

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148487.1 - phase: 1
(63 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
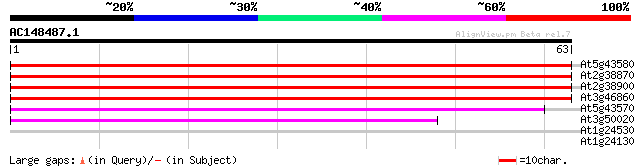
At5g43580 unknown protein 73 2e-14
At2g38870 protease inhibitor like protein 68 7e-13
At2g38900 protease inhibitor like protein 57 1e-09
At3g46860 proteinase inhibitor-like protein 55 7e-09
At5g43570 unknown protein 51 9e-08
At3g50020 putative protein 39 4e-04
At1g24530 unknown protein 30 0.22
At1g24130 hypothetical protein 25 9.4
>At5g43580 unknown protein
Length = 72
Score = 73.2 bits (178), Expect = 2e-14
Identities = 33/63 (52%), Positives = 46/63 (72%)
Query: 1 KSSWPELVGVEGKVAEATIQRENPLVDAIIVPEGSSVPFDFRCDRVWVWINKDEIVYQVP 60
KSSWPEL+G +G+ A+ I+RENP + A+I+ +G+ VP F C RV+VW+N IV Q+P
Sbjct: 10 KSSWPELLGAKGEDAKEVIERENPKMKAVIILDGTVVPEIFICSRVYVWVNDCGIVVQIP 69
Query: 61 TIG 63
IG
Sbjct: 70 IIG 72
>At2g38870 protease inhibitor like protein
Length = 70
Score = 68.2 bits (165), Expect = 7e-13
Identities = 31/63 (49%), Positives = 42/63 (66%)
Query: 1 KSSWPELVGVEGKVAEATIQRENPLVDAIIVPEGSSVPFDFRCDRVWVWINKDEIVYQVP 60
K+SWPEL G G A I+RENP V+A ++ +GS V DFRCDRV V+++ + IV + P
Sbjct: 8 KNSWPELTGTNGDYAAVVIERENPTVNAAVILDGSPVTADFRCDRVRVFVDGNRIVVKTP 67
Query: 61 TIG 63
G
Sbjct: 68 KSG 70
>At2g38900 protease inhibitor like protein
Length = 73
Score = 57.4 bits (137), Expect = 1e-09
Identities = 26/63 (41%), Positives = 40/63 (63%)
Query: 1 KSSWPELVGVEGKVAEATIQRENPLVDAIIVPEGSSVPFDFRCDRVWVWINKDEIVYQVP 60
K+SWPEL+G G A + I+ EN ++ ++V +G+ V D C RV VW+++ IV + P
Sbjct: 11 KNSWPELLGTNGDYAASVIKGENSSLNVVVVSDGNYVTEDLSCYRVRVWVDEIRIVVRNP 70
Query: 61 TIG 63
T G
Sbjct: 71 TAG 73
>At3g46860 proteinase inhibitor-like protein
Length = 85
Score = 55.1 bits (131), Expect = 7e-09
Identities = 28/63 (44%), Positives = 40/63 (63%)
Query: 1 KSSWPELVGVEGKVAEATIQRENPLVDAIIVPEGSSVPFDFRCDRVWVWINKDEIVYQVP 60
K WPELVG G++A+ TI+RENP V AI++ G +F C+RV+V++ + V P
Sbjct: 23 KYRWPELVGKNGQLAKMTIERENPNVLAIVLRYGEKRIENFCCNRVFVYLGSNGQVADAP 82
Query: 61 TIG 63
IG
Sbjct: 83 MIG 85
>At5g43570 unknown protein
Length = 91
Score = 51.2 bits (121), Expect = 9e-08
Identities = 28/60 (46%), Positives = 35/60 (57%)
Query: 1 KSSWPELVGVEGKVAEATIQRENPLVDAIIVPEGSSVPFDFRCDRVWVWINKDEIVYQVP 60
KSSWPELVG G+ + I REN V A I+ E + V CDRV+V +N IV + P
Sbjct: 2 KSSWPELVGRRGEEVKEIIDRENTKVTAKIISENAVVLAVVICDRVYVRVNDQGIVTRTP 61
>At3g50020 putative protein
Length = 96
Score = 39.3 bits (90), Expect = 4e-04
Identities = 20/48 (41%), Positives = 29/48 (59%)
Query: 1 KSSWPELVGVEGKVAEATIQRENPLVDAIIVPEGSSVPFDFRCDRVWV 48
K WPEL GV+G A+ I+ +NP V +I+P+ +V C+RV V
Sbjct: 30 KVDWPELNGVKGLEAKRIIEHDNPKVVVVIIPDDVAVLAVNCCNRVIV 77
>At1g24530 unknown protein
Length = 418
Score = 30.0 bits (66), Expect = 0.22
Identities = 16/55 (29%), Positives = 27/55 (49%), Gaps = 1/55 (1%)
Query: 4 WPELVGVEGKVAEATIQRENPLVDAIIVPEGSSVPFDFRCDR-VWVWINKDEIVY 57
W + G + AT+++ V+A+ + + SV F CDR + VW +D Y
Sbjct: 261 WAKPTGEKRHTLVATLEKHKSAVNALALNDDGSVLFSGSCDRSILVWEREDTSNY 315
>At1g24130 hypothetical protein
Length = 415
Score = 24.6 bits (52), Expect = 9.4
Identities = 15/51 (29%), Positives = 25/51 (48%), Gaps = 4/51 (7%)
Query: 17 ATIQRENPLVDAIIVPEGSSVPFDFRCDR-VWVW---INKDEIVYQVPTIG 63
AT+ + V+A+ + E V + CDR + VW IN D+ + +G
Sbjct: 272 ATLTKHLSAVNALAISEDGKVLYSGACDRSILVWERLINGDDEELHMSVVG 322
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.139 0.441
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,536,017
Number of Sequences: 26719
Number of extensions: 48227
Number of successful extensions: 74
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 1
Number of HSP's that attempted gapping in prelim test: 67
Number of HSP's gapped (non-prelim): 8
length of query: 63
length of database: 11,318,596
effective HSP length: 39
effective length of query: 24
effective length of database: 10,276,555
effective search space: 246637320
effective search space used: 246637320
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 52 (24.6 bits)
Medicago: description of AC148487.1


