

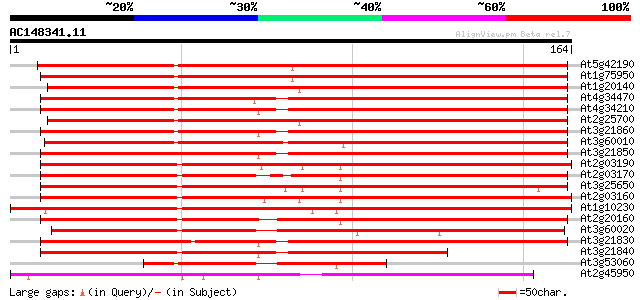
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148341.11 - phase: 0
(164 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g42190 UIP2 (gb|AAC63110.1) 224 2e-59
At1g75950 UIP1 221 1e-58
At1g20140 kinetochore protein Skp1, putative 209 7e-55
At4g34470 Skp1p - like protein 198 9e-52
At4g34210 kinetochore (SKP1p) - like protein 198 1e-51
At2g25700 putative kinetechore (Skp1p-like) protein 198 1e-51
At3g21860 kinetochore-like protein, Skp1p 184 2e-47
At3g60010 Skp1-like protein 184 2e-47
At3g21850 kinetochore-like protein, Skp1p 181 2e-46
At2g03190 putative kinetechore (Skp1p-like) protein 173 4e-44
At2g03170 putative kinetechore (Skp1p-like) protein 170 4e-43
At3g25650 kinetechore protein, putative 168 1e-42
At2g03160 putative kinetechore (Skp1p-like) protein 161 1e-40
At1g10230 putative kinetochore protein 160 3e-40
At2g20160 putative kinetechore (Skp1p-like) protein 157 3e-39
At3g60020 Skp1-like protein 156 4e-39
At3g21830 kinetochore-like protein, Skp1p 154 2e-38
At3g21840 kinetochore-like protein, Skp1p 111 2e-25
At3g53060 kinetochore - like protein 70 7e-13
At2g45950 SKP1-like protein 68 2e-12
>At5g42190 UIP2 (gb|AAC63110.1)
Length = 171
Score = 224 bits (571), Expect = 2e-59
Identities = 112/171 (65%), Positives = 134/171 (77%), Gaps = 17/171 (9%)
Query: 9 TATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHC 68
+ +KITLKSSD E FEI++AVALESQTIKH+I+D+C D+ GIPLPNVT KIL+ VIE+C
Sbjct: 2 STVRKITLKSSDGENFEIDEAVALESQTIKHMIEDDCTDN-GIPLPNVTSKILSKVIEYC 60
Query: 69 KKHVDATSSDEKP----------------SEDEINKWDTEFVKVDQDTLFDLILAANYLN 112
K+HV+A E S++++ WD+EF+KVDQ TLFDLILAANYLN
Sbjct: 61 KRHVEAAEKSETTADAAAATTTTTVASGSSDEDLKTWDSEFIKVDQGTLFDLILAANYLN 120
Query: 113 IKSLLDLTCKTVADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
IK LLDLTC+TVADMIKG+TPEEIRKTFNI ND+TPEEEEEVR E QWAF+
Sbjct: 121 IKGLLDLTCQTVADMIKGKTPEEIRKTFNIKNDFTPEEEEEVRRENQWAFE 171
>At1g75950 UIP1
Length = 160
Score = 221 bits (564), Expect = 1e-58
Identities = 107/160 (66%), Positives = 134/160 (82%), Gaps = 7/160 (4%)
Query: 10 ATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCK 69
+ KKI LKSSD E+FE+E+AVALESQTI H+++D+C D+ G+PLPNVT KILA VIE+CK
Sbjct: 2 SAKKIVLKSSDGESFEVEEAVALESQTIAHMVEDDCVDN-GVPLPNVTSKILAKVIEYCK 60
Query: 70 KHVDATSSDEKP------SEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKT 123
+HV+A +S + S+D++ WD +F+K+DQ TLF+LILAANYLNIK+LLDLTC+T
Sbjct: 61 RHVEAAASKAEAVEGAATSDDDLKAWDADFMKIDQATLFELILAANYLNIKNLLDLTCQT 120
Query: 124 VADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
VADMIKG+TPEEIR TFNI ND+TPEEEEEVR E QWAF+
Sbjct: 121 VADMIKGKTPEEIRTTFNIKNDFTPEEEEEVRRENQWAFE 160
>At1g20140 kinetochore protein Skp1, putative
Length = 163
Score = 209 bits (531), Expect = 7e-55
Identities = 105/159 (66%), Positives = 127/159 (79%), Gaps = 8/159 (5%)
Query: 12 KKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCKKH 71
K I LKSSD E+FEIE+AVA++SQTIKH+I+D+CAD+ GIPLPNVTG ILA VIE+CKKH
Sbjct: 6 KMIILKSSDGESFEIEEAVAVKSQTIKHMIEDDCADN-GIPLPNVTGAILAKVIEYCKKH 64
Query: 72 VDATSSDEKPSE-------DEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTV 124
V+A + + DE+ WD+EFVKVDQ TLFDLILAANYLNI LLDLTCK V
Sbjct: 65 VEAAAEAGGDKDFYGSAENDELKNWDSEFVKVDQPTLFDLILAANYLNIGGLLDLTCKAV 124
Query: 125 ADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
AD ++G+TPE++R FNI NDYTPEEE EVR+E +WAF+
Sbjct: 125 ADQMRGKTPEQMRAHFNIKNDYTPEEEAEVRNENKWAFE 163
>At4g34470 Skp1p - like protein
Length = 152
Score = 198 bits (504), Expect = 9e-52
Identities = 98/155 (63%), Positives = 125/155 (80%), Gaps = 5/155 (3%)
Query: 10 ATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCK 69
++K I L SSD ++FE+E+AVA++SQTI H+++D+C D GIPL NV KIL VIE+CK
Sbjct: 2 SSKMIVLMSSDGQSFEVEEAVAIQSQTIAHMVEDDCVAD-GIPLANVESKILVKVIEYCK 60
Query: 70 K-HVDATSSDEKPSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTVADMI 128
K HVD + SE+++NKWD +F+ ++Q T+F+LILAANYLNIKSL DLTC+TVADMI
Sbjct: 61 KYHVDEANPI---SEEDLNKWDEKFMDLEQSTIFELILAANYLNIKSLFDLTCQTVADMI 117
Query: 129 KGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
KG+TPEEIR TFNI ND+TPEEEE VR E QWAF+
Sbjct: 118 KGKTPEEIRSTFNIENDFTPEEEEAVRKENQWAFE 152
>At4g34210 kinetochore (SKP1p) - like protein
Length = 152
Score = 198 bits (503), Expect = 1e-51
Identities = 98/155 (63%), Positives = 125/155 (80%), Gaps = 5/155 (3%)
Query: 10 ATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCK 69
++K I L SSD ++FE+E+AVA++SQTI H+++D+C D GIPL NV KIL VIE+CK
Sbjct: 2 SSKMIVLMSSDGQSFEVEEAVAIQSQTIAHMVEDDCVAD-GIPLANVESKILVKVIEYCK 60
Query: 70 KH-VDATSSDEKPSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTVADMI 128
KH VD + SE+++N WD +F+ ++Q T+F+LILAANYLNIKSLLDLTC+TVADMI
Sbjct: 61 KHHVDEANPI---SEEDLNNWDEKFMDLEQSTIFELILAANYLNIKSLLDLTCQTVADMI 117
Query: 129 KGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
KG+TPEEIR TFNI ND+TPEEEE VR E QWAF+
Sbjct: 118 KGKTPEEIRSTFNIENDFTPEEEEAVRKENQWAFE 152
>At2g25700 putative kinetechore (Skp1p-like) protein
Length = 163
Score = 198 bits (503), Expect = 1e-51
Identities = 98/159 (61%), Positives = 122/159 (76%), Gaps = 8/159 (5%)
Query: 12 KKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCKKH 71
K I LKSSD E+FE+E+AVA+ESQTIKH+I+D+C D+ GIPLPNVTG ILA VIE+CKKH
Sbjct: 6 KMIILKSSDGESFEVEEAVAVESQTIKHMIEDDCVDN-GIPLPNVTGAILAKVIEYCKKH 64
Query: 72 VDATSSDEKPSE-------DEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTV 124
V+A + + E+ WD +FVKVD TLFDL+ AANYLNI LLDLTCK V
Sbjct: 65 VEAAAEAGGDKDFYGSTENHELKTWDNDFVKVDHPTLFDLLRAANYLNISGLLDLTCKAV 124
Query: 125 ADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
AD ++G+TP ++R+ FNI NDYTPEEE EVR+E +WAF+
Sbjct: 125 ADQMRGKTPAQMREHFNIKNDYTPEEEAEVRNENRWAFE 163
>At3g21860 kinetochore-like protein, Skp1p
Length = 152
Score = 184 bits (467), Expect = 2e-47
Identities = 95/155 (61%), Positives = 116/155 (74%), Gaps = 5/155 (3%)
Query: 10 ATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCK 69
+TKKI LKSSD +FE+E+ A + QTI H+ +D+C D+ GIPLP VTGKIL MVIE+C
Sbjct: 2 STKKIILKSSDGHSFEVEEEAACQCQTIAHMSEDDCTDN-GIPLPEVTGKILEMVIEYCN 60
Query: 70 KH-VDATSSDEKPSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTVADMI 128
KH VDA + S++++ KWD EF++ Q T+FDLI+AANYLNIKSLLDL C+TVADMI
Sbjct: 61 KHHVDAANPC---SDEDLKKWDKEFMEKYQSTIFDLIMAANYLNIKSLLDLACQTVADMI 117
Query: 129 KGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
K T E RK FNI NDYT EEEE VR E QW F+
Sbjct: 118 KDNTVEHTRKFFNIENDYTHEEEEAVRRENQWGFE 152
>At3g60010 Skp1-like protein
Length = 154
Score = 184 bits (466), Expect = 2e-47
Identities = 91/155 (58%), Positives = 120/155 (76%), Gaps = 4/155 (2%)
Query: 11 TKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCKK 70
+K + L SSD E+F++E+AVA++SQTI H+I+D+C + G+P+ NVTG IL+ VIE+CKK
Sbjct: 2 SKMVMLLSSDGESFQVEEAVAVQSQTIAHMIEDDCVAN-GVPIANVTGVILSKVIEYCKK 60
Query: 71 HVDATSSDEKPSEDEINKWDTEFVKV--DQDTLFDLILAANYLNIKSLLDLTCKTVADMI 128
HV + S E+ S+DE+ KWD EF+K TLFD++LAANYLNIK LLDL C+TVADMI
Sbjct: 61 HVVSDSPTEE-SKDELKKWDAEFMKALEQSSTLFDVMLAANYLNIKDLLDLGCQTVADMI 119
Query: 129 KGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
G+ P+EIR I ND+TPEEEEE+R E QWAF+
Sbjct: 120 TGKKPDEIRALLGIENDFTPEEEEEIRKENQWAFE 154
>At3g21850 kinetochore-like protein, Skp1p
Length = 153
Score = 181 bits (459), Expect = 2e-46
Identities = 92/155 (59%), Positives = 116/155 (74%), Gaps = 4/155 (2%)
Query: 10 ATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCK 69
+TKKI LKSSD +FE+E+ A + Q I + +N D+GIPLPNVTGKILAMVIE+C
Sbjct: 2 STKKIILKSSDGHSFEVEEEAARQCQIIIAHMSENDCTDNGIPLPNVTGKILAMVIEYCN 61
Query: 70 KH-VDATSSDEKPSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTVADMI 128
KH VDA + S+D++ KWD EF++ D T+FDLI AANYLNIKSL DL C+TVA++I
Sbjct: 62 KHHVDAANPC---SDDDLKKWDKEFMEKDTSTIFDLIKAANYLNIKSLFDLACQTVAEII 118
Query: 129 KGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
KG TPE+IR+ FNI ND TPEEE +R E +WAF+
Sbjct: 119 KGNTPEQIREFFNIENDLTPEEEAAIRRENKWAFE 153
>At2g03190 putative kinetechore (Skp1p-like) protein
Length = 170
Score = 173 bits (438), Expect = 4e-44
Identities = 90/168 (53%), Positives = 120/168 (70%), Gaps = 14/168 (8%)
Query: 10 ATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCK 69
++ KI L SSD+E+FE+E+AVA + + I H+IDD+CAD + IPL NVTG ILA+VIE+CK
Sbjct: 2 SSNKIVLTSSDDESFEVEEAVARKLKVIAHMIDDDCADKA-IPLENVTGNILALVIEYCK 60
Query: 70 KHV-----DATSSDEKPSED-------EINKWDTEFVK-VDQDTLFDLILAANYLNIKSL 116
KHV D+ S E SE+ E+ WD EF+K D +T+ LILA NYLN++ L
Sbjct: 61 KHVLDDVDDSDDSTEATSENVNEEAKNELRTWDAEFMKEFDMETVMKLILAVNYLNVQDL 120
Query: 117 LDLTCKTVADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFDE 164
L LTC+TVAD +K +PEE+R+ FNI NDYTPEEE+ +R E WAF++
Sbjct: 121 LGLTCQTVADHMKDMSPEEVRELFNIENDYTPEEEDAIRKENAWAFED 168
>At2g03170 putative kinetechore (Skp1p-like) protein
Length = 149
Score = 170 bits (430), Expect = 4e-43
Identities = 88/155 (56%), Positives = 115/155 (73%), Gaps = 8/155 (5%)
Query: 10 ATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCK 69
++ KI L SSD E+FE+E+AVA + + ++H+I+D+C +PL NVTGKIL++V+E+CK
Sbjct: 2 SSNKIVLSSSDGESFEVEEAVARKLKIVEHMIEDDCVVTE-VPLQNVTGKILSIVVEYCK 60
Query: 70 KHVDATSSDEKPSEDEINKWDTEFVK-VDQDTLFDLILAANYLNIKSLLDLTCKTVADMI 128
KHV DE+ DE WD EF+K DQ T+F L+LAANYLNIK LLDL+ +TVAD I
Sbjct: 61 KHV----VDEE--SDEFKTWDEEFMKKFDQPTVFQLLLAANYLNIKGLLDLSAQTVADRI 114
Query: 129 KGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
K +TPEEIR+ FNI ND+TPEEE VR E WAF+
Sbjct: 115 KDKTPEEIREIFNIENDFTPEEEAAVRKENAWAFE 149
>At3g25650 kinetechore protein, putative
Length = 177
Score = 168 bits (425), Expect = 1e-42
Identities = 86/167 (51%), Positives = 120/167 (71%), Gaps = 14/167 (8%)
Query: 10 ATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCK 69
++ KI L SSD E+F++E+ VA + Q +KHL++D+C + IPL NVTG IL++V+E+CK
Sbjct: 2 SSNKIVLTSSDGESFQVEEVVARKLQIVKHLLEDDCVINE-IPLQNVTGNILSIVLEYCK 60
Query: 70 KHVDATSSDE--------KPSED---EINKWDTEFVK-VDQDTLFDLILAANYLNIKSLL 117
KHVD D+ KP ++ ++ WD EF+K +D +T+F LILAANYLN++ LL
Sbjct: 61 KHVDDVVDDDASEEPKKKKPDDEAKQNLDAWDAEFMKNIDMETIFKLILAANYLNVEGLL 120
Query: 118 DLTCKTVADMIKGRTPEEIRKTFNIINDYTPEEEEE-VRSETQWAFD 163
LTC+TVAD IK +TPEE+R+ FNI ND+T EEEEE +R E WAF+
Sbjct: 121 GLTCQTVADYIKDKTPEEVRELFNIENDFTHEEEEEAIRKENAWAFE 167
>At2g03160 putative kinetechore (Skp1p-like) protein
Length = 200
Score = 161 bits (408), Expect = 1e-40
Identities = 84/191 (43%), Positives = 122/191 (62%), Gaps = 37/191 (19%)
Query: 10 ATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCK 69
++KKI L SSD E+F++E+ VA + Q + H+I+D+CA + IP+PNVTG+ILA VIE+CK
Sbjct: 2 SSKKIVLTSSDGESFKVEEVVARKLQIVGHIIEDDCATNK-IPIPNVTGEILAKVIEYCK 60
Query: 70 KHVD---------------------------------ATSSDEKPSE--DEINKWDTEFV 94
KHV+ +T D++ + +++N+WD +F+
Sbjct: 61 KHVEDDDDVVETHESSTKGDKTVEEAKKKPDDVAVPESTEGDDEAEDKKEKLNEWDAKFM 120
Query: 95 K-VDQDTLFDLILAANYLNIKSLLDLTCKTVADMIKGRTPEEIRKTFNIINDYTPEEEEE 153
K D T+FD+ILAANYLN++ L DL KT+AD IK TPEE+R+ FNI ND+TPEEEE
Sbjct: 121 KDFDIKTIFDIILAANYLNVQGLFDLCSKTIADYIKDMTPEEVRELFNIENDFTPEEEEA 180
Query: 154 VRSETQWAFDE 164
+R+E W F++
Sbjct: 181 IRNENAWTFEQ 191
>At1g10230 putative kinetochore protein
Length = 183
Score = 160 bits (405), Expect = 3e-40
Identities = 84/168 (50%), Positives = 119/168 (70%), Gaps = 5/168 (2%)
Query: 1 MSDGVESST-ATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGK 59
+ + V S T ++ KI L SSD E+FEI++AVA + I H+++DNCA ++ IPL NVTG
Sbjct: 17 IEEAVASLTMSSNKILLTSSDGESFEIDEAVARKFLIIVHMMEDNCAGEA-IPLENVTGD 75
Query: 60 ILAMVIEHCKKHVDATSSDEKPSEDEIN--KWDTEFV-KVDQDTLFDLILAANYLNIKSL 116
IL+ +IE+ K HV+ S +++ E + N WD +F+ K+D +T+F +ILAANYLN + L
Sbjct: 76 ILSKIIEYAKMHVNEPSEEDEDEEAKKNLDSWDAKFMEKLDLETIFKIILAANYLNFEGL 135
Query: 117 LDLTCKTVADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFDE 164
L +TVAD IK +TPEE+R+ FNI ND+TPEEEEE+R E W F+E
Sbjct: 136 LGFASQTVADYIKDKTPEEVREIFNIENDFTPEEEEEIRKENAWTFNE 183
>At2g20160 putative kinetechore (Skp1p-like) protein
Length = 150
Score = 157 bits (396), Expect = 3e-39
Identities = 82/155 (52%), Positives = 112/155 (71%), Gaps = 7/155 (4%)
Query: 10 ATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCK 69
++KKI L SSD+E FEI++AVA + Q + H+IDD+CAD + I L NVTGKILA++IE+CK
Sbjct: 2 SSKKIVLTSSDDECFEIDEAVARKMQMVAHMIDDDCADKA-IRLQNVTGKILAIIIEYCK 60
Query: 70 KHVDATSSDEKPSEDEINKWDTEFVK-VDQDTLFDLILAANYLNIKSLLDLTCKTVADMI 128
KHVD + +++E WD EFVK +D DTLF L+ AA+YL + L +L + +AD
Sbjct: 61 KHVD-----DVEAKNEFVTWDAEFVKNIDMDTLFKLLDAADYLIVIGLKNLIAQAIADYT 115
Query: 129 KGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
+T EIR+ FNI NDYTPEEEEE+R + +WAF+
Sbjct: 116 ADKTVNEIRELFNIENDYTPEEEEELRKKNEWAFN 150
>At3g60020 Skp1-like protein
Length = 153
Score = 156 bits (395), Expect = 4e-39
Identities = 82/156 (52%), Positives = 112/156 (71%), Gaps = 13/156 (8%)
Query: 13 KITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCKKHV 72
KI LKSSD ++FEI++ VA +S I H+++D CA D IPL NVT KIL +VI++C+KHV
Sbjct: 4 KIMLKSSDGKSFEIDEDVARKSIAINHMVEDGCATDV-IPLRNVTSKILKIVIDYCEKHV 62
Query: 73 DATSSDEKPSEDEINKWDTEFVKVDQDT-LFDLILAANYLNIKSLLDLTCKTV-----AD 126
+ E+++ +WD +F+K + T LFD+++AANYLNI+SLLDLTCKTV AD
Sbjct: 63 ------KSKEEEDLKEWDADFMKTIETTILFDVMMAANYLNIQSLLDLTCKTVSDLLQAD 116
Query: 127 MIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAF 162
++ G+TP+EIR FNI ND T EE ++R E QWAF
Sbjct: 117 LLSGKTPDEIRAHFNIENDLTAEEVAKIREENQWAF 152
>At3g21830 kinetochore-like protein, Skp1p
Length = 152
Score = 154 bits (389), Expect = 2e-38
Identities = 82/155 (52%), Positives = 111/155 (70%), Gaps = 5/155 (3%)
Query: 10 ATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCK 69
+TKKI LKSS+ +TFEIE+ A + QTI H+I+ C D+ + L +T +IL MVIE+C
Sbjct: 2 STKKIMLKSSEGKTFEIEEETARQCQTIAHMIEAECTDNVILVL-KMTSEILEMVIEYCN 60
Query: 70 KH-VDATSSDEKPSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTVADMI 128
KH VDA + S+D++ KWD EF++ D+ T+F L AAN+LN KSLL L +TVADMI
Sbjct: 61 KHHVDAANPC---SDDDLEKWDKEFMEKDKSTIFALTNAANFLNNKSLLHLAGQTVADMI 117
Query: 129 KGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
KG TP+++R+ FNI ND TPEEE +R E +WAF+
Sbjct: 118 KGNTPKQMREFFNIENDLTPEEEAAIRRENKWAFE 152
>At3g21840 kinetochore-like protein, Skp1p
Length = 125
Score = 111 bits (278), Expect = 2e-25
Identities = 63/120 (52%), Positives = 85/120 (70%), Gaps = 5/120 (4%)
Query: 10 ATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCK 69
+TKKI LKSSD + FEIE+ A + QTI H+I+ C D+ IP+ NVT +IL MVIE+C
Sbjct: 2 STKKIMLKSSDGKMFEIEEETARQCQTIAHMIEAECTDNV-IPVSNVTSEILEMVIEYCN 60
Query: 70 KH-VDATSSDEKPSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTVADMI 128
KH VDA + S++++ KWD EF++ DQ T+F L+ AA L+IKSLL L +TVADM+
Sbjct: 61 KHHVDAANPC---SDEDLKKWDKEFMEKDQYTIFHLMNAAYDLHIKSLLALAYQTVADMV 117
>At3g53060 kinetochore - like protein
Length = 85
Score = 69.7 bits (169), Expect = 7e-13
Identities = 36/72 (50%), Positives = 50/72 (69%), Gaps = 8/72 (11%)
Query: 40 LIDDNCADDSGIPLPNVTGKILAMVIEHCKKHVDATSSDEKPSEDEINKWDTEFV-KVDQ 98
+ +D+CAD+ GIPLPNVT KIL +VIE+CKKHV + E+++ KWD EF+ K++Q
Sbjct: 1 MAEDDCADN-GIPLPNVTSKILLLVIEYCKKHV------VESKEEDLKKWDAEFMKKMEQ 53
Query: 99 DTLFDLILAANY 110
LFD L A +
Sbjct: 54 SILFDAKLQARF 65
>At2g45950 SKP1-like protein
Length = 227
Score = 68.2 bits (165), Expect = 2e-12
Identities = 48/162 (29%), Positives = 88/162 (53%), Gaps = 15/162 (9%)
Query: 1 MSDG----VESSTATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDS---GIPL 53
MS+G ++ T I L+++D ++E+ VA+ I + S I L
Sbjct: 1 MSEGDLAVMKPETMKSYIWLQTADGSIQQVEQEVAMFCPMICQEVIQKGVGSSKNHAISL 60
Query: 54 PN-VTGKILAMVIEHCKKH-VDATSSDEKPSEDEINKWDTEFVKVDQDTLFDLILAANYL 111
P V + ++++++C+ H + S+ E+ + DE F+++D L +L AA+ L
Sbjct: 61 PQRVNPAMFSLILDYCRFHQLPGRSNKERKTYDE------RFIRMDTKRLCELTSAADSL 114
Query: 112 NIKSLLDLTCKTVADMIKGRTPEEIRKTFNIINDYTPEEEEE 153
+K L+DLT + +A +I+G+ PEEIR+ F++ +D T EE+ E
Sbjct: 115 QLKPLVDLTSRALARIIEGKNPEEIREIFHLPDDLTEEEKLE 156
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.310 0.129 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,749,812
Number of Sequences: 26719
Number of extensions: 152993
Number of successful extensions: 465
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 27
Number of HSP's successfully gapped in prelim test: 20
Number of HSP's that attempted gapping in prelim test: 391
Number of HSP's gapped (non-prelim): 50
length of query: 164
length of database: 11,318,596
effective HSP length: 92
effective length of query: 72
effective length of database: 8,860,448
effective search space: 637952256
effective search space used: 637952256
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC148341.11


