

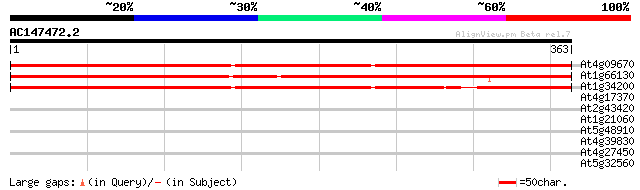
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147472.2 - phase: 0
(363 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g09670 AX110P like protein 369 e-102
At1g66130 unknown protein 353 1e-97
At1g34200 F23M19.12/F23M19.12 309 2e-84
At4g17370 unknown protein 37 0.022
At2g43420 putative sterol dehydrogenase 30 1.6
At1g21060 unknown protein 30 2.1
At5g48910 selenium-binding protein-like 29 3.5
At4g39830 putative L-ascorbate oxidase 29 4.6
At4g27450 unknown protein 28 6.0
At5g32560 putative protein 28 7.8
>At4g09670 AX110P like protein
Length = 362
Score = 369 bits (946), Expect = e-102
Identities = 186/363 (51%), Positives = 253/363 (69%), Gaps = 4/363 (1%)
Query: 1 MTEKDTVRFGILGCATIARKLARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRIY 60
M + +R G++GCA IARK++RAI L PNATIS + SRS+ KA FA N+ P S +I+
Sbjct: 1 MATETQIRIGVMGCADIARKVSRAIHLAPNATISGVASRSLEKAKAFATANNYPESTKIH 60
Query: 61 GSYDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEA 120
GSY+ ++ED +DA+Y+PLPTSLHV WA+ AA K KH+L+EKP A++V E D I++ACEA
Sbjct: 61 GSYESLLEDPEIDALYVPLPTSLHVEWAIKAAEKGKHILLEKPVAMNVTEFDKIVDACEA 120
Query: 121 NGVQFMDGCMWLHHPRTSNMKHLLDLSNSDGIGPVQFIHSTSTMPTTQEFLENNIRVKPE 180
NGVQ MDG MW+H+PRT+ +K LS+S+ G ++ + S + ++FL+N+IRVKP
Sbjct: 121 NGVQIMDGTMWVHNPRTALLKEF--LSDSERFGQLKTVQSCFSFAGDEDFLKNDIRVKPG 178
Query: 181 LDALGALGDLAWYCIGASLWAKGYKLPTTVTALPDVTRNMAGVILSITTSLQWDQPNQTV 240
LD LGALGD WY I A+L A ++LP TVTA P N AGVILS SL W+ +
Sbjct: 179 LDGLGALGDAGWYAIRATLLANNFELPKTVTAFPGAVLNEAGVILSCGASLSWE--DGRT 236
Query: 241 ANIHCSFHSHTSMDLSISGSNGSMHLKDFIIPYKETSASFDFTFGAKFVDLHIGWNVRPE 300
A I+CSF ++ +M+++ G+ G++ + DFIIPYKET ASF + A F DL W P
Sbjct: 237 ATIYCSFLANLTMEITAIGTKGTLRVHDFIIPYKETEASFTTSTKAWFNDLVTAWVSPPS 296
Query: 301 EVHVVNKLPQEALMVQEFAMLVASIRDSGSQPSIKWPEISRKTQLVVDAVKKSLELGCKP 360
E V +LPQEA MV+EFA LV I+++G++P WP ISRKTQLVVDAVK+S++ +
Sbjct: 297 EHTVKTELPQEACMVREFARLVGEIKNNGAKPDGYWPSISRKTQLVVDAVKESVDKNYQQ 356
Query: 361 VSL 363
+SL
Sbjct: 357 ISL 359
>At1g66130 unknown protein
Length = 364
Score = 353 bits (905), Expect = 1e-97
Identities = 186/368 (50%), Positives = 246/368 (66%), Gaps = 9/368 (2%)
Query: 1 MTEKDTVRFGILGCATIARKLARAITLTPNATISAIGSRSISKAAKFAVENSL-PASVRI 59
M++ TVRFG+LGC A K R +T + NA I AI S+ A FA N+L P +V I
Sbjct: 1 MSDTKTVRFGLLGCIRFASKFVRTVTESSNAVIIAISDPSLETAKTFATGNNLSPETVTI 60
Query: 60 YGSYDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACE 119
YGSY+E++ D+ VDAVY+ +P + RWAV AA KKKHVLVEKP A D EL+ I+EACE
Sbjct: 61 YGSYEELLNDANVDAVYLTMPVTQRARWAVTAAEKKKHVLVEKPPAQDATELEKIVEACE 120
Query: 120 ANGVQFMDGCMWLHHPRTSNMKHLLDLSNSDGIGPVQFIHSTSTMPTTQEFLENNIRVKP 179
NGVQFMDG +WLHH RT ++ + +S +G V+ ++ST T P ++ LE K
Sbjct: 121 YNGVQFMDGTIWLHHQRTVKIRD--TMFDSGLLGDVRHMYSTMTTPVPEQVLER--LTKE 176
Query: 180 ELDALGALGDLAWYCIGASLWAKGYKLPTTVTALP-DVTRNMAGVILSITTSLQWDQPNQ 238
+ GA+G+L WY IGA+LWA Y++P +V ALP V+ N G ILS T SLQ+
Sbjct: 177 AMGLAGAIGELGWYPIGAALWAMSYQMPISVRALPSSVSTNSVGTILSCTASLQFGSTAT 236
Query: 239 TVANIHCSFHSHTSMDLSISGSNGSMHLKDFIIPYKETSASFDFTFGAKFVDLHIGWNVR 298
T A +HCSF S S DL+ISGS GS+ + D++IPYKE A F++T GAKFVD+HIGWNV
Sbjct: 237 TTAIVHCSFLSQLSTDLAISGSKGSIQMNDYVIPYKEDKAWFEYTSGAKFVDMHIGWNVT 296
Query: 299 PEEVHV-VNKLP--QEALMVQEFAMLVASIRDSGSQPSIKWPEISRKTQLVVDAVKKSLE 355
PE V V + P QEA+M++EF LV I+ + +WP+IS+KTQL+VDAVKKS++
Sbjct: 297 PERVTVDCSGTPETQEAMMLREFTRLVEGIKRGDLEADRRWPDISKKTQLIVDAVKKSVD 356
Query: 356 LGCKPVSL 363
+GC+ V L
Sbjct: 357 IGCEVVHL 364
>At1g34200 F23M19.12/F23M19.12
Length = 352
Score = 309 bits (791), Expect = 2e-84
Identities = 165/364 (45%), Positives = 230/364 (62%), Gaps = 16/364 (4%)
Query: 1 MTEKDTVRFGILGCATIARKLARAITLTPNATISAIGSRS-ISKAAKFAVENSLPASVRI 59
M+ + +R G+L C RKL+RAI L+PNATISAI + S I +A FA N+ P + ++
Sbjct: 1 MSGDNQIRIGLLSCTNKVRKLSRAINLSPNATISAIATTSSIEEAKSFAKSNNFPPNTKL 60
Query: 60 YGSYDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACE 119
+ SY+ ++ED VDAVY P+PT LHV WA AA K KH+L++KP AL+VAE D I+EACE
Sbjct: 61 HSSYESLLEDPDVDAVYFPIPTRLHVEWATLAAIKGKHILLDKPVALNVAEFDQIVEACE 120
Query: 120 ANGVQFMDGCMWLHHPRTSNMKHLLDLSNSDGIGPVQFIHSTSTMPTTQEFLENNIRVKP 179
NGVQFMDG W+H PRT +K +++ + G ++ ++S + + +FL+++IRVKP
Sbjct: 121 VNGVQFMDGTQWMHSPRTDKIKEF--VNDLESFGQIKSVYSCFSFAASDDFLKHDIRVKP 178
Query: 180 ELDALGALGDLAWYCIGASLWAKGYKLPTTVTALPDVTRNMAGVILSITTSLQWDQPNQT 239
LD LGALGD WY I A L +KLP TVTAL N AG++LS W+ +
Sbjct: 179 GLDGLGALGDAGWYTIQAILLVNNFKLPKTVTALSGSVLNDAGIVLSCGALFDWE--DGV 236
Query: 240 VANIHCSFHSHTSMDLSISGSNGSMHLKDFIIPYKETSASFDFTFGAKFVDLHIGWNVRP 299
A I+CSF ++ +M+++ G+ GS+ + DF+IPY ET A F T G V H
Sbjct: 237 NATIYCSFLANLTMEITAIGTKGSLRVHDFVIPYMETEAVFT-TSGGGCVSEH------- 288
Query: 300 EEVHVVNKLPQEALMVQEFAMLVASIRDSGSQPSIKWPEISRKTQLVVDAVKKSLELGCK 359
V+ +L QEA MV EFA LV I++ G++P W SR TQL+VDAVK+S++ +
Sbjct: 289 ---KVMTELSQEACMVMEFARLVGEIKNRGAKPDGFWMRFSRNTQLLVDAVKESIDNNLE 345
Query: 360 PVSL 363
PV L
Sbjct: 346 PVFL 349
>At4g17370 unknown protein
Length = 368
Score = 36.6 bits (83), Expect = 0.022
Identities = 30/123 (24%), Positives = 55/123 (44%), Gaps = 7/123 (5%)
Query: 4 KDTV--RFGILGCATIARK-LARAITLTPN--ATISAIGSRSISKAAKFAVENSLPASVR 58
KD+V ++GI+G + R+ L L A + S+ + S ++
Sbjct: 5 KDSVITKYGIVGIGMMGREHLINLHHLRDQGLAVVCIADPHPPSQLLAIELAQSFGWELK 64
Query: 59 IYGSYDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKK--HVLVEKPAALDVAELDLILE 116
++ ++E+++ D + + P H + + N K HVLVEKP VA+ +LE
Sbjct: 65 VFSGHEELLKSELCDVIVVSSPNMTHHQILMDIINYSKPHHVLVEKPLCTTVADCKQVLE 124
Query: 117 ACE 119
A +
Sbjct: 125 AAK 127
>At2g43420 putative sterol dehydrogenase
Length = 437
Score = 30.4 bits (67), Expect = 1.6
Identities = 12/25 (48%), Positives = 15/25 (60%)
Query: 179 PELDALGALGDLAWYCIGASLWAKG 203
P LD L +G + WYCI AS+ G
Sbjct: 412 PSLDLLALVGFVGWYCINASMVLSG 436
>At1g21060 unknown protein
Length = 505
Score = 30.0 bits (66), Expect = 2.1
Identities = 26/106 (24%), Positives = 40/106 (37%), Gaps = 18/106 (16%)
Query: 136 RTSNMKHLLDLSNSDGIGPVQFIHSTSTMP----------------TTQEFLENNIRVKP 179
+T N+KH+ L + + ST P + +EF+ NN+R+
Sbjct: 368 KTCNVKHIYALEYPEALAHFALSSGFSTDPPVRVYTADCVFRDLRKSKEEFIRNNVRIHN 427
Query: 180 ELDALGALGDLAWYCIGASLWAKGYKLPTTVTALPDVTRNMAGVIL 225
E L L + Y + TTV LPD T+ A +L
Sbjct: 428 ETKIL--LPKIVHYYAKDMSLEPSALMETTVKCLPDSTKRTAQKLL 471
>At5g48910 selenium-binding protein-like
Length = 646
Score = 29.3 bits (64), Expect = 3.5
Identities = 21/87 (24%), Positives = 40/87 (45%), Gaps = 15/87 (17%)
Query: 262 GSMHLKDFIIPYKETSA-SFDFTFGAKFVDLHIGWNVRPEEVHVVNKLPQE-----ALMV 315
GS+ L +++ Y E S D G+ +D++ + + +HV +LP+E + M+
Sbjct: 288 GSLELGEWLHLYAEDSGIRIDDVLGSALIDMYSKCGIIEKAIHVFERLPRENVITWSAMI 347
Query: 316 QEFAM---------LVASIRDSGSQPS 333
FA+ +R +G +PS
Sbjct: 348 NGFAIHGQAGDAIDCFCKMRQAGVRPS 374
>At4g39830 putative L-ascorbate oxidase
Length = 582
Score = 28.9 bits (63), Expect = 4.6
Identities = 19/79 (24%), Positives = 30/79 (37%), Gaps = 8/79 (10%)
Query: 192 WYCIGASLWAKGY--------KLPTTVTALPDVTRNMAGVILSITTSLQWDQPNQTVANI 243
W+ G W GY + P + + +N V T+L++ N V +
Sbjct: 485 WHLHGHDFWVLGYGEGKFNESEDPKRYNRVDPIKKNTVAVQPFGWTALRFRADNPGVWSF 544
Query: 244 HCSFHSHTSMDLSISGSNG 262
HC SH M + I +G
Sbjct: 545 HCHIESHFFMGMGIVFESG 563
>At4g27450 unknown protein
Length = 250
Score = 28.5 bits (62), Expect = 6.0
Identities = 22/73 (30%), Positives = 34/73 (46%), Gaps = 12/73 (16%)
Query: 61 GSYDEVVEDSGVDAVYIPLPTSLHVR--WAVAAANKKKHVLVEKPAALDVAELDLILEAC 118
GS+ VV DS +V+ L + V+ W +AA V++ +LD+I E C
Sbjct: 133 GSFSFVVYDSKAGSVFTALGSDGGVKLYWGIAADGS---VVISD-------DLDVIKEGC 182
Query: 119 EANGVQFMDGCMW 131
+ F GCM+
Sbjct: 183 AKSFAPFPTGCMF 195
>At5g32560 putative protein
Length = 329
Score = 28.1 bits (61), Expect = 7.8
Identities = 17/43 (39%), Positives = 25/43 (57%), Gaps = 1/43 (2%)
Query: 1 MTEKDTVRFGILGCATIARKLARAITLTPNATISAIGSRSISK 43
+ +KD +GC T R+L R +TL P TI++ SRS S+
Sbjct: 137 LRDKDDETSTRVGCHT-PRRLGRGLTLPPPPTITSPSSRSGSE 178
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.132 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,814,145
Number of Sequences: 26719
Number of extensions: 319110
Number of successful extensions: 798
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 781
Number of HSP's gapped (non-prelim): 10
length of query: 363
length of database: 11,318,596
effective HSP length: 101
effective length of query: 262
effective length of database: 8,619,977
effective search space: 2258433974
effective search space used: 2258433974
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC147472.2


