives = 17/28 (59%)
Query: 184 SQEPQAQEQDPLHSTDHVTSPITPPALA 211
S P AQ+ +P+ + D VT P PP +A
Sbjct: 699 SSLPPAQDPNPIFTHDDVTEPPLPPVIA 726
>At1g51710 ubiquitin-specific protease 6 (UBP6)
Length = 482
Score = 28.5 bits (62), Expect = 5.6
Identities = 23/108 (21%), Positives = 47/108 (43%), Gaps = 8/108 (7%)
Query: 13 VLASEFEFCSDLNTTLFLKRVIQDETPDFIAFTGDNIFGPSSHDAAESMFKAFGPAMESG 72
VL ++ S L + +++ ++ + ++ P+S + A+++ FG ++S
Sbjct: 177 VLRKKYPQFSQLQNGMHMQQDAEECWTQLLYTLSQSLKAPTSSEGADAVKALFGVNLQSR 236
Query: 73 LPWAAILGNHDQESTLNREELMSLISLMDYSVSQINPSADSLTNSAKG 120
L H QES E S+ SL + ++N + L + KG
Sbjct: 237 L--------HCQESGEESSETESVYSLKCHISHEVNHLHEGLKHGLKG 276
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.137 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,154,620
Number of Sequences: 26719
Number of extensions: 346503
Number of successful extensions: 908
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 880
Number of HSP's gapped (non-prelim): 28
length of query: 343
length of database: 11,318,596
effective HSP length: 100
effective length of query: 243
effective length of database: 8,646,696
effective search space: 2101147128
effective search space used: 2101147128
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148819.7


