

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146865.2 - phase: 0
(107 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
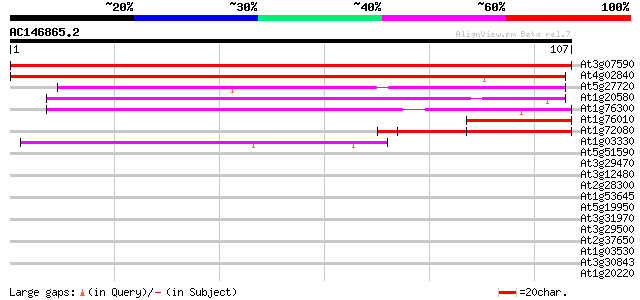
At3g07590 putative small nuclear ribonucleoprotein (Sm-D1) 204 8e-54
At4g02840 small nuclear riboprotein Sm-D1-like protein 203 1e-53
At5g27720 glycine rich protein - like 60 2e-10
At1g20580 unknown protein 60 2e-10
At1g76300 putative small nuclear ribonucleoprotein Sm D3 59 4e-10
At1g76010 unknown protein 42 5e-05
At1g72080 hypothetical protein 40 2e-04
At1g03330 unknown protein 39 6e-04
At5g51590 unknown protein 37 0.002
At3g29470 hypothetical protein 37 0.002
At3g12480 unknown protein 35 0.005
At2g28300 unknown protein 35 0.005
At1g53645 unknown protein 35 0.008
At5g19950 unknown protein 34 0.011
At3g31970 hypothetical protein 34 0.011
At3g29500 hypothetical protein 34 0.014
At2g37650 scarecrow-like 9 (SCL9) 34 0.014
At1g03530 unknown protein 34 0.014
At3g30843 hypothetical protein 33 0.024
At1g20220 unknown protein 33 0.024
>At3g07590 putative small nuclear ribonucleoprotein (Sm-D1)
Length = 114
Score = 204 bits (518), Expect = 8e-54
Identities = 97/107 (90%), Positives = 104/107 (96%)
Query: 1 MKLNNETVSIELKNGTIVHGTITGVDISMNTHLKTVKLTLKGKNPVTLDHLSVRGNNIRY 60
MKLNNETVSIELKNGT+VHGTITGVD+SMNTHLKTVK++LKGKNPVTLDHLS+RGNNIRY
Sbjct: 8 MKLNNETVSIELKNGTVVHGTITGVDVSMNTHLKTVKMSLKGKNPVTLDHLSLRGNNIRY 67
Query: 61 YILPDSLNLETLLVEEAPRVKPKKPTAGKPLGRGRGRGRGRGRGRGR 107
YILPDSLNLETLLVE+ PRVKPKKP AGK +GRGRGRGRGRGRGRGR
Sbjct: 68 YILPDSLNLETLLVEDTPRVKPKKPVAGKAVGRGRGRGRGRGRGRGR 114
>At4g02840 small nuclear riboprotein Sm-D1-like protein
Length = 116
Score = 203 bits (516), Expect = 1e-53
Identities = 100/107 (93%), Positives = 103/107 (95%), Gaps = 1/107 (0%)
Query: 1 MKLNNETVSIELKNGTIVHGTITGVDISMNTHLKTVKLTLKGKNPVTLDHLSVRGNNIRY 60
MKLNNETVSIELKNGTIVHGTITGVD+SMNTHLK VKLTLKGKNPVTLDHLSVRGNNIRY
Sbjct: 8 MKLNNETVSIELKNGTIVHGTITGVDVSMNTHLKAVKLTLKGKNPVTLDHLSVRGNNIRY 67
Query: 61 YILPDSLNLETLLVEEAPRVKPKKPTAGK-PLGRGRGRGRGRGRGRG 106
YILPDSLNLETLLVE+ PR+KPKKPTAGK P GRGRGRGRGRGRGRG
Sbjct: 68 YILPDSLNLETLLVEDTPRIKPKKPTAGKIPAGRGRGRGRGRGRGRG 114
>At5g27720 glycine rich protein - like
Length = 129
Score = 59.7 bits (143), Expect = 2e-10
Identities = 38/98 (38%), Positives = 50/98 (50%), Gaps = 3/98 (3%)
Query: 10 IELKNGTIVHGTITGVDISMNTHLKTVKLTLK-GKNPVTLDHLSVRGNNIRYYILPDSLN 68
+ELKNG +G + D MN HL+ V T K G + +RGN I+Y +PD +
Sbjct: 17 VELKNGETYNGHLVNCDTWMNIHLREVICTSKDGDRFWRMPECYIRGNTIKYLRVPDEVI 76
Query: 69 LETLLVEEAPRVKPKKPTAGKPLGRGRGRGRGRGRGRG 106
+ + EE R K P G+ GRG G RGRGRG
Sbjct: 77 DK--VQEEKTRTDRKPPGVGRGRGRGVDDGGARGRGRG 112
>At1g20580 unknown protein
Length = 131
Score = 59.7 bits (143), Expect = 2e-10
Identities = 33/102 (32%), Positives = 56/102 (54%), Gaps = 5/102 (4%)
Query: 8 VSIELKNGTIVHGTITGVDISMNTHLKTVKLTLKGKNPVTLDHLSVRGNNIRYYILPDSL 67
V++ELK+G + G++ + + N L+ + T K L+H+ +RG+ +R+ ++PD L
Sbjct: 20 VTVELKSGELYRGSMIECEDNWNCQLEDITYTAKDGKVSQLEHVFIRGSKVRFMVIPDIL 79
Query: 68 NLETLLVEEAPRVKPKKPTAGKPLGRGRGRGRGR---GRGRG 106
+ R+K K + G +GRGRG RG+ G GRG
Sbjct: 80 KHAPMFKRLDARIKGKSSSLG--VGRGRGAMRGKPAAGPGRG 119
>At1g76300 putative small nuclear ribonucleoprotein Sm D3
Length = 128
Score = 58.9 bits (141), Expect = 4e-10
Identities = 33/105 (31%), Positives = 57/105 (53%), Gaps = 9/105 (8%)
Query: 8 VSIELKNGTIVHGTITGVDISMNTHLKTVKLTLKGKNPVTLDHLSVRGNNIRYYILPDSL 67
VS+E+K+G + G++ + + N L+ + T K L+H+ +RG+ +R+ ++PD L
Sbjct: 20 VSVEMKSGELYRGSMIECEDNWNCQLENITYTAKDGKVSQLEHVFIRGSLVRFLVIPDML 79
Query: 68 NLETLLVEEAPRVKPKKPTAGKPLGRGRG-----RGRGRGRGRGR 107
+ + V+ K +A +GRGRG +G GRG G GR
Sbjct: 80 KNAPMFKD----VRGKGKSASLGVGRGRGAAMRAKGTGRGTGGGR 120
>At1g76010 unknown protein
Length = 350
Score = 42.0 bits (97), Expect = 5e-05
Identities = 18/20 (90%), Positives = 18/20 (90%)
Query: 88 GKPLGRGRGRGRGRGRGRGR 107
G P GRGRGRGRGRGRGRGR
Sbjct: 146 GSPGGRGRGRGRGRGRGRGR 165
Score = 36.6 bits (83), Expect = 0.002
Identities = 15/15 (100%), Positives = 15/15 (100%)
Query: 92 GRGRGRGRGRGRGRG 106
GRGRGRGRGRGRGRG
Sbjct: 152 GRGRGRGRGRGRGRG 166
Score = 33.9 bits (76), Expect = 0.014
Identities = 14/17 (82%), Positives = 15/17 (87%)
Query: 88 GKPLGRGRGRGRGRGRG 104
G+ GRGRGRGRGRGRG
Sbjct: 150 GRGRGRGRGRGRGRGRG 166
Score = 33.9 bits (76), Expect = 0.014
Identities = 16/18 (88%), Positives = 16/18 (88%), Gaps = 1/18 (5%)
Query: 90 PLGRGRGRGRGR-GRGRG 106
P GRGRGRGRGR GRGRG
Sbjct: 311 PQGRGRGRGRGRGGRGRG 328
Score = 30.8 bits (68), Expect = 0.12
Identities = 16/27 (59%), Positives = 18/27 (66%), Gaps = 1/27 (3%)
Query: 82 PKKPTAGKPLGRGRGRGRGRGR-GRGR 107
P + G +GRGRGRGRGR GRGR
Sbjct: 301 PSQGRGGYDGPQGRGRGRGRGRGGRGR 327
Score = 26.9 bits (58), Expect = 1.7
Identities = 17/43 (39%), Positives = 18/43 (41%), Gaps = 23/43 (53%)
Query: 88 GKPLGRGRGRG-----------------------RGRGRGRGR 107
G+ GRGRGRG RGRGRGRGR
Sbjct: 156 GRGRGRGRGRGGRGNAYVNVEHEDGGWEREQSYGRGRGRGRGR 198
>At1g72080 hypothetical protein
Length = 243
Score = 40.0 bits (92), Expect = 2e-04
Identities = 17/20 (85%), Positives = 18/20 (90%)
Query: 88 GKPLGRGRGRGRGRGRGRGR 107
G+ GRGRGRGRGRGRGRGR
Sbjct: 122 GRGRGRGRGRGRGRGRGRGR 141
Score = 40.0 bits (92), Expect = 2e-04
Identities = 17/20 (85%), Positives = 18/20 (90%)
Query: 88 GKPLGRGRGRGRGRGRGRGR 107
G+ GRGRGRGRGRGRGRGR
Sbjct: 124 GRGRGRGRGRGRGRGRGRGR 143
Score = 40.0 bits (92), Expect = 2e-04
Identities = 17/20 (85%), Positives = 18/20 (90%)
Query: 88 GKPLGRGRGRGRGRGRGRGR 107
G+ GRGRGRGRGRGRGRGR
Sbjct: 126 GRGRGRGRGRGRGRGRGRGR 145
Score = 39.7 bits (91), Expect = 3e-04
Identities = 19/37 (51%), Positives = 23/37 (61%)
Query: 71 TLLVEEAPRVKPKKPTAGKPLGRGRGRGRGRGRGRGR 107
T++ E R + + GRGRGRGRGRGRGRGR
Sbjct: 103 TIINTEERRAGDDDGSRSRGRGRGRGRGRGRGRGRGR 139
Score = 38.1 bits (87), Expect = 7e-04
Identities = 17/33 (51%), Positives = 22/33 (66%)
Query: 75 EEAPRVKPKKPTAGKPLGRGRGRGRGRGRGRGR 107
++ R + + G+ GRGRGRGRGRGRGR R
Sbjct: 115 DDGSRSRGRGRGRGRGRGRGRGRGRGRGRGRNR 147
Score = 35.4 bits (80), Expect = 0.005
Identities = 15/20 (75%), Positives = 16/20 (80%)
Query: 88 GKPLGRGRGRGRGRGRGRGR 107
G+ GRGRGRGRGRGR R R
Sbjct: 130 GRGRGRGRGRGRGRGRNRSR 149
Score = 33.1 bits (74), Expect = 0.024
Identities = 14/20 (70%), Positives = 15/20 (75%)
Query: 88 GKPLGRGRGRGRGRGRGRGR 107
G+ GRGRGRGRGR R R R
Sbjct: 132 GRGRGRGRGRGRGRNRSRSR 151
Score = 27.7 bits (60), Expect = 0.99
Identities = 12/18 (66%), Positives = 12/18 (66%)
Query: 90 PLGRGRGRGRGRGRGRGR 107
P GRGRGRGR R R R
Sbjct: 55 PSNGGRGRGRGRSRRRSR 72
>At1g03330 unknown protein
Length = 93
Score = 38.5 bits (88), Expect = 6e-04
Identities = 22/73 (30%), Positives = 43/73 (58%), Gaps = 3/73 (4%)
Query: 3 LNNETVSIELKNGTIVHGTITGVDISMNTHLKTVKLTLKGKNP--VTLDHLSVRGNNIRY 60
L + V++ELKN + GT+ VD +N L+ ++ + K P +++ + +RG+ +RY
Sbjct: 10 LVGQEVTVELKNDLAIRGTLHSVDQYLNIKLENTRVVDQDKYPHMLSVRNCFIRGSVVRY 69
Query: 61 YILP-DSLNLETL 72
LP D ++++ L
Sbjct: 70 VQLPKDGVDVDLL 82
>At5g51590 unknown protein
Length = 419
Score = 37.0 bits (84), Expect = 0.002
Identities = 15/16 (93%), Positives = 16/16 (99%)
Query: 92 GRGRGRGRGRGRGRGR 107
GRGRGRGRGRGRGRG+
Sbjct: 118 GRGRGRGRGRGRGRGQ 133
Score = 36.2 bits (82), Expect = 0.003
Identities = 15/15 (100%), Positives = 15/15 (100%)
Query: 93 RGRGRGRGRGRGRGR 107
RGRGRGRGRGRGRGR
Sbjct: 117 RGRGRGRGRGRGRGR 131
Score = 35.4 bits (80), Expect = 0.005
Identities = 14/16 (87%), Positives = 16/16 (99%)
Query: 92 GRGRGRGRGRGRGRGR 107
GRGRGRGRGRGRG+G+
Sbjct: 120 GRGRGRGRGRGRGQGQ 135
Score = 35.0 bits (79), Expect = 0.006
Identities = 14/19 (73%), Positives = 17/19 (88%)
Query: 88 GKPLGRGRGRGRGRGRGRG 106
G+ GRGRGRGRGRG+G+G
Sbjct: 118 GRGRGRGRGRGRGRGQGQG 136
Score = 33.1 bits (74), Expect = 0.024
Identities = 14/15 (93%), Positives = 14/15 (93%)
Query: 93 RGRGRGRGRGRGRGR 107
R RGRGRGRGRGRGR
Sbjct: 115 RKRGRGRGRGRGRGR 129
Score = 31.6 bits (70), Expect = 0.068
Identities = 13/22 (59%), Positives = 17/22 (77%)
Query: 83 KKPTAGKPLGRGRGRGRGRGRG 104
+K G+ GRGRGRGRG+G+G
Sbjct: 115 RKRGRGRGRGRGRGRGRGQGQG 136
>At3g29470 hypothetical protein
Length = 174
Score = 36.6 bits (83), Expect = 0.002
Identities = 15/19 (78%), Positives = 16/19 (83%)
Query: 88 GKPLGRGRGRGRGRGRGRG 106
G+ G GRGRGRGRGRGRG
Sbjct: 95 GRGCGHGRGRGRGRGRGRG 113
Score = 33.9 bits (76), Expect = 0.014
Identities = 15/21 (71%), Positives = 16/21 (75%)
Query: 87 AGKPLGRGRGRGRGRGRGRGR 107
+G GRG G GRGRGRGRGR
Sbjct: 90 SGYNRGRGCGHGRGRGRGRGR 110
Score = 33.5 bits (75), Expect = 0.018
Identities = 14/16 (87%), Positives = 14/16 (87%)
Query: 92 GRGRGRGRGRGRGRGR 107
G G GRGRGRGRGRGR
Sbjct: 97 GCGHGRGRGRGRGRGR 112
Score = 31.2 bits (69), Expect = 0.089
Identities = 14/20 (70%), Positives = 14/20 (70%)
Query: 88 GKPLGRGRGRGRGRGRGRGR 107
G GRGRGRGRGRG GR
Sbjct: 99 GHGRGRGRGRGRGRGYSGGR 118
Score = 30.4 bits (67), Expect = 0.15
Identities = 13/19 (68%), Positives = 14/19 (73%)
Query: 88 GKPLGRGRGRGRGRGRGRG 106
G+ GRGRGRGRG GRG
Sbjct: 101 GRGRGRGRGRGRGYSGGRG 119
Score = 28.1 bits (61), Expect = 0.76
Identities = 12/19 (63%), Positives = 13/19 (68%)
Query: 88 GKPLGRGRGRGRGRGRGRG 106
G+ GRGRGRG GRG G
Sbjct: 103 GRGRGRGRGRGYSGGRGSG 121
>At3g12480 unknown protein
Length = 293
Score = 35.4 bits (80), Expect = 0.005
Identities = 26/66 (39%), Positives = 36/66 (54%), Gaps = 7/66 (10%)
Query: 41 KGKNPVTLDHLSVRGNNIRYYILPDSLNLETLLVEEAPRVKPKKPTAGKPLGRGRGRGRG 100
+G VT+D S+ + R + D +N EE + K ++ + K GRG GRGRG
Sbjct: 96 QGHGDVTMDDRSI---SKRRKPISDEVNDSD---EEYKKSKTQEIGSAKTSGRG-GRGRG 148
Query: 101 RGRGRG 106
RGRGRG
Sbjct: 149 RGRGRG 154
>At2g28300 unknown protein
Length = 2218
Score = 35.4 bits (80), Expect = 0.005
Identities = 16/26 (61%), Positives = 16/26 (61%)
Query: 82 PKKPTAGKPLGRGRGRGRGRGRGRGR 107
P P GRGRGR RGRG GRGR
Sbjct: 191 PASPAPTPIRGRGRGRSRGRGAGRGR 216
Score = 29.3 bits (64), Expect = 0.34
Identities = 13/29 (44%), Positives = 15/29 (50%)
Query: 75 EEAPRVKPKKPTAGKPLGRGRGRGRGRGR 103
+ P P G+ GR RGRG GRGR
Sbjct: 188 QSLPASPAPTPIRGRGRGRSRGRGAGRGR 216
>At1g53645 unknown protein
Length = 523
Score = 34.7 bits (78), Expect = 0.008
Identities = 19/33 (57%), Positives = 22/33 (66%), Gaps = 1/33 (3%)
Query: 76 EAPRVKPKKPTAGKPLGRGRGRGRG-RGRGRGR 107
E R + + + G GRGRGRGRG RGRGRGR
Sbjct: 280 ELSRGEAEGSSVGGRGGRGRGRGRGARGRGRGR 312
Score = 32.0 bits (71), Expect = 0.052
Identities = 15/16 (93%), Positives = 15/16 (93%), Gaps = 1/16 (6%)
Query: 92 GRGRGRG-RGRGRGRG 106
GRGRGRG RGRGRGRG
Sbjct: 298 GRGRGRGARGRGRGRG 313
Score = 29.3 bits (64), Expect = 0.34
Identities = 14/18 (77%), Positives = 15/18 (82%), Gaps = 1/18 (5%)
Query: 88 GKPLGRGRG-RGRGRGRG 104
G+ GRGRG RGRGRGRG
Sbjct: 296 GRGRGRGRGARGRGRGRG 313
Score = 27.7 bits (60), Expect = 0.99
Identities = 16/36 (44%), Positives = 19/36 (52%)
Query: 72 LLVEEAPRVKPKKPTAGKPLGRGRGRGRGRGRGRGR 107
L EEA R + + G+ G G GRGRGRGR
Sbjct: 268 LSAEEAGRRARSELSRGEAEGSSVGGRGGRGRGRGR 303
>At5g19950 unknown protein
Length = 443
Score = 34.3 bits (77), Expect = 0.011
Identities = 15/20 (75%), Positives = 16/20 (80%)
Query: 88 GKPLGRGRGRGRGRGRGRGR 107
G+ GRGRGRGRG RGRGR
Sbjct: 423 GRGGGRGRGRGRGHRRGRGR 442
Score = 33.9 bits (76), Expect = 0.014
Identities = 14/16 (87%), Positives = 14/16 (87%)
Query: 92 GRGRGRGRGRGRGRGR 107
G GRGRG GRGRGRGR
Sbjct: 419 GHGRGRGGGRGRGRGR 434
Score = 33.9 bits (76), Expect = 0.014
Identities = 14/15 (93%), Positives = 14/15 (93%)
Query: 92 GRGRGRGRGRGRGRG 106
GRGRG GRGRGRGRG
Sbjct: 421 GRGRGGGRGRGRGRG 435
Score = 32.3 bits (72), Expect = 0.040
Identities = 14/20 (70%), Positives = 15/20 (75%)
Query: 88 GKPLGRGRGRGRGRGRGRGR 107
G+ G GRGRGRGRG RGR
Sbjct: 421 GRGRGGGRGRGRGRGHRRGR 440
Score = 32.0 bits (71), Expect = 0.052
Identities = 13/13 (100%), Positives = 13/13 (100%)
Query: 95 RGRGRGRGRGRGR 107
RGRGRGRGRGRGR
Sbjct: 328 RGRGRGRGRGRGR 340
Score = 32.0 bits (71), Expect = 0.052
Identities = 13/13 (100%), Positives = 13/13 (100%)
Query: 93 RGRGRGRGRGRGR 105
RGRGRGRGRGRGR
Sbjct: 328 RGRGRGRGRGRGR 340
Score = 31.2 bits (69), Expect = 0.089
Identities = 14/20 (70%), Positives = 14/20 (70%)
Query: 88 GKPLGRGRGRGRGRGRGRGR 107
G GRG GRGRGRGRG R
Sbjct: 419 GHGRGRGGGRGRGRGRGHRR 438
Score = 30.0 bits (66), Expect = 0.20
Identities = 12/12 (100%), Positives = 12/12 (100%)
Query: 92 GRGRGRGRGRGR 103
GRGRGRGRGRGR
Sbjct: 329 GRGRGRGRGRGR 340
>At3g31970 hypothetical protein
Length = 1329
Score = 34.3 bits (77), Expect = 0.011
Identities = 14/15 (93%), Positives = 14/15 (93%)
Query: 93 RGRGRGRGRGRGRGR 107
RG GRGRGRGRGRGR
Sbjct: 8 RGHGRGRGRGRGRGR 22
Score = 32.3 bits (72), Expect = 0.040
Identities = 13/14 (92%), Positives = 13/14 (92%)
Query: 92 GRGRGRGRGRGRGR 105
G GRGRGRGRGRGR
Sbjct: 9 GHGRGRGRGRGRGR 22
Score = 26.2 bits (56), Expect = 2.9
Identities = 11/14 (78%), Positives = 11/14 (78%)
Query: 88 GKPLGRGRGRGRGR 101
G GRGRGRGRGR
Sbjct: 9 GHGRGRGRGRGRGR 22
>At3g29500 hypothetical protein
Length = 341
Score = 33.9 bits (76), Expect = 0.014
Identities = 16/19 (84%), Positives = 16/19 (84%), Gaps = 1/19 (5%)
Query: 88 GKPLGRGRGRGRGRGRGRG 106
G P GRGRGRGRG GRGRG
Sbjct: 223 GNPHGRGRGRGRG-GRGRG 240
Score = 29.6 bits (65), Expect = 0.26
Identities = 19/40 (47%), Positives = 22/40 (54%), Gaps = 7/40 (17%)
Query: 74 VEEAPRVK----PKKPTAGKP---LGRGRGRGRGRGRGRG 106
V EA R + P +P A K + RG GRGRGRGRG
Sbjct: 196 VPEANRAEMAKAPNEPQATKESNYVHRGNPHGRGRGRGRG 235
Score = 27.7 bits (60), Expect = 0.99
Identities = 11/16 (68%), Positives = 13/16 (80%)
Query: 92 GRGRGRGRGRGRGRGR 107
GRGRGRGRG + RG+
Sbjct: 258 GRGRGRGRGVNKPRGK 273
>At2g37650 scarecrow-like 9 (SCL9)
Length = 718
Score = 33.9 bits (76), Expect = 0.014
Identities = 17/39 (43%), Positives = 21/39 (53%)
Query: 69 LETLLVEEAPRVKPKKPTAGKPLGRGRGRGRGRGRGRGR 107
L +L + + K GK RGRGRGRGRG G G+
Sbjct: 297 LRDVLKKGVEKKKASDAQGGKRRARGRGRGRGRGGGGGQ 335
>At1g03530 unknown protein
Length = 801
Score = 33.9 bits (76), Expect = 0.014
Identities = 16/17 (94%), Positives = 16/17 (94%), Gaps = 1/17 (5%)
Query: 92 GRGRGRGR-GRGRGRGR 107
GRGRGRGR GRGRGRGR
Sbjct: 781 GRGRGRGRFGRGRGRGR 797
Score = 33.9 bits (76), Expect = 0.014
Identities = 16/17 (94%), Positives = 16/17 (94%), Gaps = 1/17 (5%)
Query: 92 GRGRGRGRGR-GRGRGR 107
GRGRGRGRGR GRGRGR
Sbjct: 779 GRGRGRGRGRFGRGRGR 795
Score = 31.2 bits (69), Expect = 0.089
Identities = 15/19 (78%), Positives = 16/19 (83%), Gaps = 1/19 (5%)
Query: 88 GKPLGRGRGR-GRGRGRGR 105
G+ GRGRGR GRGRGRGR
Sbjct: 779 GRGRGRGRGRFGRGRGRGR 797
>At3g30843 hypothetical protein
Length = 310
Score = 33.1 bits (74), Expect = 0.024
Identities = 13/16 (81%), Positives = 15/16 (93%)
Query: 92 GRGRGRGRGRGRGRGR 107
GRGRGRGRGR RG+G+
Sbjct: 217 GRGRGRGRGRDRGQGK 232
Score = 32.7 bits (73), Expect = 0.031
Identities = 18/33 (54%), Positives = 21/33 (63%), Gaps = 2/33 (6%)
Query: 77 APRVKPKKPTAGKPLGRGRG--RGRGRGRGRGR 107
AP ++ T+G RG G RGRGRGRGRGR
Sbjct: 194 APLLEVNVATSGDDNWRGPGPCRGRGRGRGRGR 226
Score = 32.0 bits (71), Expect = 0.052
Identities = 13/15 (86%), Positives = 14/15 (92%)
Query: 93 RGRGRGRGRGRGRGR 107
RGRGRGRGRGR RG+
Sbjct: 216 RGRGRGRGRGRDRGQ 230
Score = 30.0 bits (66), Expect = 0.20
Identities = 12/16 (75%), Positives = 14/16 (87%)
Query: 92 GRGRGRGRGRGRGRGR 107
GRGRGRGR RG+G+ R
Sbjct: 219 GRGRGRGRDRGQGKRR 234
Score = 29.3 bits (64), Expect = 0.34
Identities = 12/21 (57%), Positives = 15/21 (71%)
Query: 85 PTAGKPLGRGRGRGRGRGRGR 105
P G+ GRGRGR RG+G+ R
Sbjct: 214 PCRGRGRGRGRGRDRGQGKRR 234
>At1g20220 unknown protein
Length = 315
Score = 33.1 bits (74), Expect = 0.024
Identities = 16/19 (84%), Positives = 16/19 (84%), Gaps = 1/19 (5%)
Query: 90 PLGRGRGRGR-GRGRGRGR 107
P GRGRGRGR GRGRG GR
Sbjct: 282 PQGRGRGRGRGGRGRGGGR 300
Score = 32.3 bits (72), Expect = 0.040
Identities = 16/21 (76%), Positives = 16/21 (76%), Gaps = 1/21 (4%)
Query: 88 GKPLGRGRGRGRG-RGRGRGR 107
G P GRG RGRG RGRGRGR
Sbjct: 146 GSPRGRGGRRGRGGRGRGRGR 166
Score = 31.2 bits (69), Expect = 0.089
Identities = 15/17 (88%), Positives = 15/17 (88%), Gaps = 1/17 (5%)
Query: 92 GRGRGRGRGRGR-GRGR 107
GRGRGRGRG GR GRGR
Sbjct: 228 GRGRGRGRGGGRGGRGR 244
Score = 28.9 bits (63), Expect = 0.44
Identities = 14/18 (77%), Positives = 15/18 (82%), Gaps = 1/18 (5%)
Query: 90 PLGRGRGRGRGR-GRGRG 106
P +GRGRGRGR GRGRG
Sbjct: 280 PPPQGRGRGRGRGGRGRG 297
Score = 28.9 bits (63), Expect = 0.44
Identities = 13/20 (65%), Positives = 13/20 (65%)
Query: 88 GKPLGRGRGRGRGRGRGRGR 107
G R GRGRGRGRG GR
Sbjct: 220 GMEQDRSYGRGRGRGRGGGR 239
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.138 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,520,749
Number of Sequences: 26719
Number of extensions: 102732
Number of successful extensions: 1223
Number of sequences better than 10.0: 104
Number of HSP's better than 10.0 without gapping: 86
Number of HSP's successfully gapped in prelim test: 18
Number of HSP's that attempted gapping in prelim test: 479
Number of HSP's gapped (non-prelim): 396
length of query: 107
length of database: 11,318,596
effective HSP length: 83
effective length of query: 24
effective length of database: 9,100,919
effective search space: 218422056
effective search space used: 218422056
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 52 (24.6 bits)
Medicago: description of AC146865.2


