

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146752.2 + phase: 0
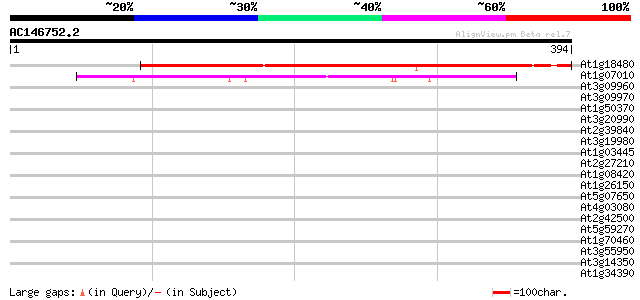
(394 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g18480 unknown protein 382 e-106
At1g07010 unknown protein 177 1e-44
At3g09960 unknown protein 39 0.005
At3g09970 unknown protein 36 0.032
At1g50370 phosphoprotein phosphatase 36 0.042
At3g20990 unknown protein 35 0.093
At2g39840 putative serine/threonine protein phosphatase PP1 isoz... 34 0.12
At3g19980 serine/threonine protein phosphatase, putative 33 0.21
At1g03445 putative serine/threonine protein phosphatase 33 0.27
At2g27210 putative phosphoprotein phosphatase 33 0.36
At1g08420 protein serine/threonine phosphatase alpha, putative 33 0.36
At1g26150 Pto kinase interactor, putative 32 0.46
At5g07650 putative protein 32 0.61
At4g03080 phospho-ser/thr phosphatase like protein 32 0.61
At2g42500 putative serine/threonine protein phosphatase catalyti... 32 0.61
At5g59270 receptor-like protein kinase 32 0.79
At1g70460 putative protein kinase 32 0.79
At3g55950 receptor kinase - like protein 31 1.0
At3g14350 SRF7 (LRR receptor protein kinase like) 31 1.0
At1g34390 auxin response factor, putative 31 1.0
>At1g18480 unknown protein
Length = 301
Score = 382 bits (982), Expect = e-106
Identities = 193/306 (63%), Positives = 234/306 (76%), Gaps = 10/306 (3%)
Query: 93 VVQIGDVLDRGGDEIKILYLLEKLKRQAAIHGGNFITMNGNHEIMNAEGDFRFATKNGVE 152
VVQ+GDVLDRGG+E+KILY LEKLKR+A GG +TMNGNHEIMN EGDFR+ TK G+E
Sbjct: 2 VVQVGDVLDRGGEELKILYFLEKLKREAERAGGKILTMNGNHEIMNIEGDFRYVTKKGLE 61
Query: 153 EFKVWLEWFRQGNKMKNLCKGLEETVVDPLENVHVAFRGVREEFHDGFRARVAALRPNGP 212
EF++W +W+ GNKMK LC GL++ DP E + ++F +R + +G RAR+AALRP+GP
Sbjct: 62 EFQIWADWYCLGNKMKTLCSGLDKPK-DPYEGIPMSFPRMRADCFEGIRARIAALRPDGP 120
Query: 213 ISKRFFTQNVTVLVVGDSIFVHGGLLKEHVDYGLEKINGEVSDWYKGLFGNRFSPPYCRG 272
I+KRF T+N TV VVGDS+FVHGGLL EH++YGLE+IN EV W G G R++P YCRG
Sbjct: 121 IAKRFLTKNQTVAVVGDSVFVHGGLLAEHIEYGLERINEEVRGWINGFKGGRYAPAYCRG 180
Query: 273 RNALVWLRKFSD---GNCDCSSLEHVLSTIPGVKRMIMGHTIQKEGINGVCENKAIRIDV 329
N++VWLRKFS+ CDC++LEH LSTIPGVKRMIMGHTIQ GINGVC +KAIRIDV
Sbjct: 181 GNSVVWLRKFSEEMAHKCDCAALEHALSTIPGVKRMIMGHTIQDAGINGVCNDKAIRIDV 240
Query: 330 GMSKGCGGGLPEVLEIDR-YGVRILTSNPLYNQMNKENVDIGKVEEGFGLLLNNQDGRPR 388
GMSKGC GLPEVLEI R GVRI+TSNPLY + +V + G GLL+ P+
Sbjct: 241 GMSKGCADGLPEVLEIRRDSGVRIVTSNPLYKENLYSHV-APDSKTGLGLLV----PVPK 295
Query: 389 QVEVKA 394
QVEVKA
Sbjct: 296 QVEVKA 301
>At1g07010 unknown protein
Length = 389
Score = 177 bits (448), Expect = 1e-44
Identities = 121/329 (36%), Positives = 169/329 (50%), Gaps = 21/329 (6%)
Query: 48 PTRLPSPSR-LIAIGDLHGDLKKSKEALSIAGLIDSSGN--YTGGSATVVQIGDVLDRGG 104
PT + +P+R ++A+GDLHGDL K+++AL +AG++ S G + G +VQ+GD+LDRG
Sbjct: 49 PTFVSAPARRIVAVGDLHGDLGKARDALQLAGVLSSDGRDQWVGQDTVLVQVGDILDRGD 108
Query: 105 DEIKILYLLEKLKRQAAIHGGNFITMNGNHEIMNAEGDFRFATKNGVEE---FKVWLEWF 161
DEI IL LL L QA +GG +NGNHE MN EGDFR+ +E F +LE +
Sbjct: 109 DEIAILSLLRSLDDQAKANGGAVFQVNGNHETMNVEGDFRYVDARAFDECTDFLDYLEDY 168
Query: 162 RQG--NKMKNLCKGLEETVVDPLENVHVAFRGVREEFHDGFRARVAALRPNGPISKRFFT 219
Q +N + D + + + G AR LRP G ++
Sbjct: 169 AQDWDKAFRNWIFESRQWKEDRRSSQTYWDQWNVVKRQKGVIARSVLLRPGGRLACELSR 228
Query: 220 QNVTVLVVGDSIFVHGGLLKEHVDYGLEKINGEVSDWYKGLFGNRFSP--PY--CRGRNA 275
V +L V + +F HGGLL HV YG+E+IN EVS W + SP P+ RG ++
Sbjct: 229 HGV-ILRVNNWLFCHGGLLPHHVAYGIERINREVSTWMRSPTNYEDSPQMPFIATRGYDS 287
Query: 276 LVWLRKFSDGNCDCSSLE--------HVLSTIPGVKRMIMGHTIQKEGINGVCENKAIRI 327
+VW R +S + + H G K M++GHT Q G+N R+
Sbjct: 288 VVWSRLYSRETSELEDYQIEQVNKILHDTLEAVGAKAMVVGHTPQLSGVNCEYGCGIWRV 347
Query: 328 DVGMSKGCGGGLPEVLEIDRYGVRILTSN 356
DVGMS G PEVLEI R++ SN
Sbjct: 348 DVGMSSGVLDSRPEVLEIRGDKARVIRSN 376
>At3g09960 unknown protein
Length = 311
Score = 38.9 bits (89), Expect = 0.005
Identities = 40/145 (27%), Positives = 65/145 (44%), Gaps = 26/145 (17%)
Query: 54 PSRLIAIGDLHGDLKK-SKEALSIAGLIDSSGNYTGGSATVVQIGDVLDRGGDEIKILYL 112
P +I +GD+HG++ K +K L++ I +S SA V+ +GD DRG + K++
Sbjct: 6 PRTVICVGDIHGNISKLNKLWLNLQSDIQNS---DFSSALVIFLGDYCDRGPETRKVIDF 62
Query: 113 LEKLKRQAAIHGGNFITMNGNHEIMNAEGDFRFATKNGV-------EEFKVWLEWFRQGN 165
L L + F+ GNH DF FA G+ EFK + + +
Sbjct: 63 LISLPEKHPDQTHVFLA--GNH-------DFAFAGFLGLLPRPSDGSEFKETWKEYSKSE 113
Query: 166 KMKNLCKGLEETVVDPLENVHVAFR 190
+ + KG + EN+H+ R
Sbjct: 114 EREGWYKG------EGFENMHLQSR 132
>At3g09970 unknown protein
Length = 309
Score = 36.2 bits (82), Expect = 0.032
Identities = 26/83 (31%), Positives = 40/83 (47%), Gaps = 6/83 (7%)
Query: 54 PSRLIAIGDLHGDLKKSKEA-LSIAGLIDSSGNYTGGSATVVQIGDVLDRGGDEIKILYL 112
P +I +GD+HG + K L++ ID S SA V+ +GD DRG + K++
Sbjct: 5 PRTVICVGDIHGYISKLNNLWLNLQSAIDPSDF---SSALVIFLGDYCDRGPETRKVIDF 61
Query: 113 LEKLKRQAAIHGGNFITMNGNHE 135
L L + F+ GNH+
Sbjct: 62 LISLPEKHPDQTHVFLA--GNHD 82
>At1g50370 phosphoprotein phosphatase
Length = 303
Score = 35.8 bits (81), Expect = 0.042
Identities = 24/84 (28%), Positives = 38/84 (44%), Gaps = 11/84 (13%)
Query: 52 PSPSRLIAIGDLHGDLKKSKEALSIAGLIDSSGNYTGGSATVVQIGDVLDRGGDEIKILY 111
P S + GD+HG + G + + NY + +GD +DRG + +++
Sbjct: 40 PVNSPVTVCGDIHGQFHDLMKLFQTGGHVPET-NY-------IFMGDFVDRGYNSLEVFT 91
Query: 112 LLEKLKRQAAIHGGNFITMNGNHE 135
+L LK A H N + GNHE
Sbjct: 92 ILLLLK---ARHPANITLLRGNHE 112
>At3g20990 unknown protein
Length = 379
Score = 34.7 bits (78), Expect = 0.093
Identities = 24/79 (30%), Positives = 39/79 (48%), Gaps = 1/79 (1%)
Query: 37 LPPPPSSPPPIPTRLPSPSRLIAIGDLHGDLKKSKEALSIAGLIDSSGNYTGGSATVVQI 96
+PP PS P + T++ P L DL K+ + L + L DS+ T +++ +
Sbjct: 41 VPPDPSKIPELATKI-QPEELSRWRDLAVKDMKALQILQSSSLTDSAFRKTLSASSAKDL 99
Query: 97 GDVLDRGGDEIKILYLLEK 115
D L++G +E L LEK
Sbjct: 100 WDSLEKGNNEQAKLRRLEK 118
>At2g39840 putative serine/threonine protein phosphatase PP1 isozyme
4 (TOPP4)
Length = 321
Score = 34.3 bits (77), Expect = 0.12
Identities = 25/88 (28%), Positives = 37/88 (41%), Gaps = 11/88 (12%)
Query: 48 PTRLPSPSRLIAIGDLHGDLKKSKEALSIAGLIDSSGNYTGGSATVVQIGDVLDRGGDEI 107
P L + + GD+HG G S NY + +GD +DRG +
Sbjct: 60 PNLLELEAPIKICGDIHGQYSDLLRLFEYGGF-PPSANY-------LFLGDYVDRGKQSL 111
Query: 108 KILYLLEKLKRQAAIHGGNFITMNGNHE 135
+ + LL K + + GNF + GNHE
Sbjct: 112 ETICLLLAYKIK---YPGNFFLLRGNHE 136
>At3g19980 serine/threonine protein phosphatase, putative
Length = 303
Score = 33.5 bits (75), Expect = 0.21
Identities = 23/84 (27%), Positives = 38/84 (44%), Gaps = 11/84 (13%)
Query: 52 PSPSRLIAIGDLHGDLKKSKEALSIAGLIDSSGNYTGGSATVVQIGDVLDRGGDEIKILY 111
P S + GD+HG + G + + NY + +GD +DRG + +++
Sbjct: 40 PVNSPVTVCGDIHGQFHDLMKLFQTGGHVPDT-NY-------IFMGDFVDRGYNSLEVFT 91
Query: 112 LLEKLKRQAAIHGGNFITMNGNHE 135
+L LK A + N + GNHE
Sbjct: 92 ILLLLK---ARYPANITLLRGNHE 112
>At1g03445 putative serine/threonine protein phosphatase
Length = 846
Score = 33.1 bits (74), Expect = 0.27
Identities = 32/119 (26%), Positives = 49/119 (40%), Gaps = 14/119 (11%)
Query: 48 PTRLPSPSRLIAIGDLHGDLKKSKEALSIAGLIDSSGNYTGGSATVVQIGDVLDRGGDEI 107
PT L + GD+HG G G+ T + +GD +DRG +
Sbjct: 549 PTLLQLKVPIKVFGDIHGQYGDLMRLFHEYGHPSVEGDIT--HIDYLFLGDYVDRGQHSL 606
Query: 108 KILYLLEKLKRQAAIHGGNFITMNGNHEIMNAEGDFRFATKNGVEE-------FKVWLE 159
+I+ LL LK + + N + GNHE + + F T+ EE F+ WL+
Sbjct: 607 EIIMLLFALKIE---YPKNIHLIRGNHESLAMNRIYGFLTE--CEERMGESYGFEAWLK 660
>At2g27210 putative phosphoprotein phosphatase
Length = 715
Score = 32.7 bits (73), Expect = 0.36
Identities = 29/102 (28%), Positives = 44/102 (42%), Gaps = 13/102 (12%)
Query: 48 PTRLPSPSRLIAIGDLHGDLKKSKEALSIAGLIDSSGN-YTGGSATVVQ---IGDVLDRG 103
PT L + + GDLHG + L D G+ T G + + +GD +DRG
Sbjct: 424 PTVLQLKAPIKIFGDLHGQFG------DLMRLFDEYGSPSTAGDISYIDYLFLGDYVDRG 477
Query: 104 GDEIKILYLLEKLKRQAAIHGGNFITMNGNHEIMNAEGDFRF 145
++ + LL LK + + N + GNHE + F F
Sbjct: 478 QHSLETITLLLALKVE---YQHNVHLIRGNHEAADINALFGF 516
>At1g08420 protein serine/threonine phosphatase alpha, putative
Length = 1018
Score = 32.7 bits (73), Expect = 0.36
Identities = 29/102 (28%), Positives = 44/102 (42%), Gaps = 13/102 (12%)
Query: 48 PTRLPSPSRLIAIGDLHGDLKKSKEALSIAGLIDSSGN-YTGGSATVVQ---IGDVLDRG 103
PT L + + GDLHG + L D G+ T G + + +GD +DRG
Sbjct: 706 PTVLQLKAPIKIFGDLHGQFG------DLMRLFDEYGSPSTAGDISYIDYLFLGDYVDRG 759
Query: 104 GDEIKILYLLEKLKRQAAIHGGNFITMNGNHEIMNAEGDFRF 145
++ + LL LK + + N + GNHE + F F
Sbjct: 760 QHSLETISLLLALKVE---YQHNVHLIRGNHEAADINALFGF 798
>At1g26150 Pto kinase interactor, putative
Length = 760
Score = 32.3 bits (72), Expect = 0.46
Identities = 11/22 (50%), Positives = 15/22 (68%)
Query: 38 PPPPSSPPPIPTRLPSPSRLIA 59
PP PS PPP+PT P P+ ++
Sbjct: 95 PPEPSPPPPLPTEAPPPANPVS 116
Score = 28.5 bits (62), Expect = 6.7
Identities = 11/22 (50%), Positives = 14/22 (63%)
Query: 37 LPPPPSSPPPIPTRLPSPSRLI 58
LPPP SP P+P+ SP L+
Sbjct: 265 LPPPKPSPDPLPSNSSSPPTLL 286
>At5g07650 putative protein
Length = 746
Score = 32.0 bits (71), Expect = 0.61
Identities = 11/16 (68%), Positives = 13/16 (80%)
Query: 37 LPPPPSSPPPIPTRLP 52
+PPPP PPP+P RLP
Sbjct: 20 VPPPPPRPPPMPRRLP 35
>At4g03080 phospho-ser/thr phosphatase like protein
Length = 881
Score = 32.0 bits (71), Expect = 0.61
Identities = 24/85 (28%), Positives = 37/85 (43%), Gaps = 5/85 (5%)
Query: 61 GDLHGDLKKSKEALSIAGLIDSSGNYTGGSATVVQIGDVLDRGGDEIKILYLLEKLKRQA 120
GDLHG G ++G+ T + +GD +DRG ++ + LL LK +
Sbjct: 583 GDLHGQFGDLMRLFDEYGFPSTAGDIT--YIDYLFLGDYVDRGQHSLETITLLLALKIE- 639
Query: 121 AIHGGNFITMNGNHEIMNAEGDFRF 145
+ N + GNHE + F F
Sbjct: 640 --YPENVHLIRGNHEAADINALFGF 662
>At2g42500 putative serine/threonine protein phosphatase catalytic
subunit, PP2A-3
Length = 313
Score = 32.0 bits (71), Expect = 0.61
Identities = 24/84 (28%), Positives = 37/84 (43%), Gaps = 11/84 (13%)
Query: 52 PSPSRLIAIGDLHGDLKKSKEALSIAGLIDSSGNYTGGSATVVQIGDVLDRGGDEIKILY 111
P S + GD+HG E I G+ + NY + +GD +DRG ++ +
Sbjct: 51 PVKSPVTICGDIHGQFHDLAELFRIGGMCPDT-NY-------LFMGDYVDRGYYSVETVT 102
Query: 112 LLEKLKRQAAIHGGNFITMNGNHE 135
LL LK + + + GNHE
Sbjct: 103 LLVALKMR---YPQRITILRGNHE 123
>At5g59270 receptor-like protein kinase
Length = 668
Score = 31.6 bits (70), Expect = 0.79
Identities = 12/17 (70%), Positives = 13/17 (75%)
Query: 38 PPPPSSPPPIPTRLPSP 54
PPPPSSPPP P P+P
Sbjct: 266 PPPPSSPPPPPPPPPTP 282
>At1g70460 putative protein kinase
Length = 710
Score = 31.6 bits (70), Expect = 0.79
Identities = 11/17 (64%), Positives = 12/17 (69%)
Query: 38 PPPPSSPPPIPTRLPSP 54
PPPP +PPPIP P P
Sbjct: 90 PPPPDAPPPIPIVFPPP 106
Score = 28.1 bits (61), Expect = 8.7
Identities = 13/20 (65%), Positives = 13/20 (65%), Gaps = 2/20 (10%)
Query: 38 PPPP--SSPPPIPTRLPSPS 55
PPPP SSPPP P P PS
Sbjct: 69 PPPPLDSSPPPPPDLTPPPS 88
>At3g55950 receptor kinase - like protein
Length = 814
Score = 31.2 bits (69), Expect = 1.0
Identities = 12/25 (48%), Positives = 15/25 (60%)
Query: 37 LPPPPSSPPPIPTRLPSPSRLIAIG 61
LPPPP PPP P+ PS+ + G
Sbjct: 370 LPPPPPPPPPSPSTSSPPSKALTRG 394
>At3g14350 SRF7 (LRR receptor protein kinase like)
Length = 717
Score = 31.2 bits (69), Expect = 1.0
Identities = 26/87 (29%), Positives = 36/87 (40%), Gaps = 8/87 (9%)
Query: 8 SNTFCNQIPNFLSSFIDTFVDFSVSGGLFLPPPPSSPPPIPTRLPSPSRLIAIGDLHGDL 67
+N F IP+ L I+ D ++ PPPP PPI P+P +GD
Sbjct: 219 NNRFTGWIPDSLKG-INLQKDGNLLNSGPAPPPPPGTPPISKSSPTPKSGNRGNRSNGDS 277
Query: 68 KKSKEALSIAGLIDSSGNYTGGSATVV 94
SK++ SG GG A +V
Sbjct: 278 SNSKDS-------SKSGLGAGGVAGIV 297
>At1g34390 auxin response factor, putative
Length = 600
Score = 31.2 bits (69), Expect = 1.0
Identities = 17/56 (30%), Positives = 26/56 (46%), Gaps = 2/56 (3%)
Query: 13 NQIPNFLSSFIDTFVDFSVSGGLFLPPPPSSP--PPIPTRLPSPSRLIAIGDLHGD 66
NQ ++SF SGG F+P + PP+ P P++ + DLHG+
Sbjct: 117 NQFRPLVNSFTKVLTASDTSGGFFVPKKHAIECLPPLDMSQPLPTQELLATDLHGN 172
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.141 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,987,528
Number of Sequences: 26719
Number of extensions: 485216
Number of successful extensions: 3809
Number of sequences better than 10.0: 81
Number of HSP's better than 10.0 without gapping: 43
Number of HSP's successfully gapped in prelim test: 43
Number of HSP's that attempted gapping in prelim test: 2954
Number of HSP's gapped (non-prelim): 672
length of query: 394
length of database: 11,318,596
effective HSP length: 101
effective length of query: 293
effective length of database: 8,619,977
effective search space: 2525653261
effective search space used: 2525653261
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146752.2


