

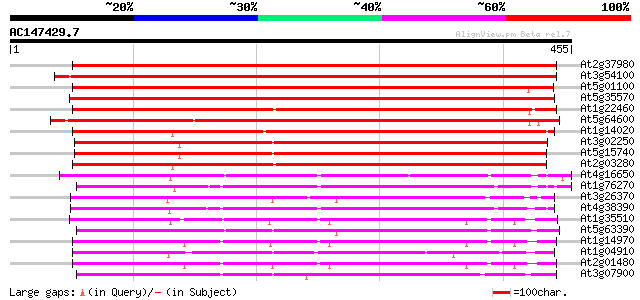
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147429.7 - phase: 0 /pseudo
(455 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g37980 similar to axi 1 protein from Nicotiana tabacum 564 e-161
At3g54100 unknown protein 560 e-160
At5g01100 unknown protein 539 e-153
At5g35570 unknown protein 520 e-148
At1g22460 Similar to auxin-independent growth promoter 388 e-108
At5g64600 auxin-independent growth promoter-like protein 384 e-107
At1g14020 growth regulator like protein 380 e-106
At3g02250 putative auxin-independent growth promoter 379 e-105
At5g15740 unknown protein 376 e-104
At2g03280 axi 1 -like protein 363 e-100
At4g16650 growth regulator like protein 291 7e-79
At1g76270 putative auxin-independent growth promoter (At1g76270) 278 6e-75
At3g26370 unknown protein 264 7e-71
At4g38390 putative growth regulator protein 262 3e-70
At1g35510 putative growth regulator protein (At1g35510) 252 3e-67
At5g63390 auxin-independent growth promoter-like protein 244 9e-65
At1g14970 auxin-independent growth promoter, putative 243 1e-64
At1g04910 Similar to auxin-independent growth promoter (axi 1) 243 2e-64
At2g01480 similar to axi 1 protein from Nicotiana tabacum 240 1e-63
At3g07900 putative auxin-independent growth promoter 234 6e-62
>At2g37980 similar to axi 1 protein from Nicotiana tabacum
Length = 638
Score = 564 bits (1454), Expect = e-161
Identities = 263/392 (67%), Positives = 322/392 (82%)
Query: 52 TNGYLLVHANGGLNQMKTGISDMVAIAKIMNATLVFPTLDHNSFWTDPSDFKEIFNWKNF 111
TNGYLLVHANGGLNQM+TGI DMVA AKIMNATLV P LDH SFWTDPS FK+IF+W++F
Sbjct: 226 TNGYLLVHANGGLNQMRTGICDMVAAAKIMNATLVLPLLDHESFWTDPSTFKDIFDWRHF 285
Query: 112 VEVLNEDVQVVESLPPELAAIKPALKAPVSWSKASYYRTDMLQLLKKHKVIKFTHTDSRL 171
+ VL +DV +VE LPP AA++P LKAPVSWSKASYYR++ML LLKKHKVIKFTHTDSRL
Sbjct: 286 MNVLKDDVDIVEYLPPRYAAMRPLLKAPVSWSKASYYRSEMLPLLKKHKVIKFTHTDSRL 345
Query: 172 VNNGLASSIQRVRCRAMYEALRFAVPIEELGKKLVNRLRENNTPYIALHLRYEKDMLAFT 231
NNGL SIQR+RCRA Y+AL ++ IE+ GK LVNRLR N+ P+IALHLRYEKDMLAFT
Sbjct: 346 ANNGLPPSIQRLRCRANYQALGYSKEIEDFGKVLVNRLRNNSEPFIALHLRYEKDMLAFT 405
Query: 232 GCSHNLTKEETQELKKMRYSVKHWKEKEIDSKSKRLKGSCPMTPREVAVFLEALGYPVDT 291
GCSHNLT E +EL+ MRY+VKHWKEKEIDS+ +R++G CPM+PRE A+FL+A+GYP T
Sbjct: 406 GCSHNLTAGEAEELRIMRYNVKHWKEKEIDSRERRIQGGCPMSPREAAIFLKAMGYPSST 465
Query: 292 KIYVAAGVIYGSEGMKPLQKKFPNLLWHSSLATKEELQPFEGHLNQLAALDYYITVESDV 351
+Y+ AG IYG M ++++PN+ HS LAT+EEL+PF+ + N+LAALDY + +ESDV
Sbjct: 466 TVYIVAGEIYGGNSMDAFREEYPNVFAHSYLATEEELEPFKPYQNRLAALDYIVALESDV 525
Query: 352 FVYSYDGNMAKAARGHRKFDGFKKTISPDKQRFVRLIDQLDNGLISWNDFSTKVKSIHAK 411
FVY+YDGNMAKA +GHR+F+GFKKTI+PD+ FVRLID LD G++SW++FS++VK +H
Sbjct: 526 FVYTYDGNMAKAVQGHRRFEGFKKTINPDRLNFVRLIDHLDEGVMSWDEFSSEVKRLHNN 585
Query: 412 KKGAPQARKIHRHPKFEETFYANPFPGCICQK 443
+ GAP AR P+ EE FYANP P CIC K
Sbjct: 586 RIGAPYARLPGEFPRLEENFYANPQPDCICNK 617
>At3g54100 unknown protein
Length = 638
Score = 560 bits (1444), Expect = e-160
Identities = 266/407 (65%), Positives = 324/407 (79%), Gaps = 1/407 (0%)
Query: 37 CITMKNFYYIGQGNNTNGYLLVHANGGLNQMKTGISDMVAIAKIMNATLVFPTLDHNSFW 96
C+T Y Q TNGYL+VHANGGLNQM+TGI DMVA+AKIMNATLV P LDH SFW
Sbjct: 208 CVTRPKNYTRLQ-RQTNGYLVVHANGGLNQMRTGICDMVAVAKIMNATLVLPLLDHESFW 266
Query: 97 TDPSDFKEIFNWKNFVEVLNEDVQVVESLPPELAAIKPALKAPVSWSKASYYRTDMLQLL 156
TDPS FK+IF+W+NF+ VL DV +VE LPP+ AA+KP LKAPVSWSKASYYR++ML LL
Sbjct: 267 TDPSTFKDIFDWRNFMNVLKHDVDIVEYLPPQYAAMKPLLKAPVSWSKASYYRSEMLPLL 326
Query: 157 KKHKVIKFTHTDSRLVNNGLASSIQRVRCRAMYEALRFAVPIEELGKKLVNRLRENNTPY 216
K+HKV+KFT TDSRL NNGL SIQR+RCRA Y+AL + IE+LGK LVNRLR N PY
Sbjct: 327 KRHKVLKFTLTDSRLANNGLPPSIQRLRCRANYQALLYTKEIEDLGKILVNRLRNNTEPY 386
Query: 217 IALHLRYEKDMLAFTGCSHNLTKEETQELKKMRYSVKHWKEKEIDSKSKRLKGSCPMTPR 276
IALHLRYEKDMLAFTGC+HNLT EE +EL+ MRYSVKHWKEKEIDS+ +R++G CPM+PR
Sbjct: 387 IALHLRYEKDMLAFTGCNHNLTTEEAEELRIMRYSVKHWKEKEIDSRERRIQGGCPMSPR 446
Query: 277 EVAVFLEALGYPVDTKIYVAAGVIYGSEGMKPLQKKFPNLLWHSSLATKEELQPFEGHLN 336
E A+FL+A+GYP T +Y+ AG IYGSE M + ++PN+ HS+LAT+EEL+PF + N
Sbjct: 447 EAAIFLKAMGYPSSTTVYIVAGEIYGSESMDAFRAEYPNVFSHSTLATEEELEPFSQYQN 506
Query: 337 QLAALDYYITVESDVFVYSYDGNMAKAARGHRKFDGFKKTISPDKQRFVRLIDQLDNGLI 396
+LAALDY + +ESDVFVY+YDGNMAKA +GHRKF+GF+K+I+PD+ FVRLID D G+I
Sbjct: 507 RLAALDYIVALESDVFVYTYDGNMAKAVQGHRKFEGFRKSINPDRLNFVRLIDHFDEGII 566
Query: 397 SWNDFSTKVKSIHAKKKGAPQARKIHRHPKFEETFYANPFPGCICQK 443
SW +FS++VK ++ + GA R P+ EE FYANP P CIC K
Sbjct: 567 SWEEFSSEVKRLNRDRIGAAYGRLPAALPRLEENFYANPQPDCICNK 613
>At5g01100 unknown protein
Length = 631
Score = 539 bits (1388), Expect = e-153
Identities = 255/393 (64%), Positives = 314/393 (79%), Gaps = 3/393 (0%)
Query: 52 TNGYLLVHANGGLNQMKTGISDMVAIAKIMNATLVFPTLDHNSFWTDPSDFKEIFNWKNF 111
TNGYLLVHANGGLNQM+TGI DMVAIAKIMNATLV P LDH+SFW+DPS FK+IF+WK+F
Sbjct: 216 TNGYLLVHANGGLNQMRTGICDMVAIAKIMNATLVLPFLDHSSFWSDPSSFKDIFDWKHF 275
Query: 112 VEVLNEDVQVVESLPPELAAIKPALKAPVSWSKASYYRTDMLQLLKKHKVIKFTHTDSRL 171
++VL EDV +VE LP E A+IKP K PVSWSK+SYYR + +LLKKHKVI F HTDSRL
Sbjct: 276 IKVLAEDVNIVEYLPQEFASIKPLEKNPVSWSKSSYYRNSISKLLKKHKVIVFNHTDSRL 335
Query: 172 VNNGLASSIQRVRCRAMYEALRFAVPIEELGKKLVNRLRENNTPYIALHLRYEKDMLAFT 231
NN SIQR+RCRA YEALR++ IE L L +RLRENN PY+ALHLRYEKDMLAFT
Sbjct: 336 ANNSPPPSIQRLRCRANYEALRYSEDIENLSNVLSSRLRENNEPYLALHLRYEKDMLAFT 395
Query: 232 GCSHNLTKEETQELKKMRYSVKHWKEKEIDSKSKRLKGSCPMTPREVAVFLEALGYPVDT 291
GC+H+L+ EE+ +L+KMR+S+ HWKEK I+ +RL+G+CPMTPRE AVFL+A+G+P T
Sbjct: 396 GCNHSLSNEESIDLEKMRFSIPHWKEKVINGTERRLEGNCPMTPREAAVFLKAMGFPSTT 455
Query: 292 KIYVAAGVIYGSEGMKPLQKKFPNLLWHSSLATKEELQPFEGHLNQLAALDYYITVESDV 351
IY+ AG IYG M ++FPN+ +H++LAT+EEL + + N+LAALDY + +ESD+
Sbjct: 456 NIYIVAGKIYGQNSMTAFHEEFPNVFFHNTLATEEELSTIKPYQNRLAALDYNLALESDI 515
Query: 352 FVYSYDGNMAKAARGHRKFDGFKKTISPDKQRFVRLIDQLDNGLISWNDFSTKVKSIHAK 411
F Y+YDGNMAKA +GHR+F+GF+KTI+PD+QRFVRLID+LD GLISW DFS+KVK +H
Sbjct: 516 FAYTYDGNMAKAVQGHRRFEGFRKTINPDRQRFVRLIDRLDAGLISWEDFSSKVKKMHQH 575
Query: 412 KKGAPQAR---KIHRHPKFEETFYANPFPGCIC 441
+ GAP R K PK EE FYANP PGC+C
Sbjct: 576 RIGAPYLRQPGKAGMSPKLEENFYANPLPGCVC 608
>At5g35570 unknown protein
Length = 652
Score = 520 bits (1339), Expect = e-148
Identities = 235/394 (59%), Positives = 304/394 (76%)
Query: 49 GNNTNGYLLVHANGGLNQMKTGISDMVAIAKIMNATLVFPTLDHNSFWTDPSDFKEIFNW 108
G NTNGYLL++ANGGLNQM+ GI DMVA+AKIM ATLV P+LDH+S+W D S FK++F+W
Sbjct: 248 GANTNGYLLINANGGLNQMRFGICDMVAVAKIMKATLVLPSLDHSSYWADDSGFKDLFDW 307
Query: 109 KNFVEVLNEDVQVVESLPPELAAIKPALKAPVSWSKASYYRTDMLQLLKKHKVIKFTHTD 168
++F+E L +D+ +VE LP ELA I+P +K P+SWSK YY+ ++L LLK+H V+ THTD
Sbjct: 308 QHFIEELKDDIHIVEMLPSELAGIEPFVKTPISWSKVGYYKKEVLPLLKQHIVMYLTHTD 367
Query: 169 SRLVNNGLASSIQRVRCRAMYEALRFAVPIEELGKKLVNRLRENNTPYIALHLRYEKDML 228
SRL NN L S+Q++RCR Y AL+++ PIEELG LV+R+R++ PY+ALHLRYEKDML
Sbjct: 368 SRLANNDLPDSVQKLRCRVNYRALKYSAPIEELGNVLVSRMRQDRGPYLALHLRYEKDML 427
Query: 229 AFTGCSHNLTKEETQELKKMRYSVKHWKEKEIDSKSKRLKGSCPMTPREVAVFLEALGYP 288
AFTGCSH+LT EE +EL++MRY V HWKEKEI+ +RL+G CP+TPRE ++ L AL +P
Sbjct: 428 AFTGCSHSLTAEEDEELRQMRYEVSHWKEKEINGTERRLQGGCPLTPRETSLLLRALEFP 487
Query: 289 VDTKIYVAAGVIYGSEGMKPLQKKFPNLLWHSSLATKEELQPFEGHLNQLAALDYYITVE 348
++IY+ AG YG+ M PL FPN+ HS LATKEEL PF H N LA LDY + ++
Sbjct: 488 SSSRIYLVAGEAYGNGSMDPLNTDFPNIFSHSILATKEELSPFNNHQNMLAGLDYIVALQ 547
Query: 349 SDVFVYSYDGNMAKAARGHRKFDGFKKTISPDKQRFVRLIDQLDNGLISWNDFSTKVKSI 408
S+VF+Y+YDGNMAKA +GHR+F+ FKKTI+PDK FV+L+D LD G ISW FS+KVK +
Sbjct: 548 SEVFLYTYDGNMAKAVQGHRRFEDFKKTINPDKMNFVKLVDALDEGRISWKKFSSKVKKL 607
Query: 409 HAKKKGAPQARKIHRHPKFEETFYANPFPGCICQ 442
H + GAP R+ PK EE+FYANP PGCIC+
Sbjct: 608 HKDRNGAPYNRESGEFPKLEESFYANPLPGCICE 641
>At1g22460 Similar to auxin-independent growth promoter
Length = 565
Score = 388 bits (997), Expect = e-108
Identities = 192/402 (47%), Positives = 274/402 (67%), Gaps = 12/402 (2%)
Query: 52 TNGYLLVHANGGLNQMKTGISDMVAIAKIMNATLVFPTLDHNSFWTDPSDFKEIFNWKNF 111
+ GYLLVH NGGLNQM+ GI DMVAIA+I+NATLV P LD SFW D S F ++F+ +F
Sbjct: 160 SRGYLLVHTNGGLNQMRAGICDMVAIARIINATLVVPELDKRSFWQDTSKFSDVFDEDHF 219
Query: 112 VEVLNEDVQVVESLPPELAAIKPALKAPVSWSKASYYRTDMLQLLKKHKVIKFTHTDSRL 171
+ L++D++V++ LP + + +K S+S SYY+ ++ + ++KVI+ +DSRL
Sbjct: 220 INALSKDIRVIKKLPKGIDGLTKVVKHFKSYSGLSYYQNEIASMWDEYKVIRAAKSDSRL 279
Query: 172 VNNGLASSIQRVRCRAMYEALRFAVPIEELGKKLVNRLRENNTPYIALHLRYEKDMLAFT 231
NN L IQ++RCRA YEALRF+ I +G+ LV+R+R YIALHLR+EK+MLAF+
Sbjct: 280 ANNNLPPDIQKLRCRACYEALRFSTKIRSMGELLVDRMRSYGL-YIALHLRFEKEMLAFS 338
Query: 232 GCSHNLTKEETQELKKMRYSVKHWKEKEIDSKSKRLKGSCPMTPREVAVFLEALGYPVDT 291
GC+H L+ E EL+++R + +WK K+ID + +RLKG CP+TP+EV + L ALGY DT
Sbjct: 339 GCNHGLSASEAAELRRIRKNTAYWKVKDIDGRVQRLKGYCPLTPKEVGILLTALGYSSDT 398
Query: 292 KIYVAAGVIYGSEG-MKPLQKKFPNLLWHSSLATKEELQPFEGHLNQLAALDYYITVESD 350
+Y+AAG IYG E + L+ +F L+ LAT+EEL+ F H Q+AALDY +++ESD
Sbjct: 399 PVYIAAGEIYGGESRLADLRSRFSMLMSKEKLATREELKTFMNHSTQMAALDYIVSIESD 458
Query: 351 VFVYSYDGNMAKAARGHRKFDGFKKTISPDKQRFVRLIDQLDNGLISWN-DFSTKVKSIH 409
VF+ SY GNMA+A GHR+F G +KTISPD++ VRL+D++ G N ++ IH
Sbjct: 459 VFIPSYSGNMARAVEGHRRFLGHRKTISPDRKAIVRLVDRIGRGAEKDNRKVYERINEIH 518
Query: 410 AKKKGAPQARK--------IHRHPKFEETFYANPFPGCICQK 443
++G+P+ RK + RH + EE+FY NP P C+CQ+
Sbjct: 519 KTRQGSPRRRKGPASGTKGLERH-RSEESFYENPLPDCLCQR 559
>At5g64600 auxin-independent growth promoter-like protein
Length = 539
Score = 384 bits (987), Expect = e-107
Identities = 202/422 (47%), Positives = 276/422 (64%), Gaps = 12/422 (2%)
Query: 34 IHLCITMKNFYYIGQGNNTNGYLLVHANGGLNQMKTGISDMVAIAKIMNATLVFPTLDHN 93
+H C+ Y + + ++ Y+ V +NGGLNQM+TGI+D+VA+A IMNATLV P LD
Sbjct: 118 LHPCVKPTPKYK--EFSESDHYITVKSNGGLNQMRTGIADIVAVAHIMNATLVIPELDKR 175
Query: 94 SFWTDPSDFKEIFNWKNFVEVLNEDVQVVESLPPELAAIKPALKAPVSWSKASYYRTDML 153
SFW D S F +IF+ + F++ L DV+V++ LP E+ ++ A K SWS YY +M
Sbjct: 176 SFWQDSSVFSDIFDEEQFIKSLRRDVKVIKKLPKEVESLPRARKHFTSWSSVGYYE-EMT 234
Query: 154 QLLKKHKVIKFTHTDSRLVNNGLASSIQRVRCRAMYEALRFAVPIEELGKKLVNRLRENN 213
L K++KVI +DSRL NN L +QR+RCR +Y L F+ IE LG+KLV RL+
Sbjct: 235 HLWKEYKVIHVAKSDSRLANNDLPIDVQRLRCRVLYRGLCFSPAIESLGQKLVERLKSRA 294
Query: 214 TPYIALHLRYEKDMLAFTGCSHNLTKEETQELKKMRYSVKHWKEKEIDSKSKRLKGSCPM 273
YIALHLRYEKDMLAFTGC++ LT E++EL+ MR S HWK K I+S +R +G CP+
Sbjct: 295 GRYIALHLRYEKDMLAFTGCTYGLTDAESEELRVMRESTSHWKIKSINSTEQREEGLCPL 354
Query: 274 TPREVAVFLEALGYPVDTKIYVAAGVIYGSEG-MKPLQKKFPNLLWHSSLATKEELQPFE 332
TP+EV +FL+ LGY T IY+AAG IYG + + L+ +FPNL++ +LA EEL+ F
Sbjct: 355 TPKEVGIFLKGLGYSQSTVIYIAAGEIYGGDDRLSELKSRFPNLVFKETLAGNEELKGFT 414
Query: 333 GHLNQLAALDYYITVESDVFVYSYDGNMAKAARGHRKFDGFKKTISPDKQRFVRLIDQLD 392
GH + AALDY I+VESDVFV S+ GNMA+A GHR+F G ++TI+PD++ V+L +++
Sbjct: 415 GHATKTAALDYIISVESDVFVPSHSGNMARAVEGHRRFLGHRRTITPDRKGLVKLFVKME 474
Query: 393 NG-LISWNDFSTKVKSIHAKKKGAPQARK-----IHRHPKF--EETFYANPFPGCICQKP 444
G L S V +H ++GAP+ RK I +F EE FY NP+P CIC
Sbjct: 475 RGQLKEGPKLSNFVNQMHKDRQGAPRRRKGPTQGIKGRARFRTEEAFYENPYPECICSSK 534
Query: 445 TH 446
H
Sbjct: 535 EH 536
>At1g14020 growth regulator like protein
Length = 499
Score = 380 bits (976), Expect = e-106
Identities = 188/396 (47%), Positives = 268/396 (67%), Gaps = 7/396 (1%)
Query: 52 TNGYLLVHANGGLNQMKTGISDMVAIAKIMNATLVFPTLDHNSFWTDPSDFKEIFNWKNF 111
+NG LLV NGGLNQM++ I DMV +A+++N TLV P LD SFW DPS F++IF+ ++F
Sbjct: 91 SNGILLVSCNGGLNQMRSAICDMVTVARLLNLTLVVPELDKTSFWADPSGFEDIFDVRHF 150
Query: 112 VEVLNEDVQVVESLPPELA---AIKPALKAPVSWSKASYYRTDMLQLLKKHKVIKFTHTD 168
++ L ++V+++ LP + + PVSWS YY +L L KHKV+ F TD
Sbjct: 151 IDSLRDEVRILRRLPKRFSRKYGYQMFEMPPVSWSDEKYYLKQVLPLFSKHKVVHFNRTD 210
Query: 169 SRLVNNGLASSIQRVRCRAMYEALRFAVPIEELGKKLVNRLRENNTPYIALHLRYEKDML 228
+RL NNGL S+Q +RCR ++ L+F +E LG KLV R+ + P++ALHLRYE DML
Sbjct: 211 TRLANNGLPLSLQWLRCRVNFQGLKFTPQLEALGSKLV-RILQQRGPFVALHLRYEMDML 269
Query: 229 AFTGCSHNLTKEETQELKKMRYSVKHWKEKEIDSKSKRLKGSCPMTPREVAVFLEALGYP 288
AF+GC+H T+EE +ELKKMRY+ W+EKEI S+ +R +G CP+TP EVA+ L+ALG+
Sbjct: 270 AFSGCTHGCTEEEAEELKKMRYTYPWWREKEIVSEERRAQGLCPLTPEEVALVLKALGFE 329
Query: 289 VDTKIYVAAGVIYGSE-GMKPLQKKFPNLLWHSSLATKEELQPFEGHLNQLAALDYYITV 347
+T+IY+AAG IYGSE + L++ FP ++ L ELQ F+ H +Q+AALD+ ++V
Sbjct: 330 KNTQIYIAAGEIYGSEHRLSVLREAFPRIVKKEMLLESAELQQFQNHSSQMAALDFMVSV 389
Query: 348 ESDVFVYSYDGNMAKAARGHRKFDGFKKTISPDKQRFVRLIDQLDNGLISWNDFSTKVKS 407
S+ F+ +YDGNMAK GHR++ G+KKTI D++R V L+D N ++W+ F+ VK
Sbjct: 390 ASNTFIPTYDGNMAKVVEGHRRYLGYKKTILLDRKRLVELLDLHHNKTLTWDQFAVAVKE 449
Query: 408 IHAKKKGAPQARK-IHRHPKFEETFYANPFPGCICQ 442
H ++ GAP R+ I PK E+ FYANP C+C+
Sbjct: 450 AHERRAGAPTHRRVISDKPKEEDYFYANP-QECLCE 484
>At3g02250 putative auxin-independent growth promoter
Length = 512
Score = 379 bits (974), Expect = e-105
Identities = 194/390 (49%), Positives = 263/390 (66%), Gaps = 7/390 (1%)
Query: 53 NGYLLVHANGGLNQMKTGISDMVAIAKIMNATLVFPTLDHNSFWTDPSDFKEIFNWKNFV 112
NGYL+V NGGLNQM+ I DMV IA+ MN TL+ P LD SFW DPS+FK+IF+ +F+
Sbjct: 107 NGYLMVSCNGGLNQMRAAICDMVTIARYMNVTLIVPELDKTSFWNDPSEFKDIFDVDHFI 166
Query: 113 EVLNEDVQVVESLPPELAA-IKPAL---KAPVSWSKASYYRTDMLQLLKKHKVIKFTHTD 168
L ++V++++ LPP L ++ L P+SWS SYY+ +L L+KK+KV+ TD
Sbjct: 167 SSLRDEVRILKELPPRLKRRVRLGLYHTMPPISWSNMSYYQDQILPLVKKYKVVHLNKTD 226
Query: 169 SRLVNNGLASSIQRVRCRAMYEALRFAVPIEELGKKLVNRLRENNTPYIALHLRYEKDML 228
+RL NN L IQ++RCRA + LRF IEELG+++V LRE P++ LHLRYE DML
Sbjct: 227 TRLANNELPVEIQKLRCRANFNGLRFTPKIEELGRRVVKILREKG-PFLVLHLRYEMDML 285
Query: 229 AFTGCSHNLTKEETQELKKMRYSVKHWKEKEIDSKSKRLKGSCPMTPREVAVFLEALGYP 288
AF+GCSH + E +EL +MRY+ WKEK IDS+ KR +G CP+TP E A+ L ALG
Sbjct: 286 AFSGCSHGCNRYEEEELTRMRYAYPWWKEKVIDSELKRKEGLCPLTPEETALTLSALGID 345
Query: 289 VDTKIYVAAGVIYGSEG-MKPLQKKFPNLLWHSSLATKEELQPFEGHLNQLAALDYYITV 347
+ +IY+AAG IYG + +K L FPN++ +L +L + H +Q+AALDY I++
Sbjct: 346 RNVQIYIAAGEIYGGKRRLKALTDVFPNVVRKETLLDSSDLSFCKNHSSQMAALDYLISL 405
Query: 348 ESDVFVYSYDGNMAKAARGHRKFDGFKKTISPDKQRFVRLIDQLDNGLISWNDFSTKVKS 407
ESD+FV +Y GNMAK GHR+F GFKKTI +++ V+LID+ GL+SW FST VK+
Sbjct: 406 ESDIFVPTYYGNMAKVVEGHRRFLGFKKTIELNRKLLVKLIDEYYEGLLSWEVFSTTVKA 465
Query: 408 IHAKKKGAPQAR-KIHRHPKFEETFYANPF 436
HA + G P+ R I PK E+ FYANP+
Sbjct: 466 FHATRMGGPKRRLVIPNKPKEEDYFYANPY 495
>At5g15740 unknown protein
Length = 508
Score = 376 bits (965), Expect = e-104
Identities = 188/389 (48%), Positives = 261/389 (66%), Gaps = 7/389 (1%)
Query: 53 NGYLLVHANGGLNQMKTGISDMVAIAKIMNATLVFPTLDHNSFWTDPSDFKEIFNWKNFV 112
NGYL+V NGGLNQM+ I DMV +A+ MN TL+ P LD SFW DPS+FK+IF+ +F+
Sbjct: 107 NGYLMVSCNGGLNQMRAAICDMVTVARYMNVTLIVPELDKTSFWNDPSEFKDIFDVDHFI 166
Query: 113 EVLNEDVQVVESLPPELAA-IKPAL---KAPVSWSKASYYRTDMLQLLKKHKVIKFTHTD 168
L ++V++++ LPP L ++ + P+SWS SYY+ +L L+KKHKV+ TD
Sbjct: 167 SSLRDEVRILKELPPRLKKRVELGVYHEMPPISWSNMSYYQNQILPLVKKHKVLHLNRTD 226
Query: 169 SRLVNNGLASSIQRVRCRAMYEALRFAVPIEELGKKLVNRLRENNTPYIALHLRYEKDML 228
+RL NNGL +Q++RCR + L+F IEELG+++V LRE P++ LHLRYE DML
Sbjct: 227 TRLANNGLPVEVQKLRCRVNFNGLKFTPQIEELGRRVVKILREKG-PFLVLHLRYEMDML 285
Query: 229 AFTGCSHNLTKEETQELKKMRYSVKHWKEKEIDSKSKRLKGSCPMTPREVAVFLEALGYP 288
AF+GCSH EE +EL +MRY+ WKEK I+S+ KR G CP+TP E A+ L ALG
Sbjct: 286 AFSGCSHGCNPEEEEELTRMRYAYPWWKEKVINSELKRKDGLCPLTPEETALTLTALGID 345
Query: 289 VDTKIYVAAGVIYGSEG-MKPLQKKFPNLLWHSSLATKEELQPFEGHLNQLAALDYYITV 347
+ +IY+AAG IYG + MK L FPN++ +L +L H +Q+AALDY + +
Sbjct: 346 RNVQIYIAAGEIYGGQRRMKALTDAFPNVVRKETLLESSDLDFCRNHSSQMAALDYLVAL 405
Query: 348 ESDVFVYSYDGNMAKAARGHRKFDGFKKTISPDKQRFVRLIDQLDNGLISWNDFSTKVKS 407
ESD+FV + DGNMA+ GHR+F GFKKTI +++ V+LID+ GL+SW+ FS+ VK+
Sbjct: 406 ESDIFVPTNDGNMARVVEGHRRFLGFKKTIQLNRRFLVKLIDEYTEGLLSWDVFSSTVKA 465
Query: 408 IHAKKKGAPQAR-KIHRHPKFEETFYANP 435
H+ + G+P+ R I PK E+ FYANP
Sbjct: 466 FHSTRMGSPKRRLVIPNRPKEEDYFYANP 494
>At2g03280 axi 1 -like protein
Length = 481
Score = 363 bits (931), Expect = e-100
Identities = 184/390 (47%), Positives = 259/390 (66%), Gaps = 7/390 (1%)
Query: 52 TNGYLLVHANGGLNQMKTGISDMVAIAKIMNATLVFPTLDHNSFWTDPSDFKEIFNWKNF 111
+NG LLV NGGLNQM+ I DMV +A+++N TLV P LD SFW D SDF++IF+ K+F
Sbjct: 88 SNGILLVSCNGGLNQMRAAICDMVTVARLLNLTLVVPELDKKSFWADTSDFEDIFDIKHF 147
Query: 112 VEVLNEDVQVVESLPPELA---AIKPALKAPVSWSKASYYRTDMLQLLKKHKVIKFTHTD 168
++ L ++V+++ LP + K PVSWS YY +L K KVI F +D
Sbjct: 148 IDSLRDEVRIIRRLPKRYSKKYGFKLFEMPPVSWSNDKYYLQQVLPRFSKRKVIHFVRSD 207
Query: 169 SRLVNNGLASSIQRVRCRAMYEALRFAVPIEELGKKLVNRLRENNTPYIALHLRYEKDML 228
+RL NNGL+ +QR+RCR ++ LRF IE LG KLV L++ + ++ALHLRYE DML
Sbjct: 208 TRLANNGLSLDLQRLRCRVNFQGLRFTPRIEALGSKLVRILQQRGS-FVALHLRYEMDML 266
Query: 229 AFTGCSHNLTKEETQELKKMRYSVKHWKEKEIDSKSKRLKGSCPMTPREVAVFLEALGYP 288
AF+GC+H T EE +ELKKMRY+ W+EKEI S+ +R++G CP+TP E + L+ALG+
Sbjct: 267 AFSGCTHGCTDEEAEELKKMRYAYPWWREKEIVSEERRVQGLCPLTPEEAVLVLKALGFQ 326
Query: 289 VDTKIYVAAGVIY-GSEGMKPLQKKFPNLLWHSSLATKEELQPFEGHLNQLAALDYYITV 347
DT+IY+AAG I+ G++ + L++ FP ++ L ELQ F+ H +Q+AALD+ ++V
Sbjct: 327 KDTQIYIAAGEIFGGAKRLALLKESFPRIVKKEMLLDPTELQQFQNHSSQMAALDFIVSV 386
Query: 348 ESDVFVYSYDGNMAKAARGHRKFDGFKKTISPDKQRFVRLIDQLDNGLISWNDFSTKVKS 407
S+ F+ +Y GNMAK GHR++ GFKKTI D++R V L+D +N +SW+ F+ VK
Sbjct: 387 ASNTFIPTYYGNMAKVVEGHRRYLGFKKTILLDRKRLVELLDLHNNKTLSWDQFAVAVKD 446
Query: 408 IH-AKKKGAPQARK-IHRHPKFEETFYANP 435
H ++ G P RK I PK E+ FYANP
Sbjct: 447 AHQGRRMGEPTHRKVISVRPKEEDYFYANP 476
>At4g16650 growth regulator like protein
Length = 549
Score = 291 bits (744), Expect = 7e-79
Identities = 175/431 (40%), Positives = 251/431 (57%), Gaps = 29/431 (6%)
Query: 41 KNFYYIGQGNNTNGYLLVHANGGLNQMKTGISDMVAIAKIMNATLVFPTLDHNSFWTDPS 100
+NF Q ++NGYLL+ A+GGLNQ +TGI+D V +A+I+NATLV P LDH+S+W D S
Sbjct: 111 RNFLPAVQEQSSNGYLLIAASGGLNQQRTGITDAVVVARILNATLVVPELDHHSYWKDDS 170
Query: 101 DFKEIFNWKNFVEVLNEDVQVVESLPPEL--AAIKPALKAPV-SWSKASYYRTDMLQLLK 157
DF +IF+ F+ L +DV +V+ +P + A KP V S YY +L +L
Sbjct: 171 DFSDIFDVNWFISSLAKDVTIVKRVPDRVMRAMEKPPYTTRVPRKSTLEYYLDQVLPILT 230
Query: 158 KHKVIKFTHTDSRLVNNGLASSIQRVRCRAMYEALRFAVPIEELGKKLVNRLRENNTPYI 217
+ V++ T D RL N+ L +Q++RCR Y ALRF I+ +G K+V R+R+ +I
Sbjct: 231 RRHVLQLTKFDYRLAND-LDEDMQKLRCRVNYHALRFTKRIQSVGMKVVKRMRKMAKRFI 289
Query: 218 ALHLRYEKDMLAFTGCSHNLTKEETQELKKMRYSVKHWKE-KEIDSKSKRLKGSCPMTPR 276
A+HLR+E DMLAF+GC ++E EL ++R K W ++D +R +G CP+TP
Sbjct: 290 AVHLRFEPDMLAFSGCDFGGGEKERAELAEIR---KRWDTLPDLDPLEERKRGKCPLTPH 346
Query: 277 EVAVFLEALGYPVDTKIYVAAGVIYGSE-GMKPLQKKFPNLLWHSSLATKEELQPFEGHL 335
EV + L ALG+ DT IYVA+G IYG E +KPL++ FPN LA +EL+P +
Sbjct: 347 EVGLMLRALGFTNDTYIYVASGEIYGGEKTLKPLRELFPNFYTKEMLA-NDELKPLLPYS 405
Query: 336 NQLAALDYYITVESDVFVYSYDGNMAKAARGHRKFDGFKKTISPDKQRFVRLIDQLDNGL 395
++LAA+DY ++ ESDVF+ + +GNMAK G R++ G K+TI P+ ++ L +D
Sbjct: 406 SRLAAIDYIVSDESDVFITNNNGNMAKILAGRRRYMGHKRTIRPNAKKLSALF--MDREK 463
Query: 396 ISWNDFSTKVKSIHAKKKGAPQARKIHRHPKFEETFYANPFPGCICQKPTHL-------- 447
+ W F+ KVKS G P K R F+ P CICQ+P
Sbjct: 464 MEWQTFAKKVKSCQRGFMGDPDEFKPGR-----GEFHEYP-QSCICQRPFSYDKTSTDDE 517
Query: 448 ---MAETNHES 455
M+E NH S
Sbjct: 518 EEDMSEENHNS 528
>At1g76270 putative auxin-independent growth promoter (At1g76270)
Length = 572
Score = 278 bits (710), Expect = 6e-75
Identities = 163/405 (40%), Positives = 239/405 (58%), Gaps = 18/405 (4%)
Query: 55 YLLVHANGGLNQMKTGISDMVAIAKIMNATLVFPTLDHNSFWTDPSDFKEIFNWKNFVEV 114
YL++ +GGLNQ +TGI D V A+I+NATLV P LD S+W D SDF IF+ F+
Sbjct: 109 YLVIATSGGLNQQRTGIVDAVVAARILNATLVVPKLDQKSYWKDASDFSHIFDVDWFISF 168
Query: 115 LNEDVQVVESLPPELAAI--KPALKAPVSWSKASYYRTDMLQLLKKHKVIKFTHTDSRLV 172
L+ DV++++ LP + ++ P ++ Y + LLK+H V + D RL
Sbjct: 169 LSGDVRIIKQLPLKGGRTWSTSRMRVPRKCNERCYINRVLPVLLKRHAV-QLNKFDYRL- 226
Query: 173 NNGLASSIQRVRCRAMYEALRFAVPIEELGKKLVNRLRENNTPYIALHLRYEKDMLAFTG 232
+N L+ +Q++RCR Y AL+F PI +G +LV R+R + +IALHLRYE DMLAF+G
Sbjct: 227 SNKLSDDLQKLRCRVNYHALKFTDPILTMGNELVRRMRLRSKHFIALHLRYEPDMLAFSG 286
Query: 233 CSHNLTKEETQELKKMRYSVKHWKEKEIDSKSK-RLKGSCPMTPREVAVFLEALGYPVDT 291
C + +E +EL +R + WK I++ K R +G CP+TP EV + L ALGY D
Sbjct: 287 CYYGGGDKERRELAAIR---RRWKTLHINNPEKQRRQGRCPLTPEEVGLMLRALGYGSDV 343
Query: 292 KIYVAAGVIYGSE-GMKPLQKKFPNLLWHSSLATKEELQPFEGHLNQLAALDYYITVESD 350
IYVA+G +YG E + PL+ FP+ ++ATKEEL+PF + +++AALD+ + ESD
Sbjct: 344 HIYVASGEVYGGEESLAPLKALFPHFYSKDTIATKEELEPFSSYSSRMAALDFLVCDESD 403
Query: 351 VFVYSYDGNMAKAARGHRKFDGFKKTISPDKQRFVRLIDQLDNGLISWNDFSTKVKSIHA 410
VFV + +GNMAK G R++ G K T+ P+ ++ RL +N +W +FS+KV+S
Sbjct: 404 VFVTNNNGNMAKILAGRRRYLGHKPTVRPNAKKLYRLFMNKEN--TTWEEFSSKVRSFQR 461
Query: 411 KKKGAPQARKIHRHPKFEETFYANPFPGCICQKPTHLMAETNHES 455
G P+ + R F+ NP CIC+ T A+ ES
Sbjct: 462 GFMGEPKEVRAGR-----GEFHENP-STCICE-DTEAKAKAQLES 499
>At3g26370 unknown protein
Length = 557
Score = 264 bits (675), Expect = 7e-71
Identities = 152/412 (36%), Positives = 231/412 (55%), Gaps = 30/412 (7%)
Query: 50 NNTNGYLLVHANGGLNQMKTGISDMVAIAKIMNATLVFPTLDHNSFWTDPSDFKEIFNWK 109
N TNGY+ +HA GGLNQ + I + VA+AKIMNATL+ P L + W D + F++IF+
Sbjct: 156 NETNGYVFIHAEGGLNQQRIAICNAVAVAKIMNATLILPVLKQDQIWKDTTKFEDIFDVD 215
Query: 110 NFVEVLNEDVQVVESLP------PEL-AAIKPALKAPVSWSKASYYRTDMLQLLKKHKVI 162
+F++ L +DV++V +P EL ++I+ +K ++ A +Y ++L +K+ K++
Sbjct: 216 HFIDYLKDDVRIVRDIPDWFTDKAELFSSIRRTVKNIPKYAAAQFYIDNVLPRIKEKKIM 275
Query: 163 KFTHTDSRLVNNGLASSIQRVRCRAMYEALRFAVPIEELGKKLVNRLRE---NNTPYIAL 219
RL + + I R+RCR Y AL+F IE++ LV+R+R N PY+AL
Sbjct: 276 ALKPFVDRLGYDNVPQEINRLRCRVNYHALKFLPEIEQMADSLVSRMRNRTGNPNPYMAL 335
Query: 220 HLRYEKDMLAFTGCSHNLTKEETQELKKMRYSVKHWKEKEIDSK-------SKRLKGSCP 272
HLR+EK M+ + C T+EE ++K Y K W + + KR +G CP
Sbjct: 336 HLRFEKGMVGLSFCDFVGTREE--KVKMAEYRQKEWPRRFKNGSHLWQLALQKRKEGRCP 393
Query: 273 MTPREVAVFLEALGYPVDTKIYVAAGVIYGSEG-MKPLQKKFPNLLWHSSLATKEELQPF 331
+ P EVAV L A+GYP +T+IYVA+G +YG + M PL+ FPNL+ LA KEEL F
Sbjct: 394 LEPGEVAVILRAMGYPKETQIYVASGQVYGGQNRMAPLRNMFPNLVTKEDLAGKEELTTF 453
Query: 332 EGHLNQLAALDYYITVESDVFVYSYDGNMAKAARGHRKFDGFK-KTISPDKQRFVRLIDQ 390
H+ LAALD+ + ++SDVFV ++ GN AK G R++ G + K+I PDK +
Sbjct: 454 RKHVTSLAALDFLVCLKSDVFVMTHGGNFAKLIIGARRYMGHRQKSIKPDKGLMSK---S 510
Query: 391 LDNGLISWNDFSTKVKSIHAKKKGAPQARKIHRHPKFEETFYANPFPGCICQ 442
+ + W F V H + G P+ P ++ + NP C+C+
Sbjct: 511 FGDPYMGWATFVEDVVVTHQTRTGLPE----ETFPNYD--LWENPLTPCMCK 556
>At4g38390 putative growth regulator protein
Length = 551
Score = 262 bits (670), Expect = 3e-70
Identities = 149/399 (37%), Positives = 234/399 (58%), Gaps = 19/399 (4%)
Query: 50 NNTNGYLLVHANGGLNQMKTGISDMVAIAKIMNATLVFPTLDHNSFWTDPSDFKEIFNWK 109
+ TN YLL+ +GGLNQ +TGI D V A I+NATLV P LD S+W D S+F++IF+
Sbjct: 121 SQTNRYLLIATSGGLNQQRTGIIDAVVAAYILNATLVVPKLDQKSYWKDTSNFEDIFDVD 180
Query: 110 NFVEVLNEDVQVVESLPPE----LAAIKPALKAPVSWSKASYYRTDMLQLLKKHKVIKFT 165
F+ L++DV++++ LP E ++ +++ P + + Y + + L KKH V++ +
Sbjct: 181 WFISHLSKDVKIIKELPKEEQSRISTSLQSMRVPRKCTPSCYLQRVLPILTKKH-VVQLS 239
Query: 166 HTDSRLVNNGLASSIQRVRCRAMYEALRFAVPIEELGKKLVNRLRENNTPYIALHLRYEK 225
D RL +N L + +Q++RCR Y A+R+ I +G+ LV+R+R+ ++ALHLR+E
Sbjct: 240 KFDYRL-SNALDTELQKLRCRVNYHAVRYTESINRMGQLLVDRMRKKAKHFVALHLRFEP 298
Query: 226 DMLAFTGCSHNLTKEETQELKKMRYSVKHWKEKEIDSKSK-RLKGSCPMTPREVAVFLEA 284
DMLAF+GC + ++E EL MR + WK + K R G CP+TP E+ + L
Sbjct: 299 DMLAFSGCYYGGGQKERLELGAMR---RRWKTLHAANPEKVRRHGRCPLTPEEIGLMLRG 355
Query: 285 LGYPVDTKIYVAAGVIYGSEG-MKPLQKKFPNLLWHSSLATKEELQPFEGHLNQLAALDY 343
LG+ + +YVA+G +YG E + PL+ FPNL +L +K+EL PF +++AALD+
Sbjct: 356 LGFGKEVHLYVASGEVYGGEDTLAPLRALFPNLHTKETLTSKKELAPFANFSSRMAALDF 415
Query: 344 YITVESDVFVYSYDGNMAKAARGHRKFDGFKKTISPDKQRFVRLIDQLDNGLISWNDFST 403
+ +SD FV + +GNMA+ G R++ G K TI P+ ++ + N ++W +FS+
Sbjct: 416 IVCDKSDAFVTNNNGNMARILAGRRRYLGHKVTIRPNAKKLYEIFKNRHN--MTWGEFSS 473
Query: 404 KVKSIHAKKKGAPQARKIHRHPKFEETFYANPFPGCICQ 442
KV+ G P K E F+ NP CIC+
Sbjct: 474 KVRRYQTGFMGEPDEMK-----PGEGEFHENP-ASCICR 506
>At1g35510 putative growth regulator protein (At1g35510)
Length = 568
Score = 252 bits (644), Expect = 3e-67
Identities = 156/420 (37%), Positives = 236/420 (56%), Gaps = 40/420 (9%)
Query: 49 GNNTNGYLLVHANGGLNQMKTGISDMVAIAKIMNATLVFPTLDHNSFWTDPSDFKEIFNW 108
G+N+NGY ++ ANGGLNQ + I D VA+A ++NATLV P NS W D S F +IF+
Sbjct: 156 GSNSNGYFIIEANGGLNQQRLSICDAVAVAGLLNATLVIPIFHLNSVWRDSSKFGDIFDE 215
Query: 109 KNFVEVLNEDVQVVESLPPEL---------AAIKPALKAPVSWSKASYYRTDMLQLLKKH 159
F+ L+++V VV+ LP ++ + + LKA WS +YY +L L +
Sbjct: 216 DFFIYALSKNVNVVKELPKDVLERYNYNISSIVNLRLKA---WSSPAYYLQKVLPQLLRL 272
Query: 160 KVIKFTHTDSRLVNNGLASSIQRVRCRAMYEALRFAVPIEELGKKLVNRL----RENNTP 215
++ +RL + + + IQ +RC A +EALRFA PI L +K+V+R+ E+
Sbjct: 273 GAVRVAPFSNRLA-HAVPAHIQGLRCLANFEALRFAEPIRLLAEKMVDRMVTKSVESGGK 331
Query: 216 YIALHLRYEKDMLAFTGCSHNLTKEETQELKKMRYSVKHWKEK------EIDSKSKRLKG 269
Y+++HLR+E DM+AF+ C ++ + E E+ R + WK K I + R+ G
Sbjct: 332 YVSVHLRFEMDMVAFSCCEYDFGQAEKLEMDMAR--ERGWKGKFRRRGRVIRPGANRIDG 389
Query: 270 SCPMTPREVAVFLEALGYPVDTKIYVAAGVIYGSEG-MKPLQKKFPNLLWHSSLATKEEL 328
CP+TP EV + L +G+ T +YVAAG IY ++ M PL++ FP L +LAT EEL
Sbjct: 390 KCPLTPLEVGMMLRGMGFNNSTLVYVAAGNIYKADKYMAPLRQMFPLLQTKDTLATPEEL 449
Query: 329 QPFEGHLNQLAALDYYITVESDVFVYSYDGNMAKAARGHRK--FDGFKKTISPDKQRFVR 386
PF+GH ++LAALDY + + S+VFV + GN GHR+ + G +TI PDK++ V+
Sbjct: 450 APFKGHSSRLAALDYTVCLHSEVFVSTQGGNFPHFLIGHRRYLYKGHAETIKPDKRKLVQ 509
Query: 387 LIDQLDNGLISWNDFSTKVKSI--HAKKKGAPQARKIHRHPKFEETFYANPFPGCICQKP 444
L LD I W+ F +++ + H KG K + Y P P C+C++P
Sbjct: 510 L---LDKPSIRWDYFKKQMQDMLRHNDAKGVELR-------KPAASLYTFPMPDCMCKEP 559
>At5g63390 auxin-independent growth promoter-like protein
Length = 559
Score = 244 bits (622), Expect = 9e-65
Identities = 143/396 (36%), Positives = 221/396 (55%), Gaps = 13/396 (3%)
Query: 55 YLLVHANGGLNQMKTGISDMVAIAKIMNATLVFPTLDHNSFWTDPSDFKEIFNWKNFVEV 114
+L+V +GGLNQ + I D V IA I+ A LV P L N W D S+F ++F+ ++F +
Sbjct: 167 FLVVVVSGGLNQQRNQIVDAVVIAMILEAALVVPVLQVNRVWGDESEFSDLFDVEHFKKT 226
Query: 115 LNEDVQVVESLPPELAAIKPALKAPVSWSKASYY-RTDMLQLLKKHKVIKFTHTDSRLVN 173
L DV++V SLP + ++ + W + + R + L + ++ DS+L
Sbjct: 227 LRSDVRIVSSLPSTHLMSRQTIENQIPWDVSPVWIRAKYFKQLNEEGLLVLKGLDSKLAK 286
Query: 174 NGLASSIQRVRCRAMYEALRFAVPIEELGKKLVNRLRENNTPYIALHLRYEKDMLAFTGC 233
N L +Q++RC+ + ALRFA PIE LG KL R+ PYIALHLR EKD+ TGC
Sbjct: 287 N-LPPDLQKLRCKVAFHALRFAAPIENLGNKLTRRMWIEG-PYIALHLRLEKDVWVRTGC 344
Query: 234 SHNLTKEETQELKKMRYSVKHWKEKEIDSK--SKRLKGSCPMTPREVAVFLEALGYPVDT 291
L E + + + R S + ++ +RL G CP+ E+A L+ALG P +
Sbjct: 345 LTGLGSEFDRIIAETRTSQPRYLTGRLNLTYTERRLAGFCPLNVYEIARLLKALGAPSNA 404
Query: 292 KIYVAAGVIYG-SEGMKPLQKKFPNLLWHSSLATKEELQPFEGHLNQLAALDYYITVESD 350
IY+A G +G S ++PL K+F NL+ +LA K EL P+ + LAA+DY +++ SD
Sbjct: 405 SIYIAGGEPFGGSRALEPLAKEFSNLVTKETLAHKGELLPYTNRSSALAAIDYIVSLSSD 464
Query: 351 VFVYSYDGNMAKAARGHRKFDGFKKTISPDKQRFVRLIDQLDNGLISWNDFSTKVKSIHA 410
VF+ S+ GNMAKA +G+R + G +K I P+K+ + L ++N +S + S + +H
Sbjct: 465 VFIPSHGGNMAKAMQGNRAYVGHRKFIMPNKRAMLPL---MENSSVSDAELSFLTRKLHR 521
Query: 411 KKKGAPQARKIHRHPKFEETFYANPFPGCICQKPTH 446
K +G P++R+ R + A P P C+C+ H
Sbjct: 522 KSQGYPESRRGRR----DRDVIAYPVPECMCRHRKH 553
>At1g14970 auxin-independent growth promoter, putative
Length = 458
Score = 243 bits (621), Expect = 1e-64
Identities = 149/413 (36%), Positives = 229/413 (55%), Gaps = 34/413 (8%)
Query: 52 TNGYLLVHANGGLNQMKTGISDMVAIAKIMNATLVFPTLDHNSFWTDPSDFKEIFNWKNF 111
+NG++ + ANGGLNQ +T I + VA+A +NATLV P ++S W DPS F +I++ + F
Sbjct: 51 SNGFIFIEANGGLNQQRTSICNAVAVAGYLNATLVIPNFHYHSIWKDPSKFGDIYDEEYF 110
Query: 112 VEVLNEDVQVVESLPPELAAIKPALKAPV------SWSKASYYRTDMLQLLKKHKVIKFT 165
++ L DV+VV+++P L V +W+ SYYR +L L + KVI+ +
Sbjct: 111 IDTLANDVRVVDTVPEYLMERFDYNLTNVYNFRVKAWAPTSYYRDSVLPKLLEEKVIRIS 170
Query: 166 HTDSRLVNNGLASSIQRVRCRAMYEALRFAVPIEELGKKLVNRLR----ENNTPYIALHL 221
+RL + ++QR RC A ALRF+ PI G+ LVN+++ N Y+++HL
Sbjct: 171 PFANRL-SFDAPRAVQRFRCLANNVALRFSKPILTQGETLVNKMKGLSANNAGKYVSVHL 229
Query: 222 RYEKDMLAFTGCSHNLTKEETQELKKMRYSVKHWKEK------EIDSKSKRLKGSCPMTP 275
R+E+DM+AF+ C + +E Q++ R + WK K I + RL G CP+TP
Sbjct: 230 RFEEDMVAFSCCVFDGGDQEKQDMIAAR--ERGWKGKFTKPGRVIRPGANRLNGKCPLTP 287
Query: 276 REVAVFLEALGYPVDTKIYVAAGVIY-GSEGMKPLQKKFPNLLWHSSLATKEELQPFEGH 334
EV + L +G+ T I++AAG IY + M PL + FPNL LA++E+L PF+
Sbjct: 288 LEVGLMLRGMGFNKSTYIFLAAGPIYSANRTMAPLLEMFPNLQTKEMLASEEDLAPFKNF 347
Query: 335 LNQLAALDYYITVESDVFVYSYDGNMAKAARGHRK--FDGFKKTISPDKQRFVRLIDQLD 392
+++AA+DY + + S+VFV + GN GHR+ F G KTI PDK++ L D
Sbjct: 348 SSRMAAIDYTVCLHSEVFVTTQGGNFPHFLMGHRRYLFGGHSKTIQPDKRKLAVL---FD 404
Query: 393 NGLISWNDFSTKVKSI--HAKKKGAPQARKIHRHPKFEETFYANPFPGCICQK 443
N + W F ++ S+ H+ KG R ++ Y P P C+C+K
Sbjct: 405 NPKLGWKSFKRQMLSMRSHSDSKGFELKRS-------SDSIYIFPCPDCMCRK 450
>At1g04910 Similar to auxin-independent growth promoter (axi 1)
Length = 519
Score = 243 bits (620), Expect = 2e-64
Identities = 151/405 (37%), Positives = 218/405 (53%), Gaps = 31/405 (7%)
Query: 52 TNGYLLVHANGGLNQMKTGISDMVAIAKIMNATLVFPTLDHNSFWTDPSDFKEIFNWKNF 111
TNGYL V NGGLNQ ++ I + V A+IMNATLV P LD NSFW D S F+ I++ ++F
Sbjct: 88 TNGYLRVRCNGGLNQQRSAICNAVLAARIMNATLVLPELDANSFWHDDSGFQGIYDVEHF 147
Query: 112 VEVLNEDVQVVESLPP----------ELAAIKPALKAPVSWSKASYYRTDMLQLLKKHKV 161
+E L DV++V +P + I+P AP+ W Y T L+ +++H
Sbjct: 148 IETLKYDVKIVGKIPDVHKNGKTKKIKAFQIRPPRDAPIEW-----YLTTALKAMREHSA 202
Query: 162 IKFTHTDSRLVNNGLASSIQRVRCRAMYEALRFAVPIEELGKKLVNRLRENNTPYIALHL 221
I T RL QR+RCR Y ALRF I +L + +V++LR ++++HL
Sbjct: 203 IYLTPFSHRLAEEIDNPEYQRLRCRVNYHALRFKPHIMKLSESIVDKLRSQG-HFMSIHL 261
Query: 222 RYEKDMLAFTGCSHNLTKEETQELKKMRYSVKHWKEKEIDSKSKRLKGSCPMTPREVAVF 281
R+E DMLAF GC EE + L+K Y +++ +K + +R G CP+TP EV +
Sbjct: 262 RFEMDMLAFAGCFDIFNPEEQKILRK--YRKENFADKRLIYNERRAIGKCPLTPEEVGLI 319
Query: 282 LEALGYPVDTKIYVAAGVIYGSEG-MKPLQKKFPNLLWHSSLATKEELQPFEGHLNQLAA 340
L A+ + T+IY+AAG ++G E MKP + FP L HSS+ EEL L +A
Sbjct: 320 LRAMRFDNSTRIYLAAGELFGGEQFMKPFRTLFPRLDNHSSVDPSEELSATSQGLIG-SA 378
Query: 341 LDYYITVESDVFVYSYDG--NMAKAARGHRKFDGFKKTISPDKQRFVRLIDQLDNGLISW 398
+DY + + SD+F+ +YDG N A GHR + GF+ TI PD++ + + G +
Sbjct: 379 VDYMVCLLSDIFMPTYDGPSNFANNLLGHRLYYGFRTTIRPDRKALAPIFIAREKGKRA- 437
Query: 399 NDFSTKVKSIHAKKK-GAPQARKIHRHPKFEETFYANPFPGCICQ 442
F V+ + K G P R E+FY N +P C CQ
Sbjct: 438 -GFEEAVRRVMLKTNFGGPHKR------VSPESFYTNSWPECFCQ 475
>At2g01480 similar to axi 1 protein from Nicotiana tabacum
Length = 567
Score = 240 bits (613), Expect = 1e-63
Identities = 148/413 (35%), Positives = 229/413 (54%), Gaps = 34/413 (8%)
Query: 52 TNGYLLVHANGGLNQMKTGISDMVAIAKIMNATLVFPTLDHNSFWTDPSDFKEIFNWKNF 111
+NGY+ V ANGGLNQ +T I + VA+A +NATLV P ++S W DPS F +I++ + F
Sbjct: 154 SNGYIYVEANGGLNQQRTSICNAVAVAGYLNATLVIPNFHYHSIWRDPSKFGDIYDEEFF 213
Query: 112 VEVLNEDVQVVESLPPELAAIKPALKAPV------SWSKASYYRTDMLQLLKKHKVIKFT 165
V L+ DV+VV+++P L V +WS YYR +L L + K+I+ +
Sbjct: 214 VSTLSNDVRVVDTIPEYLMERFDHNMTNVYNFRVKAWSPIQYYRDSILPKLLEEKIIRIS 273
Query: 166 HTDSRLVNNGLASSIQRVRCRAMYEALRFAVPIEELGKKLVNRLRE----NNTPYIALHL 221
+RL + ++QR+RC A YEAL+F+ I LG+ LV R++E + Y+++HL
Sbjct: 274 PFANRL-SFDAPQAVQRLRCLANYEALKFSKTILTLGETLVKRMKEQSANHGAKYVSVHL 332
Query: 222 RYEKDMLAFTGCSHNLTKEETQELKKMRYSVKHWKEK------EIDSKSKRLKGSCPMTP 275
R+E+DM+AF+ C + +E Q++ R + WK K I + R G CP+TP
Sbjct: 333 RFEEDMVAFSCCIFDGGNQEKQDMIAAR--ERGWKGKFTKPGRVIRPGAIRQNGKCPLTP 390
Query: 276 REVAVFLEALGYPVDTKIYVAAGVIY-GSEGMKPLQKKFPNLLWHSSLATKEELQPFEGH 334
EV + L +G+ T I++A+G IY + M PL + FPNL LA++EEL P++
Sbjct: 391 LEVGLMLRGMGFNKSTYIFLASGEIYDANRTMAPLLEMFPNLQTKEMLASEEELAPYKNF 450
Query: 335 LNQLAALDYYITVESDVFVYSYDGNMAKAARGHRK--FDGFKKTISPDKQRFVRLIDQLD 392
+++AA+DY + + S+VFV + GN GHR+ F G KTI PDK++ L D
Sbjct: 451 SSRMAAIDYTVCLHSEVFVTTQGGNFPHFLMGHRRYMFGGHSKTIRPDKRKLAIL---FD 507
Query: 393 NGLISWNDFSTKVKSI--HAKKKGAPQARKIHRHPKFEETFYANPFPGCICQK 443
N I W F ++ ++ H+ KG R ++ Y P P C+ ++
Sbjct: 508 NPNIGWRSFKRQMLNMRSHSDSKGFELKRP-------NDSIYTFPCPDCMSRR 553
>At3g07900 putative auxin-independent growth promoter
Length = 579
Score = 234 bits (598), Expect = 6e-62
Identities = 138/393 (35%), Positives = 214/393 (54%), Gaps = 13/393 (3%)
Query: 55 YLLVHANGGLNQMKTGISDMVAIAKIMNATLVFPTLDHNSFWTDPSDFKEIFNWKNFVEV 114
YL+V +GGLNQ K I D V IA+I+ A LV P L N W D S+F +IF+ + F V
Sbjct: 191 YLMVVVSGGLNQQKIQIVDAVVIARILGAVLVVPILQINLIWGDESEFSDIFDLEQFKSV 250
Query: 115 LNEDVQVVESLPPELAAIKPALKAPVSWSKA-SYYRTDMLQLLKKHKVIKFTHTDSRLVN 173
L DV++V LP +P+ + ++ + + R+ + + V+ DSRL +
Sbjct: 251 LANDVKIVSLLPASKVMTRPSEDGSMPFNASPQWIRSHYPKRFNREGVLLLRRLDSRL-S 309
Query: 174 NGLASSIQRVRCRAMYEALRFAVPIEELGKKLVNRLRENNTPYIALHLRYEKDMLAFTGC 233
L S +Q++RC+ +EAL+F+ + E+G KL R+R PYIALHLR EKD+ TGC
Sbjct: 310 KDLPSDLQKLRCKVAFEALKFSPRVMEMGTKLAERMRSKG-PYIALHLRMEKDVWVRTGC 368
Query: 234 SHNLTK--EETQELKKMRYSVKHWKEKEIDSKSKRLKGSCPMTPREVAVFLEALGYPVDT 291
L+ +E +++++ + + S ++L G CP+ +EV L ALG P D
Sbjct: 369 LSGLSSKYDEIVNIERIKRPELLTAKSSMTSNERKLAGLCPLNAKEVTRLLRALGAPRDA 428
Query: 292 KIYVAAGV-IYGSEGMKPLQKKFPNLLWHSSLATKEELQPFEGHLNQLAALDYYITVESD 350
+IY A G + G E +KPL +FP+L +A EL+PF + +AA+DY + ESD
Sbjct: 429 RIYWAGGEPLGGKEALKPLTSEFPHLYNKYDIALPLELKPFAKRASIMAAIDYIVCKESD 488
Query: 351 VFVYSYDGNMAKAARGHRKFDGFKKTISPDKQRFVRLIDQLDNGLISWNDFSTKVKSIHA 410
VF+ S+ GNM A +GHR ++G KK I+P+K+ ++ N ++ +F +K +H
Sbjct: 489 VFMASHGGNMGHAIQGHRAYEGHKKIITPNKR---HMLPYFVNSSLTETEFEKMIKKLHR 545
Query: 411 KKKGAPQARKIHRHPKFEETFYANPFPGCICQK 443
+ G P+ R K P P C+C +
Sbjct: 546 QSLGQPEL----RISKAGRDVTKYPVPECMCNQ 574
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,575,083
Number of Sequences: 26719
Number of extensions: 462991
Number of successful extensions: 1383
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 32
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 1218
Number of HSP's gapped (non-prelim): 56
length of query: 455
length of database: 11,318,596
effective HSP length: 103
effective length of query: 352
effective length of database: 8,566,539
effective search space: 3015421728
effective search space used: 3015421728
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147429.7


