

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148718.7 + phase: 0
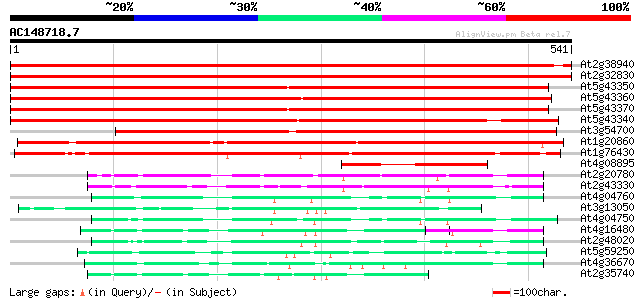
(541 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g38940 phosphate transporter (AtPT2) 852 0.0
At2g32830 phosphate transporter like protein 851 0.0
At5g43350 phosphate transporter (gb|AAB17265.1) 826 0.0
At5g43360 inorganic phosphate transporter (dbj|BAA24281.1) 822 0.0
At5g43370 inorganic phosphate transporter (dbj|BAA24282.1) 820 0.0
At5g43340 inorganic phosphate transporter (dbj|BAA34390.1) 702 0.0
At3g54700 phosphate transport protein 664 0.0
At1g20860 putative inorganic phosphate transporter protein 490 e-139
At1g76430 putative phosphate transporter 483 e-136
At4g08895 putative protein 151 1e-36
At2g20780 putative sugar transporter 82 8e-16
At2g43330 membrane transporter like protein 75 7e-14
At4g04760 putative sugar transporter 69 9e-12
At3g13050 putative transporter 65 1e-10
At4g04750 putative sugar transporter 60 3e-09
At4g16480 membrane transporter like protein 59 7e-09
At2g48020 sugar transporter like protein 59 7e-09
At5g59250 D-xylose-H+ symporter - like protein 58 1e-08
At4g36670 sugar transporter like protein 56 4e-08
At2g35740 putative sugar transporter 56 4e-08
>At2g38940 phosphate transporter (AtPT2)
Length = 534
Score = 852 bits (2201), Expect = 0.0
Identities = 411/541 (75%), Positives = 469/541 (85%), Gaps = 8/541 (1%)
Query: 1 MAREQLGVLTALDAAKTQWYHFTAIIIAGMGFFTDAYDLFCIPNVTKLLGRIYYTHPGAL 60
MAREQL VL ALD AKTQWYHFTAIIIAGMGFFTDAYDLFCI VTKLLGRIYY GA
Sbjct: 1 MAREQLQVLNALDVAKTQWYHFTAIIIAGMGFFTDAYDLFCISLVTKLLGRIYYHVEGAQ 60
Query: 61 KPGTLPPNVSAAVNGVALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFG 120
KPGTLPPNV+AAVNGVA CGTLAGQLFFGWLGDK+GRKKVYG+TL +MV S+ASGLSFG
Sbjct: 61 KPGTLPPNVAAAVNGVAFCGTLAGQLFFGWLGDKLGRKKVYGMTLMVMVLCSIASGLSFG 120
Query: 121 HSAKGTIGTLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGG 180
H K + TLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAF+++VFAMQGFGI+AGG
Sbjct: 121 HEPKAVMATLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFVSAVFAMQGFGIMAGG 180
Query: 181 IVSLVVSTAFDHAFKAPPYKVDAAASLVPEADYVWRIILMFGAVPAGLTYYWRMKMPETA 240
I ++++S+AF+ F +P Y DA S +P+AD VWRIILM GA+PA +TYY R KMPETA
Sbjct: 181 IFAIIISSAFEAKFPSPAYADDALGSTIPQADLVWRIILMAGAIPAAMTYYSRSKMPETA 240
Query: 241 RYTALVAKNAKQAAQDMSSVLQVEIEAEQEKVDKIGVQDKNSFGLFSKEFLRRHGLHLLG 300
RYTALVAK+AKQAA DMS VLQVEIE EQ+K+++I + +FGLFSKEF+ RHGLHLLG
Sbjct: 241 RYTALVAKDAKQAASDMSKVLQVEIEPEQQKLEEISKEKSKAFGLFSKEFMSRHGLHLLG 300
Query: 301 TTSTWFLLDIAYYSSNLFQKDIYSSIGWLPPAQDMNAIHEVFKVARATTLIALCGTVPGY 360
TTSTWFLLDIA+YS NLFQKDI+S+IGW+PPAQ MNAI EVFK+ARA TLIALC TVPGY
Sbjct: 301 TTSTWFLLDIAFYSQNLFQKDIFSAIGWIPPAQSMNAIQEVFKIARAQTLIALCSTVPGY 360
Query: 361 WFTVAFIDVIGRFAIQLMGFFFMTVFMFALAIPYDHWTKRENRIGFLVMYAMTFFFANFG 420
WFTVAFIDVIGRFAIQ+MGFFFMTVFMFALAIPY+HWT +ENRIGF++MY++TFFFANFG
Sbjct: 361 WFTVAFIDVIGRFAIQMMGFFFMTVFMFALAIPYNHWTHKENRIGFVIMYSLTFFFANFG 420
Query: 421 PNSTTFVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQSKDPKKRDAGYPAGI 480
PN+TTFVVPAEIFPAR RSTCHGIS+A+GK GA++GAFGFLY +Q+ D K DAGYP GI
Sbjct: 421 PNATTFVVPAEIFPARFRSTCHGISAASGKLGAMVGAFGFLYLAQNPDKDKTDAGYPPGI 480
Query: 481 GMKNTLILLAVVNCLGIFFTFLVPEANGKSLEEMSGENEEEEETEAAVVDPQASSNRTVP 540
G++N+LI+L VVN LGI FTFLVPE+ GKSLEEMSGENE+ E + + +RTVP
Sbjct: 481 GVRNSLIVLGVVNFLGILFTFLVPESKGKSLEEMSGENEDNENSN--------NDSRTVP 532
Query: 541 V 541
+
Sbjct: 533 I 533
>At2g32830 phosphate transporter like protein
Length = 542
Score = 851 bits (2199), Expect = 0.0
Identities = 416/542 (76%), Positives = 469/542 (85%), Gaps = 1/542 (0%)
Query: 1 MAREQLGVLTALDAAKTQWYHFTAIIIAGMGFFTDAYDLFCIPNVTKLLGRIYYTHPGAL 60
MA++ VL ALDAAKTQ YHFTAI+IAGMGFFTDAYDLF I VTKLLGRIYY +
Sbjct: 1 MAKKGKEVLNALDAAKTQMYHFTAIVIAGMGFFTDAYDLFSISLVTKLLGRIYYHVDSSK 60
Query: 61 KPGTLPPNVSAAVNGVALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFG 120
KPGTLPPNV+AAVNGVA CGTLAGQLFFGWLGDK+GRKKVYG+TL LMV SL SGLSFG
Sbjct: 61 KPGTLPPNVAAAVNGVAFCGTLAGQLFFGWLGDKLGRKKVYGITLMLMVLCSLGSGLSFG 120
Query: 121 HSAKGTIGTLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGG 180
HSA G + TLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIA+VFAMQGFGILAGG
Sbjct: 121 HSANGVMATLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIAAVFAMQGFGILAGG 180
Query: 181 IVSLVVSTAFDHAFKAPPYKVDAAASLVPEADYVWRIILMFGAVPAGLTYYWRMKMPETA 240
IVSL+VS+ FDHAFKAP Y+VD S VP+ADYVWRI+LMFGA+PA LTYYWRMKMPETA
Sbjct: 181 IVSLIVSSTFDHAFKAPTYEVDPVGSTVPQADYVWRIVLMFGAIPALLTYYWRMKMPETA 240
Query: 241 RYTALVAKNAKQAAQDMSSVLQVEIEAEQEKVDKIGVQDKN-SFGLFSKEFLRRHGLHLL 299
RYTALVA+N KQAA DMS VLQV++ AE+E + N +FGLF++EF RRHGLHLL
Sbjct: 241 RYTALVARNTKQAASDMSKVLQVDLIAEEEAQSNSNSSNPNFTFGLFTREFARRHGLHLL 300
Query: 300 GTTSTWFLLDIAYYSSNLFQKDIYSSIGWLPPAQDMNAIHEVFKVARATTLIALCGTVPG 359
GTT+TWFLLDIAYYSSNLFQKDIY++IGW+P A+ MNAIHEVF V++A TLIALCGTVPG
Sbjct: 301 GTTTTWFLLDIAYYSSNLFQKDIYTAIGWIPAAETMNAIHEVFTVSKAQTLIALCGTVPG 360
Query: 360 YWFTVAFIDVIGRFAIQLMGFFFMTVFMFALAIPYDHWTKRENRIGFLVMYAMTFFFANF 419
YWFTVAFID++GRF IQLMGF FMT+FMFALAIPYDHW RENRIGFL+MY++T FFANF
Sbjct: 361 YWFTVAFIDILGRFFIQLMGFIFMTIFMFALAIPYDHWRHRENRIGFLIMYSLTMFFANF 420
Query: 420 GPNSTTFVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQSKDPKKRDAGYPAG 479
GPN+TTFVVPAEIFPARLRSTCHGIS+A+GKAGAI+GAFGFLYA+QS D +K DAGYP G
Sbjct: 421 GPNATTFVVPAEIFPARLRSTCHGISAASGKAGAIVGAFGFLYAAQSSDSEKTDAGYPPG 480
Query: 480 IGMKNTLILLAVVNCLGIFFTFLVPEANGKSLEEMSGENEEEEETEAAVVDPQASSNRTV 539
IG++N+L++LA VN LGI FT LVPE+ GKSLEE+S E+EE+ + V A+S R V
Sbjct: 481 IGVRNSLLMLACVNFLGIVFTLLVPESKGKSLEEISREDEEQSGGDTVVEMTVANSGRKV 540
Query: 540 PV 541
PV
Sbjct: 541 PV 542
>At5g43350 phosphate transporter (gb|AAB17265.1)
Length = 524
Score = 826 bits (2133), Expect = 0.0
Identities = 400/519 (77%), Positives = 451/519 (86%), Gaps = 1/519 (0%)
Query: 1 MAREQLGVLTALDAAKTQWYHFTAIIIAGMGFFTDAYDLFCIPNVTKLLGRIYYTHPGAL 60
MA +QLGVL ALD AKTQ YHFTAI+IAGMGFFTDAYDLFC+ VTKLLGRIYY +P +
Sbjct: 1 MAEQQLGVLKALDVAKTQLYHFTAIVIAGMGFFTDAYDLFCVSLVTKLLGRIYYFNPESA 60
Query: 61 KPGTLPPNVSAAVNGVALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFG 120
KPG+LPP+V+AAVNGVALCGTL+GQLFFGWLGDK+GRKKVYGLTL +M+ S+ASGLSFG
Sbjct: 61 KPGSLPPHVAAAVNGVALCGTLSGQLFFGWLGDKLGRKKVYGLTLVMMILCSVASGLSFG 120
Query: 121 HSAKGTIGTLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGG 180
H AKG + TLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIA+VFAMQG GILAGG
Sbjct: 121 HEAKGVMTTLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIAAVFAMQGVGILAGG 180
Query: 181 IVSLVVSTAFDHAFKAPPYKVDAAASLVPEADYVWRIILMFGAVPAGLTYYWRMKMPETA 240
V+L VS+ FD F AP Y V+ A S P+ DY+WRII+MFGA+PA LTYYWRMKMPETA
Sbjct: 181 FVALAVSSIFDKKFPAPTYAVNRALSTPPQVDYIWRIIVMFGALPAALTYYWRMKMPETA 240
Query: 241 RYTALVAKNAKQAAQDMSSVLQVEIEAEQEKVDKIGVQDKNSFGLFSKEFLRRHGLHLLG 300
RYTALVAKN KQA DMS VLQ +IE E E+V+ K ++GLFSKEFLRRHGLHLLG
Sbjct: 241 RYTALVAKNIKQATADMSKVLQTDIELE-ERVEDDVKDPKQNYGLFSKEFLRRHGLHLLG 299
Query: 301 TTSTWFLLDIAYYSSNLFQKDIYSSIGWLPPAQDMNAIHEVFKVARATTLIALCGTVPGY 360
TTSTWFLLDIA+YS NLFQKDI+S+IGW+P A MNA HEVF++ARA TLIALC TVPGY
Sbjct: 300 TTSTWFLLDIAFYSQNLFQKDIFSAIGWIPKAATMNATHEVFRIARAQTLIALCSTVPGY 359
Query: 361 WFTVAFIDVIGRFAIQLMGFFFMTVFMFALAIPYDHWTKRENRIGFLVMYAMTFFFANFG 420
WFTVAFID IGRF IQL GFF MTVFMFA+A PY+HW K ENRIGF+VMY++TFFFANFG
Sbjct: 360 WFTVAFIDTIGRFKIQLNGFFMMTVFMFAIAFPYNHWIKPENRIGFVVMYSLTFFFANFG 419
Query: 421 PNSTTFVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQSKDPKKRDAGYPAGI 480
PN+TTF+VPAEIFPARLRSTCHGIS+AAGKAGAI+GAFGFLYA+QS+D K DAGYP GI
Sbjct: 420 PNATTFIVPAEIFPARLRSTCHGISAAAGKAGAIVGAFGFLYAAQSQDKAKVDAGYPPGI 479
Query: 481 GMKNTLILLAVVNCLGIFFTFLVPEANGKSLEEMSGENE 519
G+KN+LI+L V+N +G+ FTFLVPE GKSLEE+SGE E
Sbjct: 480 GVKNSLIMLGVLNFIGMLFTFLVPEPKGKSLEELSGEAE 518
>At5g43360 inorganic phosphate transporter (dbj|BAA24281.1)
Length = 521
Score = 822 bits (2123), Expect = 0.0
Identities = 396/522 (75%), Positives = 453/522 (85%), Gaps = 1/522 (0%)
Query: 1 MAREQLGVLTALDAAKTQWYHFTAIIIAGMGFFTDAYDLFCIPNVTKLLGRIYYTHPGAL 60
MA +QLGVL ALD AKTQ YHFTAI+IAGMGFFTDAYDLFC+ VTKLLGR+YY +P +
Sbjct: 1 MADQQLGVLKALDVAKTQLYHFTAIVIAGMGFFTDAYDLFCVSLVTKLLGRLYYFNPTSA 60
Query: 61 KPGTLPPNVSAAVNGVALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFG 120
KPG+LPP+V+AAVNGVALCGTLAGQLFFGWLGDK+GRKKVYG+TL +M+ S+ASGLS G
Sbjct: 61 KPGSLPPHVAAAVNGVALCGTLAGQLFFGWLGDKLGRKKVYGITLIMMILCSVASGLSLG 120
Query: 121 HSAKGTIGTLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGG 180
+SAKG + TLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIA+VFAMQG GILAGG
Sbjct: 121 NSAKGVMTTLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIAAVFAMQGVGILAGG 180
Query: 181 IVSLVVSTAFDHAFKAPPYKVDAAASLVPEADYVWRIILMFGAVPAGLTYYWRMKMPETA 240
V+L VS+ FD F +P Y+ D S P+ADY+WRII+MFGA+PA LTYYWRMKMPETA
Sbjct: 181 FVALAVSSIFDKKFPSPTYEQDRFLSTPPQADYIWRIIVMFGALPAALTYYWRMKMPETA 240
Query: 241 RYTALVAKNAKQAAQDMSSVLQVEIEAEQEKVDKIGVQDKNSFGLFSKEFLRRHGLHLLG 300
RYTALVAKN KQA DMS VLQ ++E E+ D + KN +GLFSKEFLRRHGLHLLG
Sbjct: 241 RYTALVAKNIKQATADMSKVLQTDLELEERVEDDVKDPKKN-YGLFSKEFLRRHGLHLLG 299
Query: 301 TTSTWFLLDIAYYSSNLFQKDIYSSIGWLPPAQDMNAIHEVFKVARATTLIALCGTVPGY 360
TTSTWFLLDIA+YS NLFQKDI+S+IGW+P A MNAIHEVFK+ARA TLIALC TVPGY
Sbjct: 300 TTSTWFLLDIAFYSQNLFQKDIFSAIGWIPKAATMNAIHEVFKIARAQTLIALCSTVPGY 359
Query: 361 WFTVAFIDVIGRFAIQLMGFFFMTVFMFALAIPYDHWTKRENRIGFLVMYAMTFFFANFG 420
WFTVAFID+IGRFAIQLMGFF MTVFMFA+A PY+HW +NRIGF+VMY++TFFFANFG
Sbjct: 360 WFTVAFIDIIGRFAIQLMGFFMMTVFMFAIAFPYNHWILPDNRIGFVVMYSLTFFFANFG 419
Query: 421 PNSTTFVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQSKDPKKRDAGYPAGI 480
PN+TTF+VPAEIFPARLRSTCHGIS+A GKAGAI+GAFGFLYA+Q +D K DAGYP GI
Sbjct: 420 PNATTFIVPAEIFPARLRSTCHGISAATGKAGAIVGAFGFLYAAQPQDKTKTDAGYPPGI 479
Query: 481 GMKNTLILLAVVNCLGIFFTFLVPEANGKSLEEMSGENEEEE 522
G+KN+LI+L V+N +G+ FTFLVPE GKSLEE+SGE E ++
Sbjct: 480 GVKNSLIMLGVINFVGMLFTFLVPEPKGKSLEELSGEAEVDK 521
>At5g43370 inorganic phosphate transporter (dbj|BAA24282.1)
Length = 524
Score = 820 bits (2119), Expect = 0.0
Identities = 398/519 (76%), Positives = 451/519 (86%), Gaps = 1/519 (0%)
Query: 1 MAREQLGVLTALDAAKTQWYHFTAIIIAGMGFFTDAYDLFCIPNVTKLLGRIYYTHPGAL 60
MA +QLGVL ALD AKTQ YHFTAI+IAGMGFFTDAYDLFC+ VTKLLGRIYY +P +
Sbjct: 1 MAEQQLGVLKALDVAKTQLYHFTAIVIAGMGFFTDAYDLFCVSLVTKLLGRIYYFNPESA 60
Query: 61 KPGTLPPNVSAAVNGVALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFG 120
KPG+LPP+V+AAVNGVALCGTL+GQLFFGWLGDK+GRKKVYGLTL +M+ S+ASGLSFG
Sbjct: 61 KPGSLPPHVAAAVNGVALCGTLSGQLFFGWLGDKLGRKKVYGLTLIMMILCSVASGLSFG 120
Query: 121 HSAKGTIGTLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGG 180
+ AKG + TLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIA+VFAMQG GILAGG
Sbjct: 121 NEAKGVMTTLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIAAVFAMQGVGILAGG 180
Query: 181 IVSLVVSTAFDHAFKAPPYKVDAAASLVPEADYVWRIILMFGAVPAGLTYYWRMKMPETA 240
V+L VS+ FD F AP Y V+ A S P+ DY+WRII+MFGA+PA LTYYWRMKMPETA
Sbjct: 181 FVALAVSSIFDKKFPAPTYAVNRALSTPPQVDYIWRIIVMFGALPAALTYYWRMKMPETA 240
Query: 241 RYTALVAKNAKQAAQDMSSVLQVEIEAEQEKVDKIGVQDKNSFGLFSKEFLRRHGLHLLG 300
RYTALVAKN KQA DMS VLQ +IE E E+V+ + ++GLFSKEFLRRHGLHLLG
Sbjct: 241 RYTALVAKNIKQATADMSKVLQTDIELE-ERVEDDVKDPRQNYGLFSKEFLRRHGLHLLG 299
Query: 301 TTSTWFLLDIAYYSSNLFQKDIYSSIGWLPPAQDMNAIHEVFKVARATTLIALCGTVPGY 360
TTSTWFLLDIA+YS NLFQKDI+S+IGW+P A MNA HEVF++ARA TLIALC TVPGY
Sbjct: 300 TTSTWFLLDIAFYSQNLFQKDIFSAIGWIPKAATMNATHEVFRIARAQTLIALCSTVPGY 359
Query: 361 WFTVAFIDVIGRFAIQLMGFFFMTVFMFALAIPYDHWTKRENRIGFLVMYAMTFFFANFG 420
WFTVAFID IGRF IQL GFF MTVFMFA+A PY+HW K ENRIGF+VMY++TFFFANFG
Sbjct: 360 WFTVAFIDTIGRFKIQLNGFFMMTVFMFAIAFPYNHWIKPENRIGFVVMYSLTFFFANFG 419
Query: 421 PNSTTFVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQSKDPKKRDAGYPAGI 480
PN+TTF+VPAEIFPARLRSTCHGIS+AAGKAGAIIGAFGFLYA+Q++D K DAGYP GI
Sbjct: 420 PNATTFIVPAEIFPARLRSTCHGISAAAGKAGAIIGAFGFLYAAQNQDKAKVDAGYPPGI 479
Query: 481 GMKNTLILLAVVNCLGIFFTFLVPEANGKSLEEMSGENE 519
G+KN+LI+L V+N +G+ FTFLVPE GKSLEE+SGE E
Sbjct: 480 GVKNSLIVLGVLNFIGMLFTFLVPEPKGKSLEELSGEAE 518
>At5g43340 inorganic phosphate transporter (dbj|BAA34390.1)
Length = 516
Score = 702 bits (1812), Expect = 0.0
Identities = 345/530 (65%), Positives = 415/530 (78%), Gaps = 15/530 (2%)
Query: 1 MAREQLG-VLTALDAAKTQWYHFTAIIIAGMGFFTDAYDLFCIPNVTKLLGRIYYTHPGA 59
MA E+ G +L ALD AKTQWYH TA++++GMGFFTD+YDLF I +TKLLGRIYY PG+
Sbjct: 1 MANEEQGSILKALDVAKTQWYHVTAVVVSGMGFFTDSYDLFVISLITKLLGRIYYQVPGS 60
Query: 60 LKPGTLPPNVSAAVNGVALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSF 119
PG+LP +SAAV+GVA GT GQ+FFG LGDK+GRK+VYGLTL +M S+ SGLS
Sbjct: 61 SSPGSLPDGISAAVSGVAFAGTFIGQIFFGCLGDKLGRKRVYGLTLLIMTICSICSGLSL 120
Query: 120 GHSAKGTIGTLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAG 179
G K + TLCFFRFWLGFGIGGDYPLSATIMSEY+NK+TRGAFIA+VF MQG GILA
Sbjct: 121 GRDPKTVMVTLCFFRFWLGFGIGGDYPLSATIMSEYSNKRTRGAFIAAVFGMQGIGILAA 180
Query: 180 GIVSLVVSTAFDHAFKAPPYKVDAAASLVPEADYVWRIILMFGAVPAGLTYYWRMKMPET 239
G VSL+VS F+ F + Y +D AAS VP+ADYVWRIILM GA+PA LTYYWRMKMPET
Sbjct: 181 GAVSLLVSAVFESKFPSRAYILDGAASTVPQADYVWRIILMVGALPALLTYYWRMKMPET 240
Query: 240 ARYTALVAKNAKQAAQDMSSVLQVEIEAEQEKVDKIGVQDKNSFGLFSKEFLRRHGLHLL 299
ARYTALV+KNA+QAA DM+ VL V+IEA K D+ V + FGLFS +FLRRHGLHLL
Sbjct: 241 ARYTALVSKNAEQAALDMTKVLNVDIEASAAKNDQARV-SSDEFGLFSMKFLRRHGLHLL 299
Query: 300 GTTSTWFLLDIAYYSSNLFQKDIYSSIGWLPPAQDMNAIHEVFKVARATTLIALCGTVPG 359
GT STWFLLDIA+YS NLFQKDI+++IGWLP A+ MNAI E++ +A+A T+IA C TVPG
Sbjct: 300 GTASTWFLLDIAFYSQNLFQKDIFTTIGWLPSAKTMNAIQELYMIAKAQTIIACCSTVPG 359
Query: 360 YWFTVAFIDVIGRFAIQLMGFFFMTVFMFALAIPYDHWTKRENRIGFLVMYAMTFFFANF 419
Y+FTV FID +GR IQ+MGF MT+FM +LAIPY HWT NRIGF+V+Y+ TFFF+NF
Sbjct: 360 YFFTVGFIDYMGRKKIQIMGFAMMTIFMLSLAIPYHHWTLPANRIGFVVLYSFTFFFSNF 419
Query: 420 GPNSTTFVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQSKDPKKRDAGYPAG 479
GPN+TTF+VPAEIFPAR+RSTCHGIS+A+GKAGA++G+FGF +
Sbjct: 420 GPNATTFIVPAEIFPARIRSTCHGISAASGKAGAMVGSFGF-------------SALVKA 466
Query: 480 IGMKNTLILLAVVNCLGIFFTFLVPEANGKSLEEMSGENEEEEETEAAVV 529
+GM NTL ++A +N LG+ TF +PE NGKSLEE+SGE E E+ E VV
Sbjct: 467 LGMSNTLYIMAGINLLGLLLTFTIPETNGKSLEELSGETEPEKIKEKIVV 516
>At3g54700 phosphate transport protein
Length = 433
Score = 664 bits (1713), Expect = 0.0
Identities = 324/426 (76%), Positives = 372/426 (87%), Gaps = 6/426 (1%)
Query: 103 LTLALMVFSSLASGLSFGHSAKGTIGTLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRG 162
+TL +MV S+ASGLSFG + K + TLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRG
Sbjct: 1 MTLMVMVLCSIASGLSFGSNPKTVMTTLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRG 60
Query: 163 AFIASVFAMQGFGILAGGIVSLVVSTAFDHAFKAPPYKVDAAASLVPEADYVWRIILMFG 222
AFIA+VFAMQGFGIL GGI +++VS AF+ F AP Y++DA AS VP+ADYVWRIILM G
Sbjct: 61 AFIAAVFAMQGFGILTGGIFAIIVSAAFEAKFPAPTYQIDALASTVPQADYVWRIILMVG 120
Query: 223 AVPAGLTYYWRMKMPETARYTALVAKNAKQAAQDMSSVLQVEIEAEQEKVDKIGVQDK-N 281
A+PA +TYY R KMPETARYTALVAK+AK AA +MS VLQVEIEAEQ+ G +DK N
Sbjct: 121 ALPAAMTYYSRSKMPETARYTALVAKDAKLAASNMSKVLQVEIEAEQQ-----GTEDKSN 175
Query: 282 SFGLFSKEFLRRHGLHLLGTTSTWFLLDIAYYSSNLFQKDIYSSIGWLPPAQDMNAIHEV 341
SFGLFSKEF++RHGLHLLGTTSTWFLLDIA+YS NLFQKDI+S+IGW+PPAQ MNAI EV
Sbjct: 176 SFGLFSKEFMKRHGLHLLGTTSTWFLLDIAFYSQNLFQKDIFSAIGWIPPAQTMNAIQEV 235
Query: 342 FKVARATTLIALCGTVPGYWFTVAFIDVIGRFAIQLMGFFFMTVFMFALAIPYDHWTKRE 401
FK+ARA TLIALC TVPGYWFTVAFIDVIGRFAIQ+MGFFFMTVFMFALAIPYDHWT +E
Sbjct: 236 FKIARAQTLIALCSTVPGYWFTVAFIDVIGRFAIQMMGFFFMTVFMFALAIPYDHWTHKE 295
Query: 402 NRIGFLVMYAMTFFFANFGPNSTTFVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFGFL 461
NRIGF+ MY++TFFFANFGPN+TTFVVPAEIFPAR RSTCHGIS+A+GK GA++GAFGFL
Sbjct: 296 NRIGFVAMYSLTFFFANFGPNATTFVVPAEIFPARFRSTCHGISAASGKLGAMVGAFGFL 355
Query: 462 YASQSKDPKKRDAGYPAGIGMKNTLILLAVVNCLGIFFTFLVPEANGKSLEEMSGENEEE 521
Y +QS D K + GYP GIG+KN+LI+L VVN LG+ FT LVPE+ GKSLEEMSGENE+
Sbjct: 356 YLAQSPDKTKTEHGYPPGIGVKNSLIVLGVVNLLGMVFTLLVPESKGKSLEEMSGENEQN 415
Query: 522 EETEAA 527
+E+ ++
Sbjct: 416 DESSSS 421
>At1g20860 putative inorganic phosphate transporter protein
Length = 534
Score = 490 bits (1261), Expect = e-139
Identities = 266/533 (49%), Positives = 339/533 (62%), Gaps = 17/533 (3%)
Query: 8 VLTALDAAKTQWYHFTAIIIAGMGFFTDAYDLFCIPNVTKLLGRIYYTHPGALKPGTLPP 67
VL++LD A+TQWYHF AII+AGMG FTDAYDLFCI V K++ +YY ++
Sbjct: 5 VLSSLDVARTQWYHFKAIIVAGMGLFTDAYDLFCIAPVMKMISHVYYNGD------SINT 58
Query: 68 NVSAAVNGVALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAKGTI 127
V + +AL GT GQL FG+LGD++GR++VYGL L +M+ SS G S + + +
Sbjct: 59 AVLSTSYAIALLGTATGQLVFGYLGDRVGRRRVYGLCLIIMILSSFGCGFSVCTTRRSCV 118
Query: 128 G-TLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVSLVV 186
+L FFRF+LG GIGGDYPLSATIMSE+ANK+TRGAFIA+VF+MQG GIL V++ V
Sbjct: 119 MVSLGFFRFFLGLGIGGDYPLSATIMSEFANKRTRGAFIAAVFSMQGLGILVSSAVTMAV 178
Query: 187 STAFDHAFKAPPYKVDAAASLVPEADYVWRIILMFGAVPAGLTYYWRMKMPETARYTALV 246
AF + +VDAAA EAD WR+ILM GA+PA LT+YWRM MPETARYTALV
Sbjct: 179 CVAFKRS--GGGLEVDAAAPT--EADLAWRLILMIGALPAALTFYWRMLMPETARYTALV 234
Query: 247 AKNAKQAAQDMSSVLQVEIEAEQEKVDKIGVQDKNSFGLFSKEFLRRHGLHLLGTTSTWF 306
N QAA+DM V+ +++ D S+ LFS+ F R HG L + WF
Sbjct: 235 ENNIVQAAKDMQRVMSRSHISDEATTDPPPPPPPPSYKLFSRCFFRLHGRDLFAASFNWF 294
Query: 307 LLDIAYYSSNLFQKDIYSSIGWLPPAQDMNAIHEVFKVARATTLIALCGTVPGYWFTVAF 366
L+DI +Y+SNL I+S P + N F+VA +IA C T+PGYWFTV F
Sbjct: 295 LVDIVFYTSNLLLSHIFSHYSKKPSTAE-NVYDAAFEVAELGAIIAACSTIPGYWFTVYF 353
Query: 367 IDVIGRFAIQLMGFFFMTVFMFALAIPYD-HWTKRE-NRIGFLVMYAMTFFFANFGPNST 424
ID IGR IQ+MGFFFM V IPY +W+K E N GF+V+Y + FFF NFGPN+T
Sbjct: 354 IDKIGRVKIQIMGFFFMAVIYLVAGIPYSWYWSKHEHNNKGFMVLYGLVFFFCNFGPNTT 413
Query: 425 TFVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQSKDPKKRDAGYPAGIGMKN 484
TF++PAE FPAR RSTCHGIS AAGK GAI+G GFL+A++ + ++ YP M+
Sbjct: 414 TFIIPAEHFPARFRSTCHGISGAAGKLGAIVGTVGFLWATKKMESDDKNQIYPEVNRMRI 473
Query: 485 TLILLAVVNCLGIFFT-FLVPEANGKSLE--EMSGENEEEEETEAAVVDPQAS 534
++L V GI T F E G+SLE E +N E E E +VD Q+S
Sbjct: 474 AFLILGGVCIAGILVTYFFTKETMGRSLEENEHDQDNNAESEDEPQIVDGQSS 526
>At1g76430 putative phosphate transporter
Length = 532
Score = 483 bits (1243), Expect = e-136
Identities = 263/537 (48%), Positives = 351/537 (64%), Gaps = 26/537 (4%)
Query: 5 QLGVLTALDAAKTQWYHFTAIIIAGMGFFTDAYDLFCIPNVTKLLGRIYYTHPGALKPGT 64
+L +L+ALDAA+ QWYHF AII+AGMG FTDAYDLFCI + K++ +IYY H ++ GT
Sbjct: 3 ELSLLSALDAARIQWYHFKAIIVAGMGLFTDAYDLFCIAPIMKMISQIYY-HKDSI--GT 59
Query: 65 LPPNVSAAVNGVALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAK 124
+ S A+ AL GT GQL FG+LGD++GR+KVYGL+L +MVFSS G S + +
Sbjct: 60 ALLSTSYAI---ALLGTALGQLIFGYLGDRVGRRKVYGLSLLIMVFSSFGCGFSVCTTRR 116
Query: 125 GTIG-TLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVS 183
+ +L FFRF LG GIGGDYPLSATIMSE+ANK+TRGAFIA+VF+MQG GIL V+
Sbjct: 117 SCVMVSLGFFRFVLGLGIGGDYPLSATIMSEFANKRTRGAFIAAVFSMQGLGILMSSAVT 176
Query: 184 LVVSTAFDHAFKAPPYKVDAAASLV---PEADYVWRIILMFGAVPAGLTYYWRMKMPETA 240
+VV AF +A + K + A PE+D WR+ILM GA+PA LT+YWRM MPETA
Sbjct: 177 MVVCLAFKNAGEGSSEKTNVAGLETLAPPESDIAWRLILMIGALPAALTFYWRMLMPETA 236
Query: 241 RYTALVAKNAKQAAQDMSSVLQVEIEAEQEKVDKIGVQD---KNSFGLFSKEFLRRHGLH 297
RYTALV N QAA+DM V+ V + ++ + ++ +S+ LFS+ FL HG
Sbjct: 237 RYTALVENNVVQAAKDMQRVMSVSMISQITEDSSSELEQPPSSSSYKLFSRRFLSLHGRD 296
Query: 298 LLGTTSTWFLLDIAYYSSNLFQKDIYSSIGWLPPAQDMNAIHEVFKVARATTLIALCGTV 357
L ++ WFL+D+ +Y+SNL I++ P N F+VA+ ++A C T+
Sbjct: 297 LFAASANWFLVDVVFYTSNLLLSQIFNFSN--KPLNSTNVYDSAFEVAKLAAIVAACSTI 354
Query: 358 PGYWFTVAFIDVIGRFAIQLMGFFFMTVFMFALAIPYD-HWTKREN-RIGFLVMYAMTFF 415
PGYWFTV FID IGR IQ+MGFF M V IPY +W+K E GF+V+Y + FF
Sbjct: 355 PGYWFTVYFIDKIGRVKIQMMGFFLMAVVYLVAGIPYSWYWSKHEKTNKGFMVLYGLIFF 414
Query: 416 FANFGPNSTTFVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQSKDPKKRDAG 475
F+NFGPN+TTF++PAE+FPAR RSTCHGIS AAGK GAI+G GFL+A++ + + G
Sbjct: 415 FSNFGPNTTTFIIPAELFPARFRSTCHGISGAAGKFGAIVGTVGFLWATRHHE----EDG 470
Query: 476 YPAGIGMKNTLILLAVVNCLGIFFTFL-VPEANGKSLEEMSGENEEEEETEAAVVDP 531
+P ++ ++L V G+ T+L E G+SLE ENE+E + +A P
Sbjct: 471 FPDVKRVRIAFLILGGVCIAGMIVTYLFTRETMGRSLE----ENEDEIVSTSAGSSP 523
>At4g08895 putative protein
Length = 155
Score = 151 bits (381), Expect = 1e-36
Identities = 71/140 (50%), Positives = 92/140 (65%), Gaps = 32/140 (22%)
Query: 321 DIYSSIGWLPPAQDMNAIHEVFKVARATTLIALCGTVPGYWFTVAFIDVIGRFAIQLMGF 380
D +++ GWLP A+ MNA+ E+F + RA T+IA C TVP
Sbjct: 23 DSFTTFGWLPSAKTMNALQELFMIVRAQTIIACCSTVP---------------------- 60
Query: 381 FFMTVFMFALAIPYDHWTKRENRIGFLVMYAMTFFFANFGPNSTTFVVPAEIFPARLRST 440
A+PY HWT NRIGF++ Y+ TFFF+NFGPN+TTF+VPAEIFPAR+RST
Sbjct: 61 ----------AVPYHHWTLPANRIGFVIFYSFTFFFSNFGPNATTFIVPAEIFPARIRST 110
Query: 441 CHGISSAAGKAGAIIGAFGF 460
CHGIS+A+GKAGA++G+FGF
Sbjct: 111 CHGISAASGKAGAMVGSFGF 130
>At2g20780 putative sugar transporter
Length = 547
Score = 82.0 bits (201), Expect = 8e-16
Identities = 102/445 (22%), Positives = 186/445 (40%), Gaps = 58/445 (13%)
Query: 76 VALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAKGTIGTLCFFRF 135
++L G+LAG G D +GRK + + LA +VF + A+ ++ S + L R
Sbjct: 125 ISLFGSLAG----GRTSDSIGRK--WTMALAALVFQTGAAVMAVAPSFE----VLMIGRT 174
Query: 136 WLGFGIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVSLVVSTAFDHAFK 195
G GIG ++ ++E + RG F + GIL G + + S H
Sbjct: 175 LAGIGIGLGVMIAPVYIAEISPTVARGFFTSFPEIFINLGILLGYVSNYAFSGLSVHIS- 233
Query: 196 APPYKVDAAASLVPEADYVWRIILMFGAVPAGLTYYWRMKMPETARYTALVAKNAKQAAQ 255
WRI+L G +P+ + +PE+ R+ LV K +A+
Sbjct: 234 -------------------WRIMLAVGILPSVFIGFALCVIPESPRW--LVMKGRVDSAR 272
Query: 256 DMSSVLQVEIEAEQEKVDKIGVQDKNSFGLFSKEFLRRHGLHLLGTTSTWFLLDIAYYSS 315
++ + +E++ +I + ++ G + R LL + + I +
Sbjct: 273 EVLMKTNERDDEAEERLAEIQLAAAHTEGSEDRPVWRE----LLSPSPVVRKMLIVGFGI 328
Query: 316 NLFQK--DIYSSIGWLPPAQDMNAIHEVFKVARATTLIALCGTVPGYWFTVAFIDVIGRF 373
FQ+ I +++ + P I + K+ AT + + TV F ID +GR
Sbjct: 329 QCFQQITGIDATVYYSPEILKEAGIQDETKLLAATVAVGVTKTV-FILFATFLIDSVGRK 387
Query: 374 AIQLMGFFFMTVFMFALAIPYDHWTKRENRIGFLVMYA---MTFFFANFGPNSTTFVVPA 430
+ + MT+ +F L+ + I +++ + FF GP +V+ +
Sbjct: 388 PLLYVSTIGMTLCLFCLSFTLTFLGQGTLGITLALLFVCGNVAFFSIGMGP--VCWVLTS 445
Query: 431 EIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQSKDPKKRDAGYPAGIGMKNTLILLA 490
EIFP RLR+ + + + + + A FL S++ I + T + +
Sbjct: 446 EIFPLRLRAQASALGAVGNRVCSGLVAMSFLSVSRA-------------ITVGGTFFVFS 492
Query: 491 VVNCLGIFFTF-LVPEANGKSLEEM 514
+V+ L + F + LVPE +GKSLE++
Sbjct: 493 LVSALSVIFVYVLVPETSGKSLEQI 517
>At2g43330 membrane transporter like protein
Length = 509
Score = 75.5 bits (184), Expect = 7e-14
Identities = 101/447 (22%), Positives = 183/447 (40%), Gaps = 58/447 (12%)
Query: 76 VALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAKGTIGTLCFFRF 135
+AL G + G GW+ D GRKK A +VF++ A + +A L R
Sbjct: 79 MALVGAMIGAAAGGWINDYYGRKK--ATLFADVVFAAGAIVM----AAAPDPYVLISGRL 132
Query: 136 WLGFGIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVSLVVSTAFDHAFK 195
+G G+G + ++E + + RG +++ M I G +S +V++AF
Sbjct: 133 LVGLGVGVASVTAPVYIAEASPSEVRGGLVSTNVLM----ITGGQFLSYLVNSAF----- 183
Query: 196 APPYKVDAAASLVPEADYVWRIILMFGAVPAGLTYYWRMKMPETARYTALVAKNAKQAAQ 255
+ WR +L VPA + + + MPE+ R+ L KN K A
Sbjct: 184 -------------TQVPGTWRWMLGVSGVPAVIQFILMLFMPESPRW--LFMKNRKAEAI 228
Query: 256 DMSSVLQVEIEAEQEKVDKIGVQDKNSFGLFSKEFLRRHGLHLLGTTSTWFLLDIAYYSS 315
+ + +I ++++D + ++ K+ R G + + L +A
Sbjct: 229 QVLA-RTYDISRLEDEIDHLSAAEEE-----EKQRKRTVGYLDVFRSKELRLAFLAGAGL 282
Query: 316 NLFQK--DIYSSIGWLPPAQDMNAIHEVFKVARATTLIALCGTVPGYWFTVAFIDVIGRF 373
FQ+ I + + + P M H ++A +LI G + FID GR
Sbjct: 283 QAFQQFTGINTVMYYSPTIVQMAGFHS-NQLALFLSLIVAAMNAAGTVVGIYFIDHCGRK 341
Query: 374 AIQLMGFFFMTVFMFALAIPYDHWTKREN---RIGFLVMYAMTFFFANFGP--NSTTFVV 428
+ L F + + + L++ + ++ + G+L + + + F P + V
Sbjct: 342 KLALSSLFGVIISLLILSVSFFKQSETSSDGGLYGWLAVLGLALYIVFFAPGMGPVPWTV 401
Query: 429 PAEIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQSKDPKKRDAGYPAGIGMKNTLIL 488
+EI+P + R C G+S+ +I A FL +++ AG GM T ++
Sbjct: 402 NSEIYPQQYRGICGGMSATVNWISNLIVAQTFLTIAEA-----------AGTGM--TFLI 448
Query: 489 LAVVNCLGIFFTFL-VPEANGKSLEEM 514
LA + L + F + VPE G + E+
Sbjct: 449 LAGIAVLAVIFVIVFVPETQGLTFSEV 475
>At4g04760 putative sugar transporter
Length = 457
Score = 68.6 bits (166), Expect = 9e-12
Identities = 96/455 (21%), Positives = 170/455 (37%), Gaps = 91/455 (20%)
Query: 80 GTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAKGTIGTLCFFRFWLGF 139
G + G L G L D +GR K +T L V A + G + L R G
Sbjct: 59 GLILGALICGKLTDLVGRVKTIWITNILFVIGWFAIAFAKG------VWLLDLGRLLQGI 112
Query: 140 GIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVSLVVSTAFDHAFKAPPY 199
IG L ++E A + RGA + G GI + +V+
Sbjct: 113 SIGISVYLGPVYITEIAPRNLRGAASSFAQLFAGVGISVFYALGTIVA------------ 160
Query: 200 KVDAAASLVPEADYVWRIILMFGAVPAGLTYYWRMKMPETARYTALVAKNAKQAA----- 254
WR + + G +P+ + +PE+ R+ A V + + A
Sbjct: 161 ---------------WRNLAILGCIPSLMVLPLLFFIPESPRWLAKVGREMEVEAVLLSL 205
Query: 255 ----QDMSSVLQVEIEAEQEKVDKIGVQDKNSFGLFSKE--FLRRHGLHLLGTTSTWFLL 308
D+S +E + + + D+ F LF ++ F G+ L+ L
Sbjct: 206 RGEKSDVSDEAAEILEYTEHVKQQQDIDDRGFFKLFQRKYAFSLTIGVVLIALPQLGGLN 265
Query: 309 DIAYYSSNLF-QKDIYSSIGWLPPAQDMNAIHEVFKVARATTLIALCGTVPGYWFTVAFI 367
++Y+ ++F + S G++ +T+++ + G + G +
Sbjct: 266 GYSFYTDSIFISTGVSSDFGFI-----------------STSVVQMFGGILG----TVLV 304
Query: 368 DVIGRFAIQLMGFFFMTVFMFALAIPY----DH-WTKRENRIGFLVMYAMTFFFANFGPN 422
DV GR + L+ M + AI + +H W E L ++++ +F ++G
Sbjct: 305 DVSGRRTLLLVSQAGMFLGCLTTAISFFLKENHCW---ETGTPVLALFSVMVYFGSYGSG 361
Query: 423 --STTFVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQSKDPKKRDAGYPAGI 480
S +++ +EI+P ++ + + A + A+ F Y Q
Sbjct: 362 MGSIPWIIASEIYPVDVKGAAGTMCNLVSSISAWLVAYSFSYLLQWSST----------- 410
Query: 481 GMKNTLILLAVVNCLG-IFFTFLVPEANGKSLEEM 514
T ++ A V LG +F LVPE GKSLEE+
Sbjct: 411 ---GTFLMFATVAGLGFVFIAKLVPETKGKSLEEI 442
>At3g13050 putative transporter
Length = 500
Score = 64.7 bits (156), Expect = 1e-10
Identities = 100/481 (20%), Positives = 168/481 (34%), Gaps = 87/481 (18%)
Query: 9 LTALDAAKTQWYHFTAIIIAGMGFFTDAYDLFCIPNVTKLLGRIYYTHPGALKPGTLPPN 68
L A+ K Q Y + AGMG+ +A ++ + + P L
Sbjct: 14 LVAMGFGKFQIY---VLAYAGMGWVAEAMEMMLLS----------FVGPAVQSLWNLSAR 60
Query: 69 VSAAVNGVALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAKGTIG 128
+ + V G L G +G + DK GR+K + +T + + S S +
Sbjct: 61 QESLITSVVFAGMLIGAYSWGIVSDKHGRRKGFIITAVVTFVAGFLSAFSPNYM------ 114
Query: 129 TLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVSLVVST 188
L R +G G+GG P+ A+ E+ +RG ++ A G +
Sbjct: 115 WLIILRCLVGLGLGGG-PVLASWYLEFIPAPSRGTWMVVFSAFWTVGTI----------- 162
Query: 189 AFDHAFKAPPYKVDAAASLVPEADYVWRIILMFGAVPAGLTYYWRMKMPETARYTALVAK 248
++ A ++P WR +L F +VP+ L + E+ RY L +
Sbjct: 163 ----------FEASLAWLVMPRLG--WRWLLAFSSVPSSLLLLFYRWTSESPRYLILQGR 210
Query: 249 NAKQAA-----------QDMSSVLQVEIEAEQEKVDKIGVQDKNSFGL--------FSKE 289
A+ A Q VL E+E E E+ I ++ + SK
Sbjct: 211 KAEALAILEKIARMNKTQLPPGVLSSELETELEENKNIPTENTHLLKAGESGEAVAVSKI 270
Query: 290 FLRRH---GLHLLGTTS------------TWFLLDIAYYSSNLFQKDIYSSIGWLPPAQD 334
L+ G LL S +F AYY L ++ +S P +
Sbjct: 271 VLKADKEPGFSLLALLSPTLMKRTLLLWVVFFGNAFAYYGVVLLTTELNNSHNRCYPTEK 330
Query: 335 MNAIHEVFKVARATTLIALCGTVPGYWFTVAFIDVIGRFAIQLMGFFFMTVFMFALAIPY 394
+ V IA PG + A +D +GR A F +F+ L
Sbjct: 331 Q--LRNSNDVNYRDVFIASFAEFPGLLISAAMVDRLGRKASMASMLFTCCIFLLPLLSHQ 388
Query: 395 DHWTKRENRIGFLVMYAMTFFFANFGPNSTTFVVPAEIFPARLRSTCHGISSAAGKAGAI 454
+ G + + F + ++ EI+P +R+T G+ S+ G+ G I
Sbjct: 389 SPFITTVLLFGGRICISAAF--------TVVYIYAPEIYPTAVRTTGVGVGSSVGRIGGI 440
Query: 455 I 455
+
Sbjct: 441 L 441
>At4g04750 putative sugar transporter
Length = 461
Score = 60.1 bits (144), Expect = 3e-09
Identities = 92/473 (19%), Positives = 176/473 (36%), Gaps = 100/473 (21%)
Query: 80 GTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAKGTIGTLCFFRFWLGF 139
G + G L G L D +GR +T L++ LA ++F + L R G
Sbjct: 64 GLILGALICGKLADLVGRVYTIWITNILVLIGWLA--IAFAKDVR----LLDLGRLLQGI 117
Query: 140 GIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVSLVVSTAFDHAFKAPPY 199
+G L +SE A + RGA + + G G+ A + V+
Sbjct: 118 SVGISSYLGPIYISELAPRNLRGAASSLMQLFVGVGLSAFYALGTAVA------------ 165
Query: 200 KVDAAASLVPEADYVWRIILMFGAVPAGLTYYWRMKMPETARYTALVAKNAK-------- 251
WR + + G++P+ + +PE+ R+ A V + +
Sbjct: 166 ---------------WRSLAILGSIPSLVVLPLLFFIPESPRWLAKVGREKEVEGVLLSL 210
Query: 252 -----QAAQDMSSVLQVEIEAEQEKVDKIGVQDKNSFGLFSKEFLR--RHGLHLLGTTST 304
+ + +++L+ EQ+ +D G F LF +++ G+ L+
Sbjct: 211 RGAKSDVSDEAATILEYTKHVEQQDIDSRGF-----FKLFQRKYALPLTIGVVLISMPQL 265
Query: 305 WFLLDIAYYSSNLFQKD-IYSSIGWLPPAQDMNAIHEVFKVARATTLIALCGTVPGYWFT 363
L +Y+ +F + S IG++ T+++ + G V G
Sbjct: 266 GGLNGYTFYTDTIFTSTGVSSDIGFI-----------------LTSIVQMTGGVLG---- 304
Query: 364 VAFIDVIGRFAIQLMGFFFMTVFMFALAIPY-----DHWTKRENRIGFLVMYAMTFFFAN 418
V +D+ GR ++ L M + A AI + + W E + + ++ +F +
Sbjct: 305 VLLVDISGRRSLLLFSQAGMFLGCLATAISFFLQKNNCW---ETGTPIMALISVMVYFGS 361
Query: 419 FG--PNSTTFVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQSKDPKKRDAGY 476
+G +++ +EI+P ++ + + + + + F + Q
Sbjct: 362 YGLGMGPIPWIIASEIYPVDVKGAAGTVCNLVTSISSWLVTYSFNFLLQWSST------- 414
Query: 477 PAGIGMKNTLILLAVVNCLGIFFTF-LVPEANGKSLEEMSGENEEEEETEAAV 528
T ++ A V LG FT LVPE GKSLEE+ + ++ +
Sbjct: 415 -------GTFMMFATVMGLGFVFTAKLVPETKGKSLEEIQSAFTDSTSEDSTI 460
>At4g16480 membrane transporter like protein
Length = 582
Score = 58.9 bits (141), Expect = 7e-09
Identities = 77/371 (20%), Positives = 143/371 (37%), Gaps = 66/371 (17%)
Query: 69 VSAAVNGVALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAKGTIG 128
+ + + +A+ G + G GW+ DK GR+ + +A ++F A ++F +
Sbjct: 68 LQSTIVSMAVAGAIVGAAVGGWINDKFGRR--MSILIADVLFLIGAIVMAFAPAP----W 121
Query: 129 TLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVSLVVST 188
+ R ++GFG+G S +SE + + RGA +++ G I G S +++
Sbjct: 122 VIIVGRIFVGFGVGMASMTSPLYISEASPARIRGALVST----NGLLITGGQFFSYLINL 177
Query: 189 AFDHAFKAPPYKVDAAASLVPEADYVWRIILMFGAVPAGLTYYWRMKMPETARY------ 242
AF H WR +L VPA + + + +PE+ R+
Sbjct: 178 AFVH------------------TPGTWRWMLGVAGVPAIVQFVLMLSLPESPRWLYRKDR 219
Query: 243 ---TALVAKNAKQAAQDMSSVLQVEIEAEQEKVDKIGVQDKNSF---GLFSKEFLRR--- 293
+ + + A + + + +++ E EK D+ + D S G F +RR
Sbjct: 220 IAESRAILERIYPADEVEAEMEALKLSVEAEKADEAIIGDSFSAKLKGAFGNPVVRRGLA 279
Query: 294 HGLHLLGTTSTWFLLDIAYYSSNLFQKDIYSSIGWLPPAQDMNAIHEVFKVARATTLIAL 353
G+ + + + YYS ++ Q Y+S K A A +LI
Sbjct: 280 AGITVQVAQQFVGINTVMYYSPSIVQFAGYAS----------------NKTAMALSLITS 323
Query: 354 CGTVPGYWFTVAFIDVIGRFAIQLMGFFFMTVFMFALAIPYDHWTKRENRIGFLVMYAMT 413
G ++ F+D GR + ++ F + + LA + +I
Sbjct: 324 GLNALGSIVSMMFVDRYGRRKLMIISMFGIIACLIILATVFSQAAIHAPKID-------A 376
Query: 414 FFFANFGPNST 424
F F PN+T
Sbjct: 377 FESRTFAPNAT 387
Score = 43.5 bits (101), Expect = 3e-04
Identities = 30/116 (25%), Positives = 54/116 (45%), Gaps = 16/116 (13%)
Query: 402 NRIGFLVMYAMTFFFANFGPNSTT--FVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFG 459
++ GFL + + + + P T ++V +EI+P R R GI++ + +I +
Sbjct: 454 SKFGFLAIVFLGLYIVVYAPGMGTVPWIVNSEIYPLRYRGLGGGIAAVSNWVSNLIVSES 513
Query: 460 FLYASQSKDPKKRDAGYPAGIGMKNTLILLAVVNCLGIFFTF-LVPEANGKSLEEM 514
FL + + +G T +L A + +G+FF + LVPE G EE+
Sbjct: 514 FLSLTHA-------------LGSSGTFLLFAGFSTIGLFFIWLLVPETKGLQFEEV 556
>At2g48020 sugar transporter like protein
Length = 463
Score = 58.9 bits (141), Expect = 7e-09
Identities = 96/449 (21%), Positives = 175/449 (38%), Gaps = 82/449 (18%)
Query: 80 GTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAKGTIGTLCFFRFWLGF 139
G + G + G + D +GRK ++ A V LA + F AKG + L R G+
Sbjct: 77 GAMIGAITSGPIADLVGRKGAMRVSSAFCVVGWLA--IIF---AKGVVA-LDLGRLATGY 130
Query: 140 GIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVSLVVSTAFDHAFKAPPY 199
G+G + ++E A K RGA + I G VS ++ T
Sbjct: 131 GMGAFSYVVPIFIAEIAPKTFRGALTT----LNQILICTGVSVSFIIGTLV--------- 177
Query: 200 KVDAAASLVPEADYVWRIILMFGAVPAGLTYYWRMKMPETARYTALVAKNAK-QAAQDMS 258
WR++ + G +P ++ +PE+ R+ A V ++ + +AA
Sbjct: 178 --------------TWRVLALIGIIPCAASFLGLFFIPESPRWLAKVGRDTEFEAALRKL 223
Query: 259 SVLQVEIEAEQEKV-DKIGVQDK----NSFGLFSKEFLRR--HGLHLLGTTSTWFLLDIA 311
+ +I E ++ D I ++ LF + ++R L+ + I
Sbjct: 224 RGKKADISEEAAEIQDYIETLERLPKAKMLDLFQRRYIRSVLIAFGLMVFQQFGGINGIC 283
Query: 312 YYSSNLF-QKDIYSSIGWLPPAQDMNAIHEVFKVARATTLIALCGTVPGYWFTVAFIDVI 370
+Y+S++F Q + +G + + A+ +V A ++ G P V+ ++
Sbjct: 284 FYTSSIFEQAGFPTRLGMI-----IYAVLQVVITALNAPIVDRAGRKP--LLLVSATGLV 336
Query: 371 GRFAIQLMGFFFMTVFMFALAIPYDHWTKRENRIGFLVMYAMTFFFANF--GPNSTTFVV 428
I + F+ M A+P L + + + +F G + +VV
Sbjct: 337 IGCLIAAVSFYLKVHDMAHEAVP------------VLAVVGIMVYIGSFSAGMGAMPWVV 384
Query: 429 PAEIFPARLRSTCHGISSAAGKAG--AIIGAFGFLYASQSKDPKKRDAGYPAGIGMKNTL 486
+EIFP ++ G+++ G A+ F FL + S T
Sbjct: 385 MSEIFPINIKGVAGGMATLVNWFGAWAVSYTFNFLMSWSS----------------YGTF 428
Query: 487 ILLAVVNCLGIFFTF-LVPEANGKSLEEM 514
++ A +N L I F +VPE GK+LE++
Sbjct: 429 LIYAAINALAIVFVIAIVPETKGKTLEQI 457
>At5g59250 D-xylose-H+ symporter - like protein
Length = 558
Score = 58.2 bits (139), Expect = 1e-08
Identities = 106/479 (22%), Positives = 175/479 (36%), Gaps = 91/479 (18%)
Query: 66 PPNVSAAVNGVALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAKG 125
P + V+G +L G L G + + D +GR++ + L + SL +G +
Sbjct: 141 PVQLGLVVSG-SLYGALLGSISVYGVADFLGRRRELIIAAVLYLLGSLITGCA------P 193
Query: 126 TIGTLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVSLV 185
+ L R GFGIG + ++E + RG I+ GIL G
Sbjct: 194 DLNILLVGRLLYGFGIGLAMHGAPLYIAETCPSQIRGTLISLKELFIVLGILLG------ 247
Query: 186 VSTAFDHAFKAPPYKVDAAASLVPEADYVWRIILMFGAVPAGLTYYWRMKMPETARYTAL 245
F +++D WR + FG A L +P + R+ L
Sbjct: 248 --------FSVGSFQIDVVGG--------WRYMYGFGTPVALLMGLGMWSLPASPRWLLL 291
Query: 246 VAKNAKQAAQDMSSVLQVEI----------EAEQEKVD------KIGVQDKNSFGLFSKE 289
A K Q+ + + + ++ VD K +D+ S G F +
Sbjct: 292 RAVQGKGQLQEYKEKAMLALSKLRGRPPGDKISEKLVDDAYLSVKTAYEDEKSGGNFLEV 351
Query: 290 FLRRHGLHLLGTTSTWFLL---------DIAYYSSNLFQKDIYSSIGWLPPAQDMNAIHE 340
F G +L T L+ + YY+ ++ Q +S+ A ++ I
Sbjct: 352 F---QGPNLKALTIGGGLVLFQQITGQPSVLYYAGSILQTAGFSAAA---DATRVSVIIG 405
Query: 341 VFKVARATTLIALCGTVPGYWFTVAFIDVIGRFAIQLMGFFFMTVFMFALAIPYDHWTKR 400
VFK+ W VA +D +GR + + G + + +F L+ Y
Sbjct: 406 VFKLLMT-------------WVAVAKVDDLGRRPLLIGGVSGIALSLFLLSAYYKFLGGF 452
Query: 401 EN-RIGFLVMYAMTFFFANFGPNSTTFVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFG 459
+G L++Y + + FGP S V +EIFP R R ++ I F
Sbjct: 453 PLVAVGALLLYVGCYQIS-FGPISWLMV--SEIFPLRTRGRGISLAVLTNFGSNAIVTFA 509
Query: 460 FLYASQSKDPKKRDAGYPAGIGMKNTLILLAVVNCLGIFFTFLV-PEANGKSLEEMSGE 517
F P K +G +N +L + + + F LV PE G SLEE+ +
Sbjct: 510 F-------SPLKEF------LGAENLFLLFGGIALVSLLFVILVVPETKGLSLEEIESK 555
>At4g36670 sugar transporter like protein
Length = 493
Score = 56.2 bits (134), Expect = 4e-08
Identities = 101/465 (21%), Positives = 173/465 (36%), Gaps = 71/465 (15%)
Query: 73 VNGVALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAKGTIGTLCF 132
+ G+ L G L G D +GR+ L L + S+ G + L
Sbjct: 59 LTGILNLCALVGSLLAGRTSDIIGRRYTIVLASILFMLGSILMGWGPNYPV------LLS 112
Query: 133 FRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVSLVVSTAFDH 192
R G G+G ++ +E A RG + GIL G IV+ +
Sbjct: 113 GRCTAGLGVGFALMVAPVYSAEIATASHRGLLASLPHLCISIGILLGYIVN--------Y 164
Query: 193 AFKAPPYKVDAAASLVPEADYVWRIILMFGAVPAGLTYYWRMKMPETARYTALVAKNAKQ 252
F P + WR++L AVP+ + + +KMPE+ R+ + + K+
Sbjct: 165 FFSKLPMHIG------------WRLMLGIAAVPSLVLAFGILKMPESPRWLIMQGR-LKE 211
Query: 253 AAQDMSSVLQVEIEAE---QEKVDKIGVQDKNSFGLFSKEFLRRHGLHLLGTTSTWFLLD 309
+ + V EAE Q+ G+ K + E + HG W +
Sbjct: 212 GKEILELVSNSPEEAELRFQDIKAAAGIDPKCVDDVVKMEGKKTHG------EGVW--KE 263
Query: 310 IAYYSSNLFQKDIYSSIG--WLPPAQDMNAI----HEVFKVARATTLIALCGTVPG---- 359
+ + ++ + +++G + A + A+ +FK A TT L G
Sbjct: 264 LILRPTPAVRRVLLTALGIHFFQHASGIEAVLLYGPRIFKKAGITTKDKLFLVTIGVGIM 323
Query: 360 ---YWFTVAFI-DVIGRFAIQLMGF----FFMTVFMFALAIPYDHWTKRENRIGFLVMYA 411
+ FT + D +GR + L +T+ F L + + K + ++ A
Sbjct: 324 KTTFIFTATLLLDKVGRRKLLLTSVGGMVIALTMLGFGLTMAQNAGGKLAWALVLSIVAA 383
Query: 412 MTFF-FANFGPNSTTFVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQSKDPK 470
+F F + G T+V +E+FP +LR+ + A + + FL + +
Sbjct: 384 YSFVAFFSIGLGPITWVYSSEVFPLKLRAQGASLGVAVNRVMNATVSMSFLSLTSA---- 439
Query: 471 KRDAGYPAGIGMKNTLILLAVVNCLGI-FFTFLVPEANGKSLEEM 514
I + A V + FF FL+PE GKSLEE+
Sbjct: 440 ---------ITTGGAFFMFAGVAAVAWNFFFFLLPETKGKSLEEI 475
>At2g35740 putative sugar transporter
Length = 580
Score = 56.2 bits (134), Expect = 4e-08
Identities = 76/347 (21%), Positives = 136/347 (38%), Gaps = 65/347 (18%)
Query: 76 VALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAKGTIGTLCFFRF 135
+ + G + G GW DK GR+ + +A ++F A + H+ + R
Sbjct: 74 MTVAGAIVGAAIGGWYNDKFGRR--MSVLIADVLFLLGALVMVIAHAP----WVIILGRL 127
Query: 136 WLGFGIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVSLVVSTAFDHAFK 195
+GFG+G S +SE + + RGA +++ G I G +S +++ AF H
Sbjct: 128 LVGFGVGMASMTSPLYISEMSPARIRGALVST----NGLLITGGQFLSYLINLAFVH--- 180
Query: 196 APPYKVDAAASLVPEADYVWRIILMFGAVPAGLTYYWRMKMPETARYTALVAKNAKQAAQ 255
WR +L A+PA + + + +PE+ R+ L + K ++
Sbjct: 181 ---------------TPGTWRWMLGVSAIPAIIQFCLMLTLPESPRW--LYRNDRKAESR 223
Query: 256 DM------SSVLQVEIEAEQEKVDKIGVQDKNSFGLFSKEFLR--------RHGLHLLGT 301
D+ + +++ EI A +E V + D++ G + LR RHGL T
Sbjct: 224 DILERIYPAEMVEAEIAALKESV-RAETADEDIIGHTFSDKLRGALSNPVVRHGLAAGIT 282
Query: 302 TST----WFLLDIAYYSSNLFQKDIYSSIGWLPPAQDMNAIHEVFKVARATTLIALCGTV 357
+ + YYS + Q Y+S K A A LI
Sbjct: 283 VQVAQQFVGINTVMYYSPTILQFAGYAS----------------NKTAMALALITSGLNA 326
Query: 358 PGYWFTVAFIDVIGRFAIQLMGFFFMTVFMFALAIPYDHWTKRENRI 404
G ++ F+D GR + ++ F + + LA ++ + +I
Sbjct: 327 VGSVVSMMFVDRYGRRKLMIISMFGIITCLVILAAVFNEASNHAPKI 373
Score = 41.6 bits (96), Expect = 0.001
Identities = 28/116 (24%), Positives = 54/116 (46%), Gaps = 16/116 (13%)
Query: 402 NRIGFLVMYAMTFFFANFGPNSTT--FVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFG 459
++ G+L + + + + P T ++V +EI+P R R GI++ + ++ +
Sbjct: 453 SKFGYLAIVFLGLYIIVYAPGMGTVPWIVNSEIYPLRYRGLAGGIAAVSNWMSNLVVSET 512
Query: 460 FLYASQSKDPKKRDAGYPAGIGMKNTLILLAVVNCLGIFFTF-LVPEANGKSLEEM 514
FL + + +G T +L A + +G+FF + LVPE G EE+
Sbjct: 513 FLTLTNA-------------VGSSGTFLLFAGSSAVGLFFIWLLVPETKGLQFEEV 555
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.139 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,633,625
Number of Sequences: 26719
Number of extensions: 478328
Number of successful extensions: 2049
Number of sequences better than 10.0: 84
Number of HSP's better than 10.0 without gapping: 23
Number of HSP's successfully gapped in prelim test: 61
Number of HSP's that attempted gapping in prelim test: 1896
Number of HSP's gapped (non-prelim): 129
length of query: 541
length of database: 11,318,596
effective HSP length: 104
effective length of query: 437
effective length of database: 8,539,820
effective search space: 3731901340
effective search space used: 3731901340
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148718.7


