

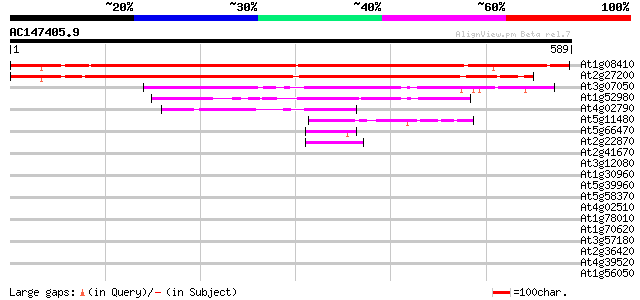
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147405.9 + phase: 0
(589 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g08410 unknown protein 733 0.0
At2g27200 putative nucleotide-binding protein 692 0.0
At3g07050 GTPase like protein 137 2e-32
At1g52980 unknown protein 137 2e-32
At4g02790 unknown protein 65 1e-10
At5g11480 putative GTP-binding protein 45 9e-05
At5g66470 GTP-binding protein-like 44 3e-04
At2g22870 putative nucleotide-binding protein 44 3e-04
At2g41670 unknown protein 38 0.018
At3g12080 unknown protein 37 0.031
At1g30960 F17F8.15 36 0.052
At5g39960 putative protein 36 0.068
At5g58370 contains similarity to GTP-binding protein CGPA 35 0.089
At4g02510 chloroplast protein import component Toc159-like 35 0.15
At1g78010 unknown protein 35 0.15
At1g70620 unknown protein 35 0.15
At3g57180 unknown protein 34 0.26
At2g36420 unknown protein 34 0.26
At4g39520 GTP-binding - like protein 33 0.34
At1g56050 like GTP-binding protein 33 0.34
>At1g08410 unknown protein
Length = 589
Score = 733 bits (1891), Expect = 0.0
Identities = 390/597 (65%), Positives = 453/597 (75%), Gaps = 21/597 (3%)
Query: 1 MGKNQKTELGRALVKQHNQMIQQTKEKGKIYK---KKFLESFTEVSDIDAIIEQADEDVD 57
MGK++KT LGR+LVK HN MIQ++K+KGK YK KK LES TEVSDIDAIIEQA+E
Sbjct: 1 MGKSEKTSLGRSLVKHHNHMIQESKDKGKYYKNLQKKVLESVTEVSDIDAIIEQAEE--- 57
Query: 58 EQQLLDIPLPPPTAL-INLDHASGSDFNGLTVEEMKKEQKIEEALHASSLRVPRRPFWSA 116
++L I T L INLD S S + EE +++QKIEEALHASSL+VPRRP W+
Sbjct: 58 AERLYTINHSSSTPLSINLDTNSSSSV--IAAEEWREQQKIEEALHASSLQVPRRPPWTP 115
Query: 117 EMSADELHANETQHFLTWRRSLARLEENKKLVLTPFEKNLDIWRQLWRVVERSDLLVMVV 176
EMS +EL ANE Q FL WRR L LEEN+KLVLTPFEKNLDIWRQLWRV+ERSDL+VMVV
Sbjct: 116 EMSVEELDANEKQAFLNWRRMLVSLEENEKLVLTPFEKNLDIWRQLWRVLERSDLIVMVV 175
Query: 177 DSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVREKWAEYFRAHDILFIFWSA 236
D+RDPLFYRCPDLEAYA+E+D HK +LLVNKADLLP VREKWAEYFR ++ILF+FWSA
Sbjct: 176 DARDPLFYRCPDLEAYAQEIDEHKKIMLLVNKADLLPTDVREKWAEYFRLNNILFVFWSA 235
Query: 237 KAATAVLEGKKLGSS--QADNMASADNPDTKIYGRDELLARLQSEAEAIVDMRRSSGSSK 294
AATA LEGK L Q DN+ D+PD IYGRDELL+RLQ EA+ IV +R S +S
Sbjct: 236 IAATATLEGKVLKEQWRQPDNLQKTDDPDIMIYGRDELLSRLQFEAQEIVKVRNSRAASV 295
Query: 295 SSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVGQKKTGVTSTPGKTKHFQTLIISEK 354
SS VVGFVGYPNVGKSSTINALVGQK+TGVTSTPGKTKHFQTLIIS++
Sbjct: 296 SSQSWTG-EYQRDQAVVGFVGYPNVGKSSTINALVGQKRTGVTSTPGKTKHFQTLIISDE 354
Query: 355 LILCDCPGLVFPSFSSSRYEMITCGVLPIDRMTQHRECVQVVANRVPRHVIEEIYNISLP 414
L+LCDCPGLVFPSFSSSRYEMI GVLPIDRMT+HRE +QVVA++VPR VIE +YNISLP
Sbjct: 355 LMLCDCPGLVFPSFSSSRYEMIASGVLPIDRMTEHREAIQVVADKVPRRVIESVYNISLP 414
Query: 415 KPKSYESQSRPPLASELLRTYCASRGQTTSSGLPDETRASRQILKDYIDGKLPHYEMPPG 474
KPK+YE QSRPP A+ELL++YCASRG SSGLPDET+A+R ILKDYI GKLPHY MPPG
Sbjct: 415 KPKTYERQSRPPHAAELLKSYCASRGYVASSGLPDETKAARLILKDYIGGKLPHYAMPPG 474
Query: 475 LSTQELASEDSNEHDQVNPHVSDASDIEDSSV---VETELAPKLEHVLDDLSSFDMANGL 531
+ A E E Q + + S+ +DS+V E E P ++ VLDDLSSFD+ANGL
Sbjct: 475 MPQ---ADEPDIEDTQELEDILEGSESDDSAVGDETENEQVPGIDDVLDDLSSFDLANGL 531
Query: 532 -ASNKVAPKKTKESQKHHRKPPRTKNRSWRAGNAGKDDTDGMPIARFHQKPVNSGPL 587
+S KV KK S K H+KP R K+R+WR N +D DGMP + QKP N+GPL
Sbjct: 532 KSSKKVTAKKQTASHKQHKKPQRKKDRTWRVQNT--EDGDGMPSVKVFQKPANTGPL 586
>At2g27200 putative nucleotide-binding protein
Length = 537
Score = 692 bits (1787), Expect = 0.0
Identities = 370/558 (66%), Positives = 427/558 (76%), Gaps = 30/558 (5%)
Query: 1 MGKNQKTELGRALVKQHNQMIQQTKEKGKIYK---KKFLESFTEVSDIDAIIEQADEDVD 57
MGKN+KT LGRALVK HN MIQ+TKEKGK YK KK LES TEVSDIDAIIEQA+E
Sbjct: 1 MGKNEKTSLGRALVKHHNHMIQETKEKGKSYKDQHKKVLESVTEVSDIDAIIEQAEE--- 57
Query: 58 EQQLLDIPLPPPTAL-INLDHASGSDFNGLTVEEMKKEQKIEEALHASSLRVPRRPFWSA 116
++L I T + IN+D +GS +G+T +E K+++ EEALHASSL+VPRRP W+
Sbjct: 58 AERLFAIHHDSATPVPINMD--TGSSSSGITAKEWKEQRMREEALHASSLQVPRRPHWTP 115
Query: 117 EMSADELHANETQHFLTWRRSLARLEENKKLVLTPFEKNLDIWRQLWRVVERSDLLVMVV 176
+M+ ++L ANE Q FLTWRR LA LEEN+KLVLTPFEKNLDIWRQLWRV+ERSDL+VMVV
Sbjct: 116 KMNVEKLDANEKQAFLTWRRKLASLEENEKLVLTPFEKNLDIWRQLWRVLERSDLIVMVV 175
Query: 177 DSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVREKWAEYFRAHDILFIFWSA 236
D+RDPLFYRCPDLEAYA+E+D HK T+LLVNKADLLP+ VREKWAEYF ++ILF+FWSA
Sbjct: 176 DARDPLFYRCPDLEAYAQEIDEHKKTMLLVNKADLLPSYVREKWAEYFSRNNILFVFWSA 235
Query: 237 KAATAVLEGKKLGSS--QADNMASADNPDTKIYGRDELLARLQSEAEAIVDMRRSSGSSK 294
KAATA LEGK L D DNP K+YGRD+LL RL+ EA IV MR+S G S
Sbjct: 236 KAATATLEGKPLKEQWRAPDTTQKTDNPAVKVYGRDDLLDRLKLEALEIVKMRKSRGVSA 295
Query: 295 SSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVGQKKTGVTSTPGKTKHFQTLIISEK 354
+S + S VVVGFVGYPNVGKSSTINALVGQK+TGVTSTPGKTKHFQTLIISE
Sbjct: 296 TSTE-----SHCEQVVVGFVGYPNVGKSSTINALVGQKRTGVTSTPGKTKHFQTLIISED 350
Query: 355 LILCDCPGLVFPSFSSSRYEMITCGVLPIDRMTQHRECVQVVANRVPRHVIEEIYNISLP 414
L+LCDCPGLVFPSFSSSRYEM+ GVLPIDRMT+H E ++VVA VPRH IE++YNISLP
Sbjct: 351 LMLCDCPGLVFPSFSSSRYEMVASGVLPIDRMTEHLEAIKVVAELVPRHAIEDVYNISLP 410
Query: 415 KPKSYESQSRPPLASELLRTYCASRGQTTSSGLPDETRASRQILKDYIDGKLPHYEMPPG 474
KPKSYE QSRPPLASELLRTYC SRG SSGLPDETRA+RQILKDYI+GKLPH+ MPP
Sbjct: 411 KPKSYEPQSRPPLASELLRTYCLSRGYVASSGLPDETRAARQILKDYIEGKLPHFAMPP- 469
Query: 475 LSTQELASEDSNE--HDQVNPHVSDASDIEDSSVVETELAPKLEHVLDDLSSFDMANGLA 532
E+ +D NE D + + S E L L+ VLDDLSSFD+ANGL
Sbjct: 470 ----EITRDDENETADDTLGAETREGSQTEKKGEEAPSLG--LDQVLDDLSSFDLANGLV 523
Query: 533 SNKVAPKKTKESQKHHRK 550
S+ KTK+ +K HRK
Sbjct: 524 SS-----KTKQHKKSHRK 536
>At3g07050 GTPase like protein
Length = 582
Score = 137 bits (344), Expect = 2e-32
Identities = 127/475 (26%), Positives = 200/475 (41%), Gaps = 80/475 (16%)
Query: 141 LEENKKLVLTPFEKNLDIWRQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHK 200
+E+ K+V +++L +V+E SD+++ V+D+RDPL RC D+E + +K
Sbjct: 109 IEDLPKVVNVRDNSERAFYKELVKVIELSDVILEVLDARDPLGTRCTDMERMVMQAGPNK 168
Query: 201 NTLLLVNKADLLPASVREKWAEYFRAHDILFIF-WSAKAATAVLEGKKLGSSQADNMASA 259
+ +LL+NK DL+P EKW Y R F S + + L K +S+ NM
Sbjct: 169 HLVLLLNKIDLVPREAAEKWLMYLREEFPAVAFKCSTQEQRSNLGWKSSKASKPSNMLQT 228
Query: 260 DNPDTKIYGRDELLARLQSEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNV 319
+ G D L+ L++ + RS KS + VG +G PNV
Sbjct: 229 SD----CLGADTLIKLLKNYS-------RSHELKKS-------------ITVGIIGLPNV 264
Query: 320 GKSSTINALVGQKKTGVTSTPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCG 379
GKSS IN+L V +TPG T+ Q + + + + L DCPG+V S + +
Sbjct: 265 GKSSLINSLKRAHVVNVGATPGLTRSLQEVHLDKNVKLLDCPGVVMLKSSGNDASIALRN 324
Query: 380 VLPIDRMTQHRECVQVVANRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASR 439
I+++ V+ + P+ ++ +Y I S+E+ + L R
Sbjct: 325 CKRIEKLDDPVSPVKEILKLCPKDMLVTLYKI-----PSFEA------VDDFLYKVATVR 373
Query: 440 GQTTSSGLPDETRASRQILKDYIDGKLPHYEMPP------------------GLSTQELA 481
G+ GL D A+R +L D+ +GK+P+Y MPP + E+
Sbjct: 374 GKLKKGGLVDIDAAARIVLHDWNEGKIPYYTMPPKRDQGGHAESKIVTELAKDFNIDEVY 433
Query: 482 SEDS---------NEHDQV-----NPHVSDASDIEDSSVVETELAPKLEHVLDDLSSFDM 527
S +S NE + V P D + IED S +TE + EH DD S
Sbjct: 434 SGESSFIGSLKTVNEFNPVIIPSNGPLNFDETMIEDESKTQTE--EEAEHESDDDESMGG 491
Query: 528 ANGLASNKVAPKK----------TKESQKHHRKPPRTKNRSWRAGNAGKDDTDGM 572
+ K K ES + +K K + +A AG D+ D M
Sbjct: 492 EEEEEAGKTKEKSETGRQNVKLYAAESMLNTKKQKAEKKKRKKAKKAGADEEDLM 546
>At1g52980 unknown protein
Length = 576
Score = 137 bits (344), Expect = 2e-32
Identities = 101/336 (30%), Positives = 156/336 (46%), Gaps = 60/336 (17%)
Query: 150 TPFEKNLD--IWRQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVN 207
T FEK IW +L++V++ SD++V V+D+RDP RC LE KE HK+ +LL+N
Sbjct: 195 TMFEKGQSKRIWGELYKVIDSSDVIVQVIDARDPQGTRCHHLEKTLKEHHKHKHMILLLN 254
Query: 208 KADLLPASVREKWAEYFRAHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIY 267
K DL+PA W+ K VL S + +A + + K +
Sbjct: 255 KCDLVPA-------------------WATKGWLRVL------SKEYPTLAFHASVN-KSF 288
Query: 268 GRDELLARLQSEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINA 327
G+ LL+ L+ A + S + VGFVGYPNVGKSS IN
Sbjct: 289 GKGSLLSVLRQFAR--------------------LKSDKQAISVGFVGYPNVGKSSVINT 328
Query: 328 LVGQKKTGVTSTPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLPIDRMT 387
L + V PG+TK +Q + +++++ L DCPG+V+ S + +++ GV+ + +
Sbjct: 329 LRTKNVCKVAPIPGETKVWQYITLTKRIFLIDCPGVVYQS-RDTETDIVLKGVVRVTNLE 387
Query: 388 QHRECVQVVANRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRGQTTSSGL 447
E + V RV + ++ Y I K +E + L C S G+ G
Sbjct: 388 DASEHIGEVLRRVKKEHLQRAYKI-----KDWEDD------HDFLLQLCKSSGKLLKGGE 436
Query: 448 PDETRASRQILKDYIDGKLPHYEMPPGLSTQELASE 483
PD ++ IL D+ G++P + PP L SE
Sbjct: 437 PDLMTGAKMILHDWQRGRIPFFVPPPKLDNVASESE 472
>At4g02790 unknown protein
Length = 372
Score = 65.1 bits (157), Expect = 1e-10
Identities = 51/205 (24%), Positives = 86/205 (41%), Gaps = 43/205 (20%)
Query: 160 RQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVREK 219
++L ++ D+++ V D+R PL P ++A+ ++ +L++N+ D++ R
Sbjct: 108 KELREQLKLMDVVIEVRDARIPLSTTHPKMDAWLG----NRKRILVLNREDMISNDDRND 163
Query: 220 WAEYFRAHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDELLARLQSE 279
WA YF I IF + K ++ +L S A ++
Sbjct: 164 WARYFAKQGIKVIFTNGKLGMGAMKLGRLAKSLAGDVNGK-------------------- 203
Query: 280 AEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVGQKKTGVTST 339
RR G S V G +GYPNVGKSS IN L+ +K
Sbjct: 204 -------RREKGLLPRS------------VRAGIIGYPNVGKSSLINRLLKRKICAAAPR 244
Query: 340 PGKTKHFQTLIISEKLILCDCPGLV 364
PG T+ + + + + L L D PG++
Sbjct: 245 PGVTREMKWVKLGKDLDLLDSPGML 269
>At5g11480 putative GTP-binding protein
Length = 318
Score = 45.4 bits (106), Expect = 9e-05
Identities = 49/181 (27%), Positives = 84/181 (46%), Gaps = 25/181 (13%)
Query: 314 VGYPNVGKSSTINALVGQKKTGVTS-TPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSR 372
VG NVGKSS +N+LV +K+ +TS PGKT+ I++K L D PG ++S+
Sbjct: 141 VGRSNVGKSSLLNSLVRRKRLALTSKKPGKTQCINHFRINDKWYLVDLPGY---GYASAP 197
Query: 373 YEMITCGVLPIDRMTQHRECVQVVANRVPRHVIEEIYNISLP-KP-----KSYESQSRPP 426
+E+ + ++ NR + + + S+P KP S+ Q++ P
Sbjct: 198 HEL---------KQDWNKFTKDYFLNRSTLVSVFLLVDASIPVKPIDLEYASWLGQNQVP 248
Query: 427 LASELLRTYCASRGQTTSSGLPDETRASRQILKDYIDGKLPHYEMPPGLSTQELASEDSN 486
+ L+ T C R + + G E A+ + +D I G PP + T + ++ +
Sbjct: 249 MT--LIFTKCDKRKKKKNGGKKPE--ANIKEFQDLIQGFFE--TTPPWIMTSSVTNQGRD 302
Query: 487 E 487
E
Sbjct: 303 E 303
>At5g66470 GTP-binding protein-like
Length = 427
Score = 43.9 bits (102), Expect = 3e-04
Identities = 25/57 (43%), Positives = 34/57 (58%), Gaps = 3/57 (5%)
Query: 311 VGFVGYPNVGKSSTINALVGQKKTGVTSTPGKTKHFQTLIISE---KLILCDCPGLV 364
V VG PNVGKS+ N ++GQK + VT P T+H I S ++IL D PG++
Sbjct: 132 VAVVGMPNVGKSTLSNQMIGQKISIVTDKPQTTRHRILGICSSPEYQMILYDTPGVI 188
>At2g22870 putative nucleotide-binding protein
Length = 300
Score = 43.9 bits (102), Expect = 3e-04
Identities = 23/62 (37%), Positives = 35/62 (56%), Gaps = 1/62 (1%)
Query: 311 VGFVGYPNVGKSSTINALVGQKKTGVTS-TPGKTKHFQTLIISEKLILCDCPGLVFPSFS 369
+ +G NVGKSS IN LV +K+ +TS PGKT+ ++++ + D PG F S
Sbjct: 123 IAILGRSNVGKSSLINCLVRKKEVALTSKKPGKTQLINHFLVNKSWYIVDLPGYGFAKVS 182
Query: 370 SS 371
+
Sbjct: 183 DA 184
>At2g41670 unknown protein
Length = 386
Score = 37.7 bits (86), Expect = 0.018
Identities = 49/212 (23%), Positives = 82/212 (38%), Gaps = 63/212 (29%)
Query: 169 SDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVREKWAEYFRAHD 228
SDL++ V D+R PL DL++ K ++ +NK DL +V KW +F
Sbjct: 47 SDLVIEVRDARIPLSSANEDLQSQMSA----KRRIIALNKKDLANPNVLNKWTRHFE--- 99
Query: 229 ILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDELLARLQSEAEAIVDMRR 288
SS+ D +A + R ++ L +V+++
Sbjct: 100 ---------------------SSKQDCIA------INAHSRSSVMKLLD-----LVELKL 127
Query: 289 SSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINAL-----------VGQKKTGVT 337
V + ++V VG PNVGKS+ IN++ K+ V
Sbjct: 128 K-----------EVIAREPTLLVMVVGVPNVGKSALINSIHQIAAARFPVQERLKRATVG 176
Query: 338 STPGKTKHFQTLIISEK--LILCDCPGLVFPS 367
PG T+ I+ + + + D PG++ PS
Sbjct: 177 PLPGVTQDIAGFKIAHRPSIYVLDSPGVLVPS 208
>At3g12080 unknown protein
Length = 663
Score = 37.0 bits (84), Expect = 0.031
Identities = 17/34 (50%), Positives = 21/34 (61%)
Query: 311 VGFVGYPNVGKSSTINALVGQKKTGVTSTPGKTK 344
V VG PNVGKS+ N LVG+ + V PG T+
Sbjct: 161 VAIVGRPNVGKSALFNRLVGENRAIVVDEPGVTR 194
Score = 36.2 bits (82), Expect = 0.052
Identities = 16/34 (47%), Positives = 23/34 (67%)
Query: 311 VGFVGYPNVGKSSTINALVGQKKTGVTSTPGKTK 344
+ +G PNVGKSS +NALV + +T V+ G T+
Sbjct: 371 IAIIGRPNVGKSSILNALVREDRTIVSPVSGTTR 404
>At1g30960 F17F8.15
Length = 437
Score = 36.2 bits (82), Expect = 0.052
Identities = 30/115 (26%), Positives = 49/115 (42%), Gaps = 12/115 (10%)
Query: 253 ADNMASADNPDTKIYGRDELLARLQSEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVG 312
A+ +S D+ D+ R +LA+ EA +D S++ VG
Sbjct: 107 AEEESSEDDEDS--VDRSRILAKALLEAALESPDEELGEGEVREEDQKSLN-------VG 157
Query: 313 FVGYPNVGKSSTINALVGQKKTGVTSTPGKTKHFQTLIISE---KLILCDCPGLV 364
+G PN GKSS N +VG K + T H ++++ ++ D PGL+
Sbjct: 158 IIGPPNAGKSSLTNFMVGTKVAAASRKTNTTTHEVLGVLTKGDTQVCFFDTPGLM 212
>At5g39960 putative protein
Length = 614
Score = 35.8 bits (81), Expect = 0.068
Identities = 21/63 (33%), Positives = 34/63 (53%), Gaps = 2/63 (3%)
Query: 284 VDMRRSSGSSKSSDDNAS--VSSSSSHVVVGFVGYPNVGKSSTINALVGQKKTGVTSTPG 341
V+M S +D+N S + S + + VG PNVGKS+ +NAL+ +++ V G
Sbjct: 284 VEMLNGSQDDVLTDENLSDEIDESKLPLQLAIVGKPNVGKSTLLNALLEEERVLVGPEAG 343
Query: 342 KTK 344
T+
Sbjct: 344 LTR 346
>At5g58370 contains similarity to GTP-binding protein CGPA
Length = 465
Score = 35.4 bits (80), Expect = 0.089
Identities = 23/56 (41%), Positives = 28/56 (49%), Gaps = 1/56 (1%)
Query: 311 VGFVGYPNVGKSSTINALVGQKKTGVTS-TPGKTKHFQTLIISEKLILCDCPGLVF 365
+ F G NVGKSS +NAL Q TS PG T+ + K+ L D PG F
Sbjct: 291 IAFAGRSNVGKSSLLNALTRQWGVVRTSDKPGLTQTINFFGLGPKVRLVDLPGYGF 346
>At4g02510 chloroplast protein import component Toc159-like
Length = 1503
Score = 34.7 bits (78), Expect = 0.15
Identities = 29/121 (23%), Positives = 56/121 (45%), Gaps = 12/121 (9%)
Query: 246 KKLGSSQADNMASADNPDTKIYGRDELLARLQSEAEAIVDMRRSSGSSKSSDDNASVSSS 305
++LG S D++A+ ++ R LLA Q+ +D + ++ N + S
Sbjct: 802 QRLGHSAEDSIAA------QVLYRLALLAGRQAGQLFSLDAAKKKAVESEAEGNEELIFS 855
Query: 306 SSHVVVGFVGYPNVGKSSTINALVGQKKTGVTS---TPGKTKHFQTLIISEKLILCDCPG 362
+ +V+G G VGKS+TIN+++G + + + + + + K+ D PG
Sbjct: 856 LNILVLGKAG---VGKSATINSILGNQIASIDAFGLSTTSVREISGTVNGVKITFIDTPG 912
Query: 363 L 363
L
Sbjct: 913 L 913
>At1g78010 unknown protein
Length = 613
Score = 34.7 bits (78), Expect = 0.15
Identities = 35/119 (29%), Positives = 55/119 (45%), Gaps = 13/119 (10%)
Query: 235 SAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDELLARLQSEAEAI---VDMRRSSG 291
S+ AA A LEG + G S A + E+ ARL E E ++ +
Sbjct: 275 SSAAADAALEGIQGGFSSLVKSLRAQCIELLT----EIEARLDFEDEMPPLDIESVINKI 330
Query: 292 SSKSSDDNASVSSSS------SHVVVGFVGYPNVGKSSTINALVGQKKTGVTSTPGKTK 344
+S S D +++ +++ S + + VG PNVGKSS +NA ++ VT G T+
Sbjct: 331 TSMSQDVESALDTANYDKLLQSGLQIAIVGRPNVGKSSLLNAWSKSERAIVTEVAGTTR 389
>At1g70620 unknown protein
Length = 897
Score = 34.7 bits (78), Expect = 0.15
Identities = 35/139 (25%), Positives = 56/139 (40%), Gaps = 13/139 (9%)
Query: 442 TTSSGLPDETRASRQILKDYIDGKLPHYEMPPGLSTQELASEDSNEHDQVNPHVSDASD- 500
+ SSGL D+T SR+ D D P + + S+ + D P+ D SD
Sbjct: 629 SASSGLDDDTSGSRKEHPDRTDSDKDAILDEPHVKNSGVKSDCNLRQDSNKPYGKDLSDE 688
Query: 501 --IEDSSVVET---------ELAPKLEHVLDDLSSFDMANGLASNKVA-PKKTKESQKHH 548
+ S +VET + K +DL S + G+ SNK + K+ +
Sbjct: 689 VSTDRSRIVETKGGKEKGDSQNDSKDRMKENDLKSAEKVKGVESNKKSTDPHVKKDSRDV 748
Query: 549 RKPPRTKNRSWRAGNAGKD 567
+P RT ++ R K+
Sbjct: 749 ERPHRTNSKEDRGKRKEKE 767
>At3g57180 unknown protein
Length = 637
Score = 33.9 bits (76), Expect = 0.26
Identities = 27/69 (39%), Positives = 34/69 (49%), Gaps = 7/69 (10%)
Query: 311 VGFVGYPNVGKSSTINALV---GQKKTGVTS--TPGKTKHFQTL--IISEKLILCDCPGL 363
V +G N GKS+ INAL G K T +T PG T + I+S K + D PGL
Sbjct: 372 VWVIGAQNAGKSTLINALSKKDGAKVTRLTEAPVPGTTLGILKIGGILSAKAKMYDTPGL 431
Query: 364 VFPSFSSSR 372
+ P S R
Sbjct: 432 LHPYLMSLR 440
>At2g36420 unknown protein
Length = 439
Score = 33.9 bits (76), Expect = 0.26
Identities = 28/121 (23%), Positives = 53/121 (43%), Gaps = 16/121 (13%)
Query: 452 RASRQILKDYIDGKLPHYEMPPGLSTQELASEDSNEHDQVNPHVSDASDIEDSSVVETEL 511
RA R++LK KL +E GL EL + S E D+ ++ + ++ + +++
Sbjct: 288 RAKRRLLK-----KLRRFEKLAGLDPVELEGKMSEEEDEEEEEYEESEEDDNIRIYDSD- 341
Query: 512 APKLEHVLDDLSSFDMANGLASNKVAPKKTKESQKHHRKPPRTKNRSWRAGNAGKDDTDG 571
++ D A S +K K++ + +K R N +WR G ++D D
Sbjct: 342 --------EEYEDVDEAMARESRCAEDEKRKKNDERQKKW-RMMN-AWRVGLGAEEDVDA 391
Query: 572 M 572
+
Sbjct: 392 V 392
>At4g39520 GTP-binding - like protein
Length = 369
Score = 33.5 bits (75), Expect = 0.34
Identities = 29/99 (29%), Positives = 44/99 (44%), Gaps = 17/99 (17%)
Query: 273 LARLQSEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVG-- 330
LA+L+ + A G+ + D S S VG VG+P+VGKS+ +N L G
Sbjct: 35 LAKLRRDLLAPPTKGGGGGAGEGFDVTKSGDSR-----VGLVGFPSVGKSTLLNKLTGTF 89
Query: 331 -----QKKTGVTSTPGKTKHFQTLIISEKLILCDCPGLV 364
+ T +T PG + K+ L D PG++
Sbjct: 90 SEVASYEFTTLTCIPGVITY-----RGAKIQLLDLPGII 123
>At1g56050 like GTP-binding protein
Length = 421
Score = 33.5 bits (75), Expect = 0.34
Identities = 23/79 (29%), Positives = 36/79 (45%), Gaps = 10/79 (12%)
Query: 262 PDTKIYGRDELLARLQSEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGK 321
P + R++LL S+ ++ +++ SSK S + G VG PNVGK
Sbjct: 17 PSKSLLCRNQLLFSGNSKFVGVLTLQKRCFSSKVS----------MSLKAGIVGLPNVGK 66
Query: 322 SSTINALVGQKKTGVTSTP 340
S+ NA+V K + P
Sbjct: 67 STLFNAVVENGKAQAANFP 85
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.130 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,504,153
Number of Sequences: 26719
Number of extensions: 584797
Number of successful extensions: 1823
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 35
Number of HSP's that attempted gapping in prelim test: 1765
Number of HSP's gapped (non-prelim): 74
length of query: 589
length of database: 11,318,596
effective HSP length: 105
effective length of query: 484
effective length of database: 8,513,101
effective search space: 4120340884
effective search space used: 4120340884
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC147405.9


