

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148651.2 - phase: 0
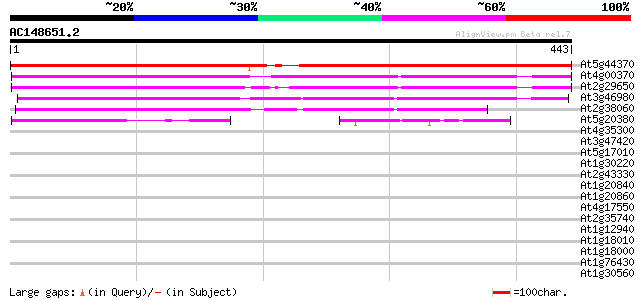
(443 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g44370 Na+-dependent inorganic phosphate cotransporter-like p... 655 0.0
At4g00370 unknown protein 289 2e-78
At2g29650 Na+-dependent inorganic phosphate cotransporter like p... 279 2e-75
At3g46980 unknown protein 208 5e-54
At2g38060 putative Na+-dependent inorganic phosphate cotransporter 187 1e-47
At5g20380 putative protein 75 9e-14
At4g35300 sugar transporter like protein 41 0.002
At3g47420 putative sugar transporter protein 40 0.003
At5g17010 sugar transporter - like protein 37 0.017
At1g30220 unknown protein 36 0.049
At2g43330 membrane transporter like protein 35 0.064
At1g20840 putative sugar transporter protein 35 0.064
At1g20860 putative inorganic phosphate transporter protein 35 0.083
At4g17550 putative protein 33 0.24
At2g35740 putative sugar transporter 33 0.24
At1g12940 nitrate transporter, putative 33 0.24
At1g18010 hypothetical protein 33 0.32
At1g18000 hypothetical protein 33 0.32
At1g76430 putative phosphate transporter 33 0.41
At1g30560 unknown protein, 5' partial 33 0.41
>At5g44370 Na+-dependent inorganic phosphate cotransporter-like
protein
Length = 432
Score = 655 bits (1691), Expect = 0.0
Identities = 328/449 (73%), Positives = 375/449 (83%), Gaps = 25/449 (5%)
Query: 1 MQNFPVRYLIVLLTFICTSVCYIERVGFSIAYTVAADAAGINQSTKGTILSTFYYGYACS 60
+ N P RY+IV LTF+ T VCYIERVGFSIAYTVAADAAGINQS+KGTILSTF+ GYACS
Sbjct: 3 LSNIPQRYVIVFLTFLSTCVCYIERVGFSIAYTVAADAAGINQSSKGTILSTFFVGYACS 62
Query: 61 QVPGGYFAQKIGGRKILLFSFLLWSLTCALLPLDPNKTLILVTARLLVGVAQGFIFPSIH 120
QVPGG+ AQKIGGRK+LL SF+LWS TC L+PLDPN+ +LV ARLLVGVAQGFIFPSIH
Sbjct: 63 QVPGGWAAQKIGGRKVLLLSFVLWSSTCFLVPLDPNRVGLLVVARLLVGVAQGFIFPSIH 122
Query: 121 TVLAQWVPPHERSRSVSLTTSGMYLGAAFGMLFLPSLVKFKGPQSVFVAESFLGFVWCFI 180
TVLAQWVPPHERSR VS+TTSGMYLGAA GM LP+LV+ +GP+SVF+AE+ G +W +
Sbjct: 123 TVLAQWVPPHERSRLVSITTSGMYLGAALGMWLLPALVELRGPESVFLAEALAGVIWSLL 182
Query: 181 WFRYSSDP------KSSASGAGESLLPVNKKIDRRVSDLKVGVEKNGGEGKKSGVGIPWV 234
W RY++DP K++A+G G +LLP N V+ KV IPW
Sbjct: 183 WIRYATDPPRSEHPKAAAAGFGGALLPTN------VNHHKV-------------THIPWK 223
Query: 235 KILTSLPVWAIVVNNFTFHYALYVLMNWLPTYFELGLKLSLHEMGSSKMLPYLNMFVFSN 294
KI+ SLPVWAIVVNNFTFHYALYVLMNWLPTYFELGL++SL M SSKM+PYLNMFVFS
Sbjct: 224 KIMLSLPVWAIVVNNFTFHYALYVLMNWLPTYFELGLQISLQGMDSSKMVPYLNMFVFSI 283
Query: 295 IGGVVADYLITRRVMSVTRTRKFLNTIGFLVASFALIVIPSFRTSGGAVFCSSVALGFLA 354
+GG +ADYLIT+R++SVTRTRKFLNT+GFL+AS AL+V+P FRT G + CSSVALGFLA
Sbjct: 284 VGGFIADYLITKRILSVTRTRKFLNTVGFLIASAALMVLPMFRTENGVILCSSVALGFLA 343
Query: 355 LGRAGFAVNHMDVAPRYAGIVMGVSNTAGTLAGIVGVDLTGKLLEAAKASDSDLSTPESW 414
LGRAGFAVNHMD+APRYAGIVMGVSNTAGTLAGI+GVDLTGKLLEA+K SDLS PESW
Sbjct: 344 LGRAGFAVNHMDIAPRYAGIVMGVSNTAGTLAGIIGVDLTGKLLEASKLVYSDLSHPESW 403
Query: 415 RLVFFIPGLLCVFSSLVFLLFSTGERIFD 443
R+VFFIPGLLC+FSS+VFLLFSTGERIFD
Sbjct: 404 RVVFFIPGLLCIFSSVVFLLFSTGERIFD 432
>At4g00370 unknown protein
Length = 541
Score = 289 bits (739), Expect = 2e-78
Identities = 160/444 (36%), Positives = 252/444 (56%), Gaps = 30/444 (6%)
Query: 2 QNFPVRYLIVLLTFICTSVCYIERVGFSIAYTVAADAAGINQSTKGTILSTFYYGYACSQ 61
Q FP R++IVLL F +C ++RV SIA + + +T G I S+F++GY +Q
Sbjct: 126 QQFPRRWVIVLLCFSSFLLCNMDRVNMSIAILPMSQEYNWSSATVGLIQSSFFWGYLLTQ 185
Query: 62 VPGGYFAQKIGGRKILLFSFLLWSLTCALLPLDPNKTL-ILVTARLLVGVAQGFIFPSIH 120
+ GG +A K GG+ +L F + WS + P+ L L+ R +G+ +G P+++
Sbjct: 186 ILGGIWADKFGGKVVLGFGVVWWSFATIMTPIAARLGLPFLLVVRAFMGIGEGVAMPAMN 245
Query: 121 TVLAQWVPPHERSRSVSLTTSGMYLGAAFGMLFLPSLVKFKGPQSVFVAESFLGFVWCFI 180
+L++W+P ERSRS++L SGMYLG+ G+ F P L+ G SVF + LG +W +
Sbjct: 246 NMLSKWIPVSERSRSLALVYSGMYLGSVTGLAFSPMLITKFGWPSVFYSFGSLGSIWFLL 305
Query: 181 WFRYS-SDPKSSASGAGESLLPVNKKIDRRVSDLKVGVEKNGGEGKKSGVGIPWVKILTS 239
W +++ S PK D +S+ + V G + ++ IPW IL+
Sbjct: 306 WLKFAYSSPKD----------------DPDLSEEEKKVILGGSKPREPVTVIPWKLILSK 349
Query: 240 LPVWAIVVNNFTFHYALYVLMNWLPTYFELGLKLSLHEMGSSKMLPYLNMFVFSNIGGVV 299
PVWA+++++F ++ ++L+ W+PTY+ LK +L E G +LP+L M VF+NIGG +
Sbjct: 350 PPVWALIISHFCHNWGTFILLTWMPTYYNQVLKFNLTESGLLCVLPWLTMAVFANIGGWI 409
Query: 300 ADYLITRRVMSVTRTRKFLNTIGFLVASFALIVIPSFRTSGGAVFCSSVALGFLALGRAG 359
AD L++R +S+T RK + +IGFL +F L + +T AV C + + G A ++G
Sbjct: 410 ADTLVSRG-LSITNVRKIMQSIGFLGPAFFLSQLSHVKTPAMAVLCMACSQGSDAFSQSG 468
Query: 360 FAVNHMDVAPRYAGIVMGVSNTAGTLAGIVGVDLTGKLLEAAKASDSDLSTPESWRLVFF 419
NH D+ PRYAG+++G+SNTAG LAG+ G TG +L+ SW VF
Sbjct: 469 LYSNHQDIGPRYAGVLLGLSNTAGVLAGVFGTAATGYILQRG-----------SWDDVFK 517
Query: 420 IPGLLCVFSSLVFLLFSTGERIFD 443
+ L + +LV+ LF+TGE+I D
Sbjct: 518 VAVALYLIGTLVWNLFATGEKILD 541
>At2g29650 Na+-dependent inorganic phosphate cotransporter like
protein
Length = 512
Score = 279 bits (714), Expect = 2e-75
Identities = 160/443 (36%), Positives = 249/443 (56%), Gaps = 28/443 (6%)
Query: 2 QNFPVRYLIVLLTFICTSVCYIERVGFSIAYTVAADAAGINQSTKGTILSTFYYGYACSQ 61
+ FP R++IVLL F +C ++RV SIA + G N +T G I S+F++GY +Q
Sbjct: 97 EEFPKRWVIVLLCFSAFLLCNMDRVNMSIAILPMSAEYGWNPATVGLIQSSFFWGYLLTQ 156
Query: 62 VPGGYFAQKIGGRKILLFSFLLWSLTCALLPLDPNKTL-ILVTARLLVGVAQGFIFPSIH 120
+ GG +A +GG+++L F + WS+ L P+ L L+ R +GV +G P+++
Sbjct: 157 IAGGIWADTVGGKRVLGFGVIWWSIATILTPVAAKLGLPYLLVVRAFMGVGEGVAMPAMN 216
Query: 121 TVLAQWVPPHERSRSVSLTTSGMYLGAAFGMLFLPSLVKFKGPQSVFVAESFLGFVWCFI 180
+L++WVP ERSRS++L SGMYLG+ G+ F P L+ G SVF + LG VW +
Sbjct: 217 NILSKWVPVQERSRSLALVYSGMYLGSVTGLAFSPFLIHQFGWPSVFYSFGSLGTVWLTL 276
Query: 181 WFRYSSDPKSSASGAGESLLPVNKKIDRRVSDLKVGVEKNGGEGKKSGVGIPWVKILTSL 240
W + S+ +LLP +K+ ++D K+ IPW IL+
Sbjct: 277 WLTKAE----SSPLEDPTLLPEERKL---IAD--------NCASKEPVKSIPWRLILSKP 321
Query: 241 PVWAIVVNNFTFHYALYVLMNWLPTYFELGLKLSLHEMGSSKMLPYLNMFVFSNIGGVVA 300
PVWA++ +F ++ ++L+ W+PTY+ LK +L E G + P++ M + +N GG +A
Sbjct: 322 PVWALISCHFCHNWGTFILLTWMPTYYHQVLKFNLMESGLLSVFPWMTMAISANAGGWIA 381
Query: 301 DYLITRRVMSVTRTRKFLNTIGFLVASFALIVIPSFRTSGGAVFCSSVALGFLALGRAGF 360
D L++R SVT RK + TIGFL +F L + + AV C + + G A ++G
Sbjct: 382 DTLVSRG-FSVTNVRKIMQTIGFLGPAFFLTQLKHIDSPTMAVLCMACSQGTDAFSQSGL 440
Query: 361 AVNHMDVAPRYAGIVMGVSNTAGTLAGIVGVDLTGKLLEAAKASDSDLSTPESWRLVFFI 420
NH D+APRY+G+++G+SNTAG LAG++G TG +L+ SW VF I
Sbjct: 441 YSNHQDIAPRYSGVLLGLSNTAGVLAGVLGTAATGHILQHG-----------SWDDVFTI 489
Query: 421 PGLLCVFSSLVFLLFSTGERIFD 443
L + ++++ LFSTGE+I D
Sbjct: 490 SVGLYLVGTVIWNLFSTGEKIID 512
>At3g46980 unknown protein
Length = 533
Score = 208 bits (529), Expect = 5e-54
Identities = 128/436 (29%), Positives = 219/436 (49%), Gaps = 20/436 (4%)
Query: 7 RYLIVLLTFICTSVCYIERVGFSIAYTVAADAAGINQSTKGTILSTFYYGYACSQVPGGY 66
R +V + + ++C +RV S+A + + G ++S G + S+F +GY S + GG
Sbjct: 111 RVKVVAMLALALALCNADRVVMSVAIVPLSLSRGWSKSFSGIVQSSFLWGYLISPIAGGT 170
Query: 67 FAQKIGGRKILLFSFLLWSLTCALLPLDPNKTL-ILVTARLLVGVAQGFIFPSIHTVLAQ 125
+ GG+ ++ + LWSL L P + +L L+ AR +VGVA+G P ++ ++A+
Sbjct: 171 LVDRYGGKVVMAWGVALWSLATFLTPWAADSSLWALLAARAMVGVAEGVALPCMNNMVAR 230
Query: 126 WVPPHERSRSVSLTTSGMYLGAAFGMLFLPSLVKFKGPQSVFVAESFLGFVWCFIWFRYS 185
W PP ERSR+V + +G LG G++ P L+ G FV GF+W +W
Sbjct: 231 WFPPTERSRAVGIAMAGFQLGNVVGLMLSPILMSQGGIYGPFVIFGLSGFLWLLVWL--- 287
Query: 186 SDPKSSASGAGESLLPVNKKIDRRVSDLKVGVEKNGGEGKKSGVGIPWVKILTSLPVWAI 245
S+ S A + + K + K SG+ P+ ++L+ +P WA+
Sbjct: 288 ----SATSSAPDRHPQITKSELEYIKQKKQISTMENKRISTSGIP-PFGRLLSKMPTWAV 342
Query: 246 VVNNFTFHYALYVLMNWLPTYFELGLKLSLHEMGSSKMLPYLNMFVFSNIGGVVADYLIT 305
+V N + +V+++W+P YF ++L + +P+ M I G +D LI
Sbjct: 343 IVANSMHSWGFFVILSWMPIYFNSVYHVNLKQAAWFSAVPWSMMAFTGYIAGFWSDLLI- 401
Query: 306 RRVMSVTRTRKFLNTIGFLVASFALIVIPSFRTSGGAVFCSSVALGFLALGRAGFAVNHM 365
RR S+T TRK + +IGF+ ALI + + + A S+A+G + GF +N
Sbjct: 402 RRGTSITLTRKIMQSIGFIGPGIALIGLTTAKQPLVASAWLSLAVGLKSFSHLGFLINLQ 461
Query: 366 DVAPRYAGIVMGVSNTAGTLAGIVGVDLTGKLLEAAKASDSDLSTPESWRLVFFIPGLLC 425
++AP Y+G++ G+ TAGTLA IVG G +E S++ + +L
Sbjct: 462 EIAPEYSGVLHGMCLTAGTLAAIVGTVGAGFFVELL----------GSFQGFILLTAILY 511
Query: 426 VFSSLVFLLFSTGERI 441
+ S+L + +++TGER+
Sbjct: 512 LLSALFYNIYATGERV 527
>At2g38060 putative Na+-dependent inorganic phosphate cotransporter
Length = 561
Score = 187 bits (475), Expect = 1e-47
Identities = 113/376 (30%), Positives = 192/376 (51%), Gaps = 17/376 (4%)
Query: 5 PVRYLIVLLTFICTSVCYIERVGFSIAYTVAADAAGINQSTKGTILSTFYYGYACSQVPG 64
P R +V+LT +C +RV S+A AD G + S G + S+F +GY S V G
Sbjct: 96 PERIKVVILTACMMCLCNADRVVMSVAVVPLADKLGWSSSFLGVVQSSFLWGYIFSSVIG 155
Query: 65 GYFAQKIGGRKILLFSFLLWSLTCALLP-LDPNKTLILVTARLLVGVAQGFIFPSIHTVL 123
G + GG+++L + LWSL L P + TL L+ R G+A+G PS+ T+L
Sbjct: 156 GALVDRYGGKRVLAWGVALWSLATLLTPWAAAHSTLALLCVRAFFGLAEGVAMPSMTTLL 215
Query: 124 AQWVPPHERSRSVSLTTSGMYLGAAFGMLFLPSLVKFKGPQSVFVAESFLGFVWCFIWFR 183
++W P ER+ +V ++ +G ++G G+L P ++ G F+ + LG +W W
Sbjct: 216 SRWFPMDERASAVGISMAGFHMGNVVGLLLTPLMLSSIGISGPFILFASLGLLWVSTWSS 275
Query: 184 -YSSDPKSSASGAGESLLPVNKKIDRRVSDLKVGVEKNGGEGKKSGVGIPWVKILTS-LP 241
+++P+ S P + + R+ V+ + K + P +++L S LP
Sbjct: 276 GVTNNPQDS---------PFITRSELRLIQAGKPVQPSTISPKPN----PSLRLLLSKLP 322
Query: 242 VWAIVVNNFTFHYALYVLMNWLPTYFELGLKLSLHEMGSSKMLPYLNMFVFSNIGGVVAD 301
WAI+ N T ++ +VL++W+P YF+ ++L + LP+ M + G +D
Sbjct: 323 TWAIIFANVTNNWGYFVLLSWMPVYFQTVFNVNLKQAAWFSALPWATMAISGYYAGAASD 382
Query: 302 YLITRRVMSVTRTRKFLNTIGFLVASFALIVIPSFRTSGGAVFCSSVALGFLALGRAGFA 361
+LI R SVT RK + +IGF+ +L+ + ++ A ++AL + +AGF
Sbjct: 383 FLI-RTGHSVTSVRKIMQSIGFMGPGLSLLCLNFAKSPSCAAVFMTIALSLSSFSQAGFL 441
Query: 362 VNHMDVAPRYAGIVMG 377
+N D+AP+YAG + G
Sbjct: 442 LNMQDIAPQYAGFLHG 457
>At5g20380 putative protein
Length = 383
Score = 74.7 bits (182), Expect = 9e-14
Identities = 48/173 (27%), Positives = 77/173 (43%), Gaps = 42/173 (24%)
Query: 2 QNFPVRYLIVLLTFICTSVCYIERVGFSIAYTVAADAAGINQSTKGTILSTFYYGYACSQ 61
+N P RY ++ T + +C +++V SIA + G + S G + S+F++GYA SQ
Sbjct: 93 KNIPQRYKLIGATSLAFVICNMDKVNLSIAIIPMSHQFGWSSSVAGLVQSSFFWGYALSQ 152
Query: 62 VPGGYFAQKIGGRKILLFSFLLWSLTCALLPLDPNKTLILVTARLLVGVAQGFIFPSIHT 121
+PGG+ ++ GGRK+L WS AL+PL
Sbjct: 153 LPGGWLSKIFGGRKVLEIGVFTWSFATALVPL---------------------------- 184
Query: 122 VLAQWVPPHERSRSVSLTTSGMYLGAAFGMLFLPSLVKFKGPQSVFVAESFLG 174
LA ++P G+ LG+ G+L P +++ +SVF LG
Sbjct: 185 -LAGFMP-------------GLSLGSVMGLLLAPPIIETFNWESVFYLFGLLG 223
Score = 72.8 bits (177), Expect = 4e-13
Identities = 50/149 (33%), Positives = 73/149 (48%), Gaps = 19/149 (12%)
Query: 261 NWLPTYFELGL--------KLSLHEMGSSKMLPYLNMFVFSNIGGVVADYLITRRVMSVT 312
NW ++ GL L+L E +LP L V +++ ADYLIT V T
Sbjct: 211 NWESVFYLFGLLGVGCEALSLNLTEAAWVSILPPLASIVVTSLASQFADYLITNGV-DTT 269
Query: 313 RTRKFLNTIGFLVASFAL------IVIPSFRTSGGAVFCSSVALGFLALGRAGFAVNHMD 366
RK TI F+ + + I +P + G + + +AL AL +G H D
Sbjct: 270 TVRKICQTIAFVAPAICMTLSSVDIGLPPWEIVG--ILTAGLALSSFAL--SGLYCTHQD 325
Query: 367 VAPRYAGIVMGVSNTAGTLAGIVGVDLTG 395
++P YA I++G++NT G + GIVGV LTG
Sbjct: 326 ISPEYASILLGITNTVGAVPGIVGVALTG 354
>At4g35300 sugar transporter like protein
Length = 739
Score = 40.8 bits (94), Expect = 0.002
Identities = 32/121 (26%), Positives = 54/121 (44%), Gaps = 7/121 (5%)
Query: 42 NQSTKGTILSTFYYGYACSQVPGGYFAQKIGGRKILLFSFLLWSLTCALLPLDPNKTLIL 101
N S +G I++ G G A +G R +L+ S +L+ + ++ PN +L
Sbjct: 41 NPSVEGLIVAMSLIGATLITTCSGGVADWLGRRPMLILSSILYFVGSLVMLWSPN-VYVL 99
Query: 102 VTARLLVGVAQGFIFPSIHTVLAQWVPPHERSRSVSLT----TSGMYLG--AAFGMLFLP 155
+ RLL G G + + +++ PP R +L + GM+L FGM +P
Sbjct: 100 LLGRLLDGFGVGLVVTLVPIYISETAPPEIRGLLNTLPQFTGSGGMFLSYCMVFGMSLMP 159
Query: 156 S 156
S
Sbjct: 160 S 160
>At3g47420 putative sugar transporter protein
Length = 523
Score = 39.7 bits (91), Expect = 0.003
Identities = 75/347 (21%), Positives = 130/347 (36%), Gaps = 53/347 (15%)
Query: 105 RLLVGVAQGFIFPSIHTVLAQWVPPHERSRSVSLTTSGMYLGAAFGMLFLPSLVKFKGPQ 164
++L G+ Q +PS+ V+ W +R + + + +G G L +++++
Sbjct: 167 QMLAGLFQSSGWPSVVAVVGNWFNKKKRGLIMGIWNAHTSVGNITGSLIAAAMLRYGWGW 226
Query: 165 SVFVAESFLGFVWCFIWFRYSSDPKSSASGAGESLLPVNKKIDRRVSD-LKVGVEKNGGE 223
S V + + + P++ + E L ++KI V++ L + + +
Sbjct: 227 SFVVPGVIIVVIGLVNYAFLPVSPENVGAERDEVLDSSSEKIGNSVNEPLLLSSSDSETD 286
Query: 224 GKKSGVGIPWVKILTSLPVWAIVVNNFTFHYALYVLMNWLPTYFE----LGLKLSLHEMG 279
KK VG + + +A+ + F Y + WLP Y G LS G
Sbjct: 287 DKKRAVGFIEAWRIPGVAPFALCL--FFAKLVAYTFLYWLPFYVSHTAIEGEYLSDETAG 344
Query: 280 SSKMLPYLNMFVFSNIGGVVADYLITRRVMSVTRTRKFLNTIGFLVASFALIVIPS---F 336
+ + V +GG++A Y+ R + ASF IP+ +
Sbjct: 345 NLSTM----FDVGGVVGGIMAGYISDR-----------IGARAITAASFMYCSIPALFFY 389
Query: 337 RTSGGAVFCSSVALGFLALGRAGFAVNHMDVAPRYAGIVMGVSNTAGTLAGIVG------ 390
R+ G ++ +L FL G VN YA I VS GT + + G
Sbjct: 390 RSYGHVSLLANASLMFL----TGMLVN-----GPYALITTAVSADLGTHSSLKGNSRALA 440
Query: 391 -----VDLTGKLLEA-AKASDSDLSTPESWRLVF-------FIPGLL 424
+D TG + A +S+ SW VF F+ GLL
Sbjct: 441 TVTAIIDGTGSVGAAVGPLLTGYISSRGSWTAVFTMLMGAAFVAGLL 487
Score = 30.8 bits (68), Expect = 1.6
Identities = 22/89 (24%), Positives = 40/89 (44%), Gaps = 8/89 (8%)
Query: 28 FSIAYTVAADAAGINQSTKGTILSTFYYGYACSQVPGGYFAQKIGGRKILLFSFLLWSLT 87
F +++T A + ++ T G + + F G + GY + +IG R I SF+ S+
Sbjct: 326 FYVSHT-AIEGEYLSDETAGNLSTMFDVGGVVGGIMAGYISDRIGARAITAASFMYCSIP 384
Query: 88 CAL-------LPLDPNKTLILVTARLLVG 109
+ L N +L+ +T L+ G
Sbjct: 385 ALFFYRSYGHVSLLANASLMFLTGMLVNG 413
>At5g17010 sugar transporter - like protein
Length = 503
Score = 37.4 bits (85), Expect = 0.017
Identities = 28/104 (26%), Positives = 46/104 (43%), Gaps = 1/104 (0%)
Query: 47 GTILSTFYYGYACSQVPGGYFAQKIGGRKILLFSFLLWSLTCALLPLDPNKTLILVTARL 106
G + S YG + A IG RK L+ + LL+ + + L P + +L+ R+
Sbjct: 94 GLVTSGSLYGALFGSIVAFTIADVIGRRKELILAALLYLVGALVTALAPTYS-VLIIGRV 152
Query: 107 LVGVAQGFIFPSIHTVLAQWVPPHERSRSVSLTTSGMYLGAAFG 150
+ GV+ G + +A+ P R + VSL + LG G
Sbjct: 153 IYGVSVGLAMHAAPMYIAETAPSPIRGQLVSLKEFFIVLGMVGG 196
>At1g30220 unknown protein
Length = 580
Score = 35.8 bits (81), Expect = 0.049
Identities = 29/109 (26%), Positives = 46/109 (41%), Gaps = 5/109 (4%)
Query: 49 ILSTFYYGYACSQVPGGYFAQKIGGRKILLFSFLLWSLTCALLPLDPNKTLILVTARLLV 108
I+S G GG+ K+G R +L + L+ L ++ PN +L LV R+ V
Sbjct: 73 IVSMAVAGAIVGAAIGGWANDKLGRRSAILMADFLFLLGAIIMAAAPNPSL-LVVGRVFV 131
Query: 109 GVAQGFIFPSIHTVLAQWVPPHERSRSVS----LTTSGMYLGAAFGMLF 153
G+ G + +++ P R VS L T G +L + F
Sbjct: 132 GLGVGMASMTAPLYISEASPAKIRGALVSTNGFLITGGQFLSYLINLAF 180
>At2g43330 membrane transporter like protein
Length = 509
Score = 35.4 bits (80), Expect = 0.064
Identities = 26/102 (25%), Positives = 47/102 (45%), Gaps = 5/102 (4%)
Query: 48 TILSTFYYGYACSQVPGGYFAQKIGGRKILLFSFLLWSLTCALLPLDPNKTLILVTARLL 107
TI+S G GG+ G +K LF+ ++++ ++ P+ +L++ RLL
Sbjct: 75 TIVSMALVGAMIGAAAGGWINDYYGRKKATLFADVVFAAGAIVMAAAPDP-YVLISGRLL 133
Query: 108 VGVAQGFIFPSIHTVLAQWVPPHER----SRSVSLTTSGMYL 145
VG+ G + +A+ P R S +V + T G +L
Sbjct: 134 VGLGVGVASVTAPVYIAEASPSEVRGGLVSTNVLMITGGQFL 175
>At1g20840 putative sugar transporter protein
Length = 734
Score = 35.4 bits (80), Expect = 0.064
Identities = 29/118 (24%), Positives = 53/118 (44%), Gaps = 7/118 (5%)
Query: 44 STKGTILSTFYYGYACSQVPGGYFAQKIGGRKILLFSFLLWSLTCALLPLDPNKTLILVT 103
S +G +++ G G + +G R +L+ S +++ C L+ L +L
Sbjct: 41 SVQGLVVAMSLIGATVITTCSGPISDWLGRRPMLILSSVMY-FVCGLIMLWSPNVYVLCF 99
Query: 104 ARLLVGVAQGFIFPSIHTVLAQWVPPHERSRSVSLT----TSGMYLGAAFGMLFLPSL 157
ARLL G G + +++ PP R + +L + GM+L ++ M+F SL
Sbjct: 100 ARLLNGFGAGLAVTLVPVYISETAPPEIRGQLNTLPQFLGSGGMFL--SYCMVFTMSL 155
>At1g20860 putative inorganic phosphate transporter protein
Length = 534
Score = 35.0 bits (79), Expect = 0.083
Identities = 33/136 (24%), Positives = 57/136 (41%), Gaps = 19/136 (13%)
Query: 44 STKGTILSTFY----YGYACSQVPGGYFAQKIGGRKI---LLFSFLLWSLTCALLPLDPN 96
S +LST Y G A Q+ GY ++G R++ L +L S C
Sbjct: 55 SINTAVLSTSYAIALLGTATGQLVFGYLGDRVGRRRVYGLCLIIMILSSFGCGFSVCTTR 114
Query: 97 KTLILVTA---RLLVGVAQGFIFPSIHTVLAQWVPPHERSRSVSLTTSGMYLGAAFGMLF 153
++ ++V+ R +G+ G +P T+++++ + T G ++ A F M
Sbjct: 115 RSCVMVSLGFFRFFLGLGIGGDYPLSATIMSEF---------ANKRTRGAFIAAVFSMQG 165
Query: 154 LPSLVKFKGPQSVFVA 169
L LV +V VA
Sbjct: 166 LGILVSSAVTMAVCVA 181
Score = 29.6 bits (65), Expect = 3.5
Identities = 25/72 (34%), Positives = 32/72 (43%), Gaps = 8/72 (11%)
Query: 370 RYAGIVMGVSNTAGTLAGIVGVDLTGKLLEAAKASDSDLST---PE--SWRLVFFIPGLL 424
R+ G+S AG L IVG T L A K +SD PE R+ F I G +
Sbjct: 425 RFRSTCHGISGAAGKLGAIVG---TVGFLWATKKMESDDKNQIYPEVNRMRIAFLILGGV 481
Query: 425 CVFSSLVFLLFS 436
C+ LV F+
Sbjct: 482 CIAGILVTYFFT 493
>At4g17550 putative protein
Length = 534
Score = 33.5 bits (75), Expect = 0.24
Identities = 77/355 (21%), Positives = 132/355 (36%), Gaps = 58/355 (16%)
Query: 105 RLLVGVAQGFIFPSIHTVLAQWVPPHERSRSVSLTTSGMYLGAAFGMLFLPSLVKFKGPQ 164
++ G+ Q +PS+ V+ W +R + + + +G G L ++++
Sbjct: 179 QMAAGLFQATGWPSVVAVVGNWFGKRKRGLIMGIWNAHTSVGNICGSLIAAGVLEYGWGW 238
Query: 165 SVFVAESF---LGFVWCFIWF-RYSSDP-----KSSASGAGESLLPVNKKIDRRVSDLKV 215
S F+A F LG V +++ Y D S++ + V ++ + DL
Sbjct: 239 S-FIAPGFVMSLGGVLVYLFLAAYPEDVGFPDINSNSGKFIKRKRDVEEEEEEVEEDLGT 297
Query: 216 GVEKNG--------GEGKKSGVGIPWVKILTSLPVWAIVVNNFTFHYALYVLMNWLPTYF 267
VE +G G K VG+ ++ + +A+ + F Y + WLP Y
Sbjct: 298 DVEGDGEGSSGSGSGYENKRSVGLLQACMIPGVIPFALCL--FFSKLVAYTFLYWLPFYL 355
Query: 268 EL----GLKLSLHEMGSSKMLPYLNMFVFSNIGGVVADYLITRRVMSVTRTRKFLNTIGF 323
G +S+ G+ L V +GG++ Y+ + T F+
Sbjct: 356 SQTTIGGEYVSVKTAGNLSTL----FDVGGIVGGILCGYISDKFKARATTAAAFM----- 406
Query: 324 LVASFALIVIPSFRTSGGAVFCSSVALGFLALGRAGFAVNHMDVAPRYAGIVMGVSNTAG 383
A A++V S+ GG S + L + AG VN YA I VS G
Sbjct: 407 YAAIPAMLVYHSY---GGV----SQTVNILLMMVAGLFVN-----GPYALITTAVSADLG 454
Query: 384 TLAGIVG-----------VDLTGKLLEAAKASDSDLSTPESWRLVFF--IPGLLC 425
T + G +D TG A + + W+ VF+ + G LC
Sbjct: 455 THKSLQGDSRALATVTAIIDGTGSAGAALGPLLTGFLSTLGWQAVFYMLVVGALC 509
>At2g35740 putative sugar transporter
Length = 580
Score = 33.5 bits (75), Expect = 0.24
Identities = 40/181 (22%), Positives = 75/181 (41%), Gaps = 17/181 (9%)
Query: 49 ILSTFYYGYACSQVPGGYFAQKIGGRKILLFSFLLWSLTCALLPLDPNKTLILVTARLLV 108
I+S G GG++ K G R +L + +L+ L AL+ + + +++ RLLV
Sbjct: 71 IVSMTVAGAIVGAAIGGWYNDKFGRRMSVLIADVLFLLG-ALVMVIAHAPWVIILGRLLV 129
Query: 109 GVAQGFIFPSIHTVLAQWVPPHERSRSVS----LTTSGMYLGAAFGMLFLPSLVKFKGPQ 164
G G + +++ P R VS L T G +L + F+ + ++
Sbjct: 130 GFGVGMASMTSPLYISEMSPARIRGALVSTNGLLITGGQFLSYLINLAFVHTPGTWRWML 189
Query: 165 SVFVAESFLGFVWCFI--------WFRYSSDPKSSASGAGESLLPVNKKIDRRVSDLKVG 216
V + + F C + W Y +D K+ + E + P + ++ ++ LK
Sbjct: 190 GVSAIPAIIQF--CLMLTLPESPRWL-YRNDRKAESRDILERIYPA-EMVEAEIAALKES 245
Query: 217 V 217
V
Sbjct: 246 V 246
>At1g12940 nitrate transporter, putative
Length = 502
Score = 33.5 bits (75), Expect = 0.24
Identities = 22/86 (25%), Positives = 38/86 (43%), Gaps = 2/86 (2%)
Query: 352 FLALGRAGFAVNHMDVAPRYAGIVMGVSNTAGTLAGIVGVDLTGKLLEAAKASDSDLSTP 411
F A F ++ ++G V+G +N G +G T ++ + ++
Sbjct: 142 FAGFSLATFVSTQFWMSSMFSGPVVGSANGIAAGWGNLGGGATQLIMPIVFSLIRNMGAT 201
Query: 412 E--SWRLVFFIPGLLCVFSSLVFLLF 435
+ +WR+ FFIPGL S+ LLF
Sbjct: 202 KFTAWRIAFFIPGLFQTLSAFAVLLF 227
>At1g18010 hypothetical protein
Length = 459
Score = 33.1 bits (74), Expect = 0.32
Identities = 19/80 (23%), Positives = 37/80 (45%), Gaps = 1/80 (1%)
Query: 102 VTARLLVGVAQGFIFPSIHTVLAQWVPPHERSRSVSLTTSGMYLGAAFGMLFLPSLVKFK 161
+ A L+G G ++ V+ + PPH + ++L S LG G L +P ++ ++
Sbjct: 125 IVAGALLGCGAGLLWAGEGAVMTSYPPPHRKGTYIALFWSIFNLGGVIGGL-IPFILNYQ 183
Query: 162 GPQSVFVAESFLGFVWCFIW 181
+ V +S CF++
Sbjct: 184 RSSAASVNDSTYIAFMCFMF 203
>At1g18000 hypothetical protein
Length = 459
Score = 33.1 bits (74), Expect = 0.32
Identities = 19/80 (23%), Positives = 37/80 (45%), Gaps = 1/80 (1%)
Query: 102 VTARLLVGVAQGFIFPSIHTVLAQWVPPHERSRSVSLTTSGMYLGAAFGMLFLPSLVKFK 161
+ A L+G G ++ V+ + PPH + ++L S LG G L +P ++ ++
Sbjct: 125 IVAGALLGCGAGLLWAGEGAVMTSYPPPHRKGTYIALFWSIFNLGGVIGGL-IPFILNYQ 183
Query: 162 GPQSVFVAESFLGFVWCFIW 181
+ V +S CF++
Sbjct: 184 RSSAASVNDSTYIAFMCFMF 203
>At1g76430 putative phosphate transporter
Length = 532
Score = 32.7 bits (73), Expect = 0.41
Identities = 26/108 (24%), Positives = 48/108 (44%), Gaps = 10/108 (9%)
Query: 49 ILSTFY----YGYACSQVPGGYFAQKIGGRKILLFSFLLW---SLTCALLPLDPNKTLIL 101
+LST Y G A Q+ GY ++G RK+ S L+ S C ++ ++
Sbjct: 61 LLSTSYAIALLGTALGQLIFGYLGDRVGRRKVYGLSLLIMVFSSFGCGFSVCTTRRSCVM 120
Query: 102 VTA---RLLVGVAQGFIFPSIHTVLAQWVPPHERSRSVSLTTSGMYLG 146
V+ R ++G+ G +P T+++++ R ++ S LG
Sbjct: 121 VSLGFFRFVLGLGIGGDYPLSATIMSEFANKRTRGAFIAAVFSMQGLG 168
>At1g30560 unknown protein, 5' partial
Length = 513
Score = 32.7 bits (73), Expect = 0.41
Identities = 76/402 (18%), Positives = 142/402 (34%), Gaps = 39/402 (9%)
Query: 47 GTILSTFYYGYACSQVPGGYFAQKIGGRKILLFSFLLWSLTCALLPL----DPNKTLILV 102
G I F YA G+ ++ R L + + AL + + + +
Sbjct: 97 GQIDLAFLSVYAVGMFVAGHLGDRLDLRTFLTIGMVGTGVCTALFGVAFWANIHAFYYFL 156
Query: 103 TARLLVGVAQGFIFPSIHTVLAQWVPPHERSRSVSLTTSGMYLGAAFGMLFLPSLVKFKG 162
+ L G Q +P + VL W R + + ++ LG G L L+KF
Sbjct: 157 AIQTLAGWFQSIGWPCVVAVLGNWFDKKRRGVIMGVWSAHTSLGNIIGTLIATGLLKFGW 216
Query: 163 PQSVFVAESFLGFVWCFIWFRYSSDPKSSASGAGESLLPVNKKIDRRVSDLKVGVEKNGG 222
S + F+ ++ +P + + S + ++ +++ + + G
Sbjct: 217 GWSFVGPALLITFLGIVVYLFLPVNPHAVEAERDGSEVDSTMRLGDTITESFLSSRTSTG 276
Query: 223 EGKKSGVGIPWVKILTSLPVWAIVVNNFTFHYALYVLMNWLPTYFELGLKLSLHEMGSSK 282
+++ VG + + +A + F Y + WLP Y +S E+G +
Sbjct: 277 FDRRA-VGFLAAWKIPGVAPFAFCL--FFTKLVSYTFLYWLPFY------VSQTEIGGEQ 327
Query: 283 MLPYL--NMFVFSNIGGVVADYLITRRVMSVTRTRKFLNTIGFLVASFALIVIPSFRTSG 340
+ N+ ++GGVV I S + + GF+ + + + +R G
Sbjct: 328 LSQETSGNLSTLFDVGGVVGG--ILAGYFSDQLDGRAITAGGFIYLTIPALFL--YRIYG 383
Query: 341 GAVFCSSVALGFLALGRAGFAVNHMDVAPRYAGIVMGVSNTAGTLAGIVG---------- 390
++ L F+ AG VN YA I V+ GT + G
Sbjct: 384 HVSMTINIILMFV----AGLFVN-----GPYALITTAVAADLGTHKSLKGNARALATVTA 434
Query: 391 -VDLTGKLLEAAKASDSDLSTPESWRLVFFIPGLLCVFSSLV 431
+D TG + A + SW VF++ + S L+
Sbjct: 435 IIDGTGSVGAAIGPVLTGYIAAISWDAVFYMLMTAALISGLL 476
Score = 28.5 bits (62), Expect = 7.8
Identities = 13/51 (25%), Positives = 24/51 (46%)
Query: 41 INQSTKGTILSTFYYGYACSQVPGGYFAQKIGGRKILLFSFLLWSLTCALL 91
++Q T G + + F G + GYF+ ++ GR I F+ ++ L
Sbjct: 328 LSQETSGNLSTLFDVGGVVGGILAGYFSDQLDGRAITAGGFIYLTIPALFL 378
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.141 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,554,614
Number of Sequences: 26719
Number of extensions: 396550
Number of successful extensions: 1254
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 24
Number of HSP's that attempted gapping in prelim test: 1204
Number of HSP's gapped (non-prelim): 55
length of query: 443
length of database: 11,318,596
effective HSP length: 102
effective length of query: 341
effective length of database: 8,593,258
effective search space: 2930300978
effective search space used: 2930300978
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148651.2


