

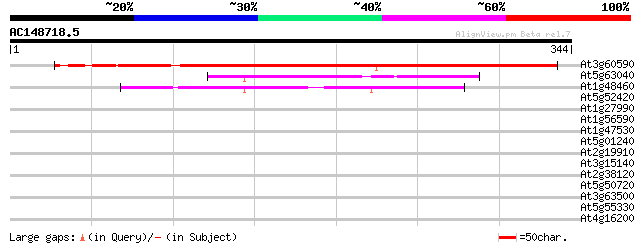
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148718.5 + phase: 0
(344 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g60590 unknown protein 372 e-103
At5g63040 unknown protein 65 5e-11
At1g48460 unknown protein 59 3e-09
At5g52420 unknown protein 37 0.012
At1g27990 hypothetical protein 35 0.046
At1g56590 clathrin coat assembly like protein 32 0.66
At1g47530 unknown protein 30 2.5
At5g01240 LAX1 / AUX1 -like permease 29 4.3
At2g19910 hypothetical protein 29 4.3
At3g15140 unknown protein 28 5.6
At2g38120 unknown protein 28 5.6
At5g50720 unknown protein 28 7.3
At3g63500 putative protein 28 7.3
At5g55330 wax synthase-like protein 28 9.5
At4g16200 splicing factor SF3a like protein 28 9.5
>At3g60590 unknown protein
Length = 329
Score = 372 bits (956), Expect = e-103
Identities = 182/312 (58%), Positives = 232/312 (74%), Gaps = 17/312 (5%)
Query: 28 LKSGKPFQSRRFNFPSIRLNQSFICCTKLTPWEPSPGVAYAPTDNQSDNFLQSTASVFET 87
LK+G S F F ++R+ +C KL+ WEPSP + +A + +D L TA+VFE+
Sbjct: 31 LKTG----SCNFRFRNLRV----LCTPKLSQWEPSPFI-HASAEEAADIVLDKTANVFES 81
Query: 88 LESSKVDESPTANVEGLVEEKDRPGPELQLFKWPMWLLGPSILLATGMVPTLWLPISSIF 147
+ S +E + + R ++Q+ KWP+WLLGPS+LL +GM PTLWLP+SS+F
Sbjct: 82 IVSESAEEEKVD-----MSAQQRTNSQVQVLKWPIWLLGPSVLLTSGMAPTLWLPLSSVF 136
Query: 148 LGPNIASLLSLIGLDCIFNLGATLFLLMADSCSRPKNPTQEIKSKAPFSYQFWNIVATLT 207
LG N+ SLLSLIGLDCIFNLGATLFLLMADSC+RPK+P+Q SK PFSY+FWN+ + +
Sbjct: 137 LGSNVVSLLSLIGLDCIFNLGATLFLLMADSCARPKDPSQSCNSKPPFSYKFWNMFSLII 196
Query: 208 GFIVPSLLMFGSQKGF---LQPQLPFISSAVLLGPYLLLLSVQILTELLTWYWQSPVWLV 264
GF+VP LL+FGSQ G LQPQ+PF+SSAV+L PY +LL+VQ LTE+LTW+WQSPVWLV
Sbjct: 197 GFLVPMLLLFGSQSGLLASLQPQIPFLSSAVILFPYFILLAVQTLTEILTWHWQSPVWLV 256
Query: 265 TPIIYEAYRILQLMRGLKLGAELTAPAWMMHTIRGLVCWWVLILGLQLMRVAWFAGLSAR 324
TP++YEAYRILQLMRGL L AE+ AP W++H +RGLV WWVLILG+QLMRVAWFAG ++R
Sbjct: 257 TPVVYEAYRILQLMRGLTLSAEVNAPVWVVHMLRGLVSWWVLILGMQLMRVAWFAGFASR 316
Query: 325 ARKDQSSSSEAS 336
Q S AS
Sbjct: 317 TTTGQQPQSVAS 328
>At5g63040 unknown protein
Length = 366
Score = 65.1 bits (157), Expect = 5e-11
Identities = 49/170 (28%), Positives = 84/170 (48%), Gaps = 9/170 (5%)
Query: 122 MWLLGPSILLATGMVPTLWLP--ISSIFLGPNIASLLSLIGLDCIFNLGATLFLLMADSC 179
+WL+GP++L+++ ++P ++L +S++F + L L + +F G FLL+ D
Sbjct: 151 LWLIGPAVLVSSFILPPVYLRRIVSAVFEDSLLTDFLILFFTEALFYCGVAAFLLIIDRS 210
Query: 180 SRPKNPTQEIKSKAPFSYQFWNIVATLT-GFIVPSLLMFGSQKGFLQPQLPFISSAVLLG 238
+ + + Q + VATL ++P + M GF+ P +SA L
Sbjct: 211 RKGSGKVPQNRINPSQLGQRISSVATLVLSLMIPMVTM-----GFVWPWTGPAASATL-A 264
Query: 239 PYLLLLSVQILTELLTWYWQSPVWLVTPIIYEAYRILQLMRGLKLGAELT 288
PYL+ + VQ E Y SP + PII++ YR+ QL R +L L+
Sbjct: 265 PYLVGIVVQFAFEQYARYRNSPSSPIIPIIFQVYRLHQLNRAAQLVTALS 314
>At1g48460 unknown protein
Length = 340
Score = 59.3 bits (142), Expect = 3e-09
Identities = 52/220 (23%), Positives = 93/220 (41%), Gaps = 21/220 (9%)
Query: 69 PTDNQSDNFLQSTASVFETLESSKVDESPTANVEGLVEEKDRPGPELQLFKWP-MWLLGP 127
P QSD + A +F S T LVEE Q K +W+L P
Sbjct: 70 PEKKQSDKSNYARAELFRGKSGSVSFNGLTHQ---LVEESKLVSAPFQEEKGSFLWVLAP 126
Query: 128 SILLATGMVPTLWLP--ISSIFLGPNIASLLSLIGLDCIFNLGATLFLLMADSCSRPKNP 185
+L+++ ++P +L I + F +A +++ + +F G +FL + D RP
Sbjct: 127 VVLISSLILPQFFLSGIIEATFKNDTVAEIVTSFCFETVFYAGLAIFLSVTDRVQRPY-- 184
Query: 186 TQEIKSKAPFSYQFWNIVATLTGFIVPSLLMFGSQ------KGFLQPQLPFISSAVLLGP 239
FS + W ++ L G++ + L G + ++ I + + + P
Sbjct: 185 -------LDFSSKRWGLITGLRGYLTSAFLTMGLKVVVPVFAVYMTWPALGIDALIAVLP 237
Query: 240 YLLLLSVQILTELLTWYWQSPVWLVTPIIYEAYRILQLMR 279
+L+ +VQ + E S W + PI++E YR+ Q+ R
Sbjct: 238 FLVGCAVQRVFEARLERRGSSCWPIVPIVFEVYRLYQVTR 277
>At5g52420 unknown protein
Length = 242
Score = 37.4 bits (85), Expect = 0.012
Identities = 36/137 (26%), Positives = 65/137 (47%), Gaps = 16/137 (11%)
Query: 152 IASLLSLIGL----DCIFNLGATLFLLMADSCSRP--KNPTQE--IKSKAPFSYQFWNIV 203
I +LS GL D IF + ++ P NP ++ + S ++ +
Sbjct: 50 IIIVLSASGLVTIQDFIFTILTLIYFFFLSKLIFPPHNNPNRDAPLTSSTNKIFRIYVTA 109
Query: 204 ATLTGFIVPSLLMFGSQKGFLQPQLPFISSAVLLGPYLLLLSVQILTE-LLTWY-WQSPV 261
A + G I+P +F +G ++ +S+A P++ LL+ QI E L T + + +P
Sbjct: 110 AGIVGLIIPICYIF---EGIVEDDKNGVSAAA---PHVFLLASQIFMEGLATMFGFSAPA 163
Query: 262 WLVTPIIYEAYRILQLM 278
++ PI+Y A R+L L+
Sbjct: 164 RILVPIVYNARRVLTLV 180
>At1g27990 hypothetical protein
Length = 271
Score = 35.4 bits (80), Expect = 0.046
Identities = 33/127 (25%), Positives = 60/127 (46%), Gaps = 18/127 (14%)
Query: 161 LDCIFNLGATLFLLMADSCSRPKNPTQEIKSKAPFSYQ------FWNIVATLTGFIVPSL 214
LD +F A+++++ + SR P+ + + +P ++ + I T G +P
Sbjct: 66 LDMLFPAFASIYII---ALSRLAFPSHGVSTASPEVFRGSKLFRLYVISGTTIGLFLPLA 122
Query: 215 LMFGSQKGFLQPQLPFISSAVLLGPYLLLLSVQILTELL---TWYWQSPVWLVTPIIYEA 271
+ G GF + + SA P+L LLS QILTE + + PV + P++Y
Sbjct: 123 YVLG---GFARGDDHAVRSAT---PHLFLLSCQILTENVISGLSLFSPPVRALVPLLYTV 176
Query: 272 YRILQLM 278
+RI ++
Sbjct: 177 WRIFVII 183
>At1g56590 clathrin coat assembly like protein
Length = 480
Score = 31.6 bits (70), Expect = 0.66
Identities = 24/94 (25%), Positives = 41/94 (43%), Gaps = 9/94 (9%)
Query: 30 SGKPFQSRRFNFPSIRLNQSFICCTKLTPWEPSPGVAYAPTDNQSDNFLQSTASVFETLE 89
+G P + F PSI + F C + PWE +++ P D + L S V + L+
Sbjct: 278 TGFPDLTLSFANPSILEDMRFHPCVRYRPWESHQVLSFVPPDGEFK--LMSYRCVVKKLK 335
Query: 90 SSKV-------DESPTANVEGLVEEKDRPGPELQ 116
++ V +S T + LV + PG ++
Sbjct: 336 NTPVYVKPQITSDSGTCRISVLVGIRSDPGKTIE 369
>At1g47530 unknown protein
Length = 484
Score = 29.6 bits (65), Expect = 2.5
Identities = 46/171 (26%), Positives = 79/171 (45%), Gaps = 29/171 (16%)
Query: 129 ILLATGMVPTLWLPISSIFLGPNIASLLSLIGLDCIFNLGATLFLLMADSCSRPKNPTQE 188
+++ TG++P +P+ +I + NI ++I + A + + +++
Sbjct: 280 LVVVTGLLPNPLIPVDAISICMNIEGWTAMISI----GFNAAISVRVSNELGAGN----- 330
Query: 189 IKSKAPFSYQFWNIVATLTGFIVPSLLMFGSQKGFLQPQLPFISS----------AVLLG 238
+ A FS +I +TL G IV +++ ++ F P L F SS AVLLG
Sbjct: 331 -AALAKFSVIVVSITSTLIG-IVCMIVVLATKDSF--PYL-FTSSEAVAAETTRIAVLLG 385
Query: 239 PYLLLLSVQ-ILTELLTWY-WQSPVWLVTPIIYEAYRILQLMRGLKLGAEL 287
+LL S+Q +L+ + WQ+ LV + Y I+ L GL LG L
Sbjct: 386 FTVLLNSLQPVLSGVAVGAGWQA---LVAYVNIACYYIIGLPAGLVLGFTL 433
>At5g01240 LAX1 / AUX1 -like permease
Length = 488
Score = 28.9 bits (63), Expect = 4.3
Identities = 22/84 (26%), Positives = 39/84 (46%), Gaps = 4/84 (4%)
Query: 135 MVPTLWLPISSIFLGPNIASLLSLIGLDCIFNLGATLFLLMADSCSRPKNPTQEIKSKAP 194
+VP +L I F GP +++ +L+ ++ + A +L + S +N + K P
Sbjct: 366 VVPIWFLAIIFPFFGPINSAVGALLVTFTVYIIPALAHMLTYRTASARRNAAE----KPP 421
Query: 195 FSYQFWNIVATLTGFIVPSLLMFG 218
F W V + FIV +L+ G
Sbjct: 422 FFIPSWAGVYVINAFIVVWVLVLG 445
>At2g19910 hypothetical protein
Length = 966
Score = 28.9 bits (63), Expect = 4.3
Identities = 11/40 (27%), Positives = 22/40 (54%)
Query: 84 VFETLESSKVDESPTANVEGLVEEKDRPGPELQLFKWPMW 123
+F+T++S +E P + + L +D P P+ + K+ W
Sbjct: 817 IFDTVDSHNAEEPPPSEISKLWYFEDEPVPKSHMDKFTSW 856
>At3g15140 unknown protein
Length = 337
Score = 28.5 bits (62), Expect = 5.6
Identities = 17/62 (27%), Positives = 30/62 (47%), Gaps = 2/62 (3%)
Query: 33 PFQSRRFNFPSIRL--NQSFICCTKLTPWEPSPGVAYAPTDNQSDNFLQSTASVFETLES 90
PF+ RF++P+ + I C PSP + + + S + ST S+ ET E+
Sbjct: 17 PFRDTRFSYPATLALAHTKRIMCNSSHSVSPSPSPSDFSSSSSSSSSSPSTFSLMETSEN 76
Query: 91 SK 92
++
Sbjct: 77 AR 78
>At2g38120 unknown protein
Length = 485
Score = 28.5 bits (62), Expect = 5.6
Identities = 20/84 (23%), Positives = 39/84 (45%), Gaps = 4/84 (4%)
Query: 135 MVPTLWLPISSIFLGPNIASLLSLIGLDCIFNLGATLFLLMADSCSRPKNPTQEIKSKAP 194
++P +L I F GP +++ +L+ ++ + + +L S S +N + K P
Sbjct: 360 VIPIWFLAIIFPFFGPINSAVGALLVSFTVYIIPSLAHMLTYRSASARQNAAE----KPP 415
Query: 195 FSYQFWNIVATLTGFIVPSLLMFG 218
F W + L F+V +L+ G
Sbjct: 416 FFMPSWTAMYVLNAFVVVWVLIVG 439
>At5g50720 unknown protein
Length = 116
Score = 28.1 bits (61), Expect = 7.3
Identities = 14/39 (35%), Positives = 20/39 (50%), Gaps = 3/39 (7%)
Query: 242 LLLSVQILTELLTWYWQSPVWLVTPIIYEAYRILQLMRG 280
L LS IL LL W P+W +++ A+ +L RG
Sbjct: 53 LTLSELILQSLLEWI---PIWYTAKLVFVAWLVLPQFRG 88
>At3g63500 putative protein
Length = 1162
Score = 28.1 bits (61), Expect = 7.3
Identities = 33/118 (27%), Positives = 48/118 (39%), Gaps = 12/118 (10%)
Query: 71 DNQSDNFLQSTASVFETLESSKVDESPTANVEGLVEEKDRPGPELQLFKWPMWLLGPSIL 130
D + DNF + FE SS V + G+ + KD +L L + L P +L
Sbjct: 452 DEKDDNFGGPSIRGFELFSSSPVRRAKKTEQSGVNKHKDE---KLLLEPLDLSLSLPDVL 508
Query: 131 LATGMVPT--LWLPISSIFLGPNIASLLSLIGLDCIFNLGATLFLLMADSCSRPKNPT 186
L G T L P+ S S+ SL C + G T+ + + S S NP+
Sbjct: 509 LPIGGQDTNQLGSPVRS-------GSVRSLTDTFCTNSDGFTMSMSFSGSRSFNHNPS 559
>At5g55330 wax synthase-like protein
Length = 346
Score = 27.7 bits (60), Expect = 9.5
Identities = 19/83 (22%), Positives = 36/83 (42%), Gaps = 4/83 (4%)
Query: 200 WNIVATLTGFIVPSLLMFGSQKGFLQPQLPFISSAVLLGPYLLLL----SVQILTELLTW 255
+ I L+G VP L++F +KG L P P + V + + L + + W
Sbjct: 60 FTIAFFLSGLAVPKLILFALEKGPLFPLPPNLPHFVCFACFPIKLQKKPNPENTNHFPKW 119
Query: 256 YWQSPVWLVTPIIYEAYRILQLM 278
+ V++ ++ +AY Q +
Sbjct: 120 VFALKVFIFGALLLQAYHYKQFL 142
>At4g16200 splicing factor SF3a like protein
Length = 288
Score = 27.7 bits (60), Expect = 9.5
Identities = 16/41 (39%), Positives = 22/41 (53%), Gaps = 1/41 (2%)
Query: 69 PTDNQSDNFLQSTASVFETLESSKVDESPTANVEGLVEEKD 109
PTD + NFL+STA T K+ E + N +G +E D
Sbjct: 50 PTDAKF-NFLRSTADPCHTYYKHKLAEYSSQNQDGATDESD 89
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.137 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,545,454
Number of Sequences: 26719
Number of extensions: 313907
Number of successful extensions: 762
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 750
Number of HSP's gapped (non-prelim): 16
length of query: 344
length of database: 11,318,596
effective HSP length: 100
effective length of query: 244
effective length of database: 8,646,696
effective search space: 2109793824
effective search space used: 2109793824
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148718.5


