

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146308.4 + phase: 0
(357 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
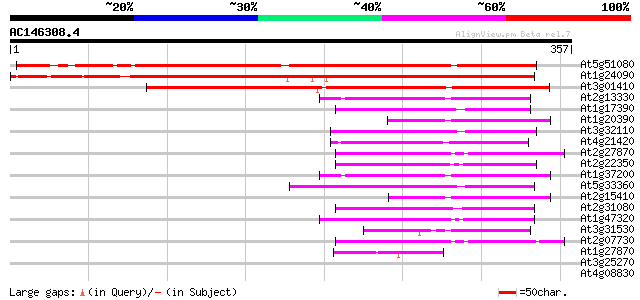
Score E
Sequences producing significant alignments: (bits) Value
At5g51080 unknown protein 307 5e-84
At1g24090 unknown protein 291 4e-79
At3g01410 putative RNase H 259 2e-69
At2g13330 F14O4.9 66 3e-11
At1g17390 hypothetical protein 65 6e-11
At1g20390 hypothetical protein 62 4e-10
At3g32110 non-LTR reverse transcriptase, putative 61 1e-09
At4g21420 hypothetical protein 60 2e-09
At2g27870 putative non-LTR retroelement reverse transcriptase 59 3e-09
At2g22350 putative non-LTR retroelement reverse transcriptase 59 5e-09
At1g37200 hypothetical protein 57 2e-08
At5g33360 putative protein 57 2e-08
At2g15410 putative retroelement pol polyprotein 54 2e-07
At2g31080 putative non-LTR retroelement reverse transcriptase 50 2e-06
At1g47320 hypothetical protein 49 4e-06
At3g31530 hypothetical protein 46 3e-05
At2g07730 putative non-LTR retroelement reverse transcriptase 44 1e-04
At1g27870 hypothetical protein 42 5e-04
At3g25270 hypothetical protein 41 0.001
At4g08830 putative protein 40 0.001
>At5g51080 unknown protein
Length = 322
Score = 307 bits (787), Expect = 5e-84
Identities = 169/331 (51%), Positives = 222/331 (67%), Gaps = 19/331 (5%)
Query: 5 FSHLSSFTSTILARTTQFVANRSLHTTPHTFSFPVKSHHSLLTSFHSQFVLTTVTRCYST 64
FS S+ S +L R + +V + P F + SL +S V + CYS+
Sbjct: 4 FSRARSYISLVLFRKSSYVTSH----IPWNQCF----YTSLKSSLKPASVSVSSVHCYSS 55
Query: 65 RKLRKTGSSKLHKVEAETTMNQEQNESDAFYVVRKGDVVGIYNTLTDSQAQVGSSVCDPP 124
R KT SK+ K + +++ E DAF+VVRKGD+VGIY L D QAQVGSSV DPP
Sbjct: 56 RS--KTAKSKMSK--SSVSVSDSDKEKDAFFVVRKGDIVGIYKDLIDCQAQVGSSVYDPP 111
Query: 125 VSVYKGYSMSNETEEYLLSHGLKNALYTIRASDLKEDLFGTLVPCPFQDPSSTQGTTSNA 184
VSVYKGYS+ +TEE L + GLK LY RA DLKED+FG L PC FQD Q +++
Sbjct: 112 VSVYKGYSLLKDTEECLSTVGLKKPLYVFRALDLKEDMFGALTPCLFQD----QLPSASM 167
Query: 185 DSSKKRALEVLEQNNVKTCIVEFDGASKGNPGKAGAGAILRSKDGNLIYRVREGVGIATN 244
K LE + +TCI+EFDGASKGNPG +GA A+L+++DG+LI+++R+G+GIATN
Sbjct: 168 SVEKLAELEPSADTSYETCIIEFDGASKGNPGLSGAAAVLKTEDGSLIFKMRQGLGIATN 227
Query: 245 NVAEYRAMILGMRHALKKGFTSICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKEL 304
N AEY +ILG++HA++KG+T I ++ DSKLVCMQMK G WKV +E LS L+K AK+L
Sbjct: 228 NAAEYHGLILGLKHAIEKGYTKIKVKTDSKLVCMQMK---GQWKVNHEVLSKLHKEAKQL 284
Query: 305 KDKFVSFKISHVLRDLNSEADAQANLAVNLA 335
DK +SF+ISHVLR LNS+AD QAN+A L+
Sbjct: 285 SDKCLSFEISHVLRSLNSDADEQANMAARLS 315
>At1g24090 unknown protein
Length = 535
Score = 291 bits (745), Expect = 4e-79
Identities = 173/358 (48%), Positives = 232/358 (64%), Gaps = 34/358 (9%)
Query: 1 MNALFSHLSSFTSTILARTTQFVANRSLHTTPHTFSFPVKSHHSLLTSFHSQFVLTTVTR 60
MN L SH S+ + L + + +V+ S+ F P KS + + S F + +V
Sbjct: 1 MNCL-SHARSYIALGLLKRSSYVS--SIPWNECFFYMPSKSCLKPV-AVSSVFGICSVHS 56
Query: 61 CYSTRKLRKTGSSKLHKVEAETTMNQEQNESDAFYVVRKGDVVGIYNTLTDSQAQVGSSV 120
S K K+ K+ + T ++ E DAF+VVRKGDV+GIY L+D QAQVGSSV
Sbjct: 57 YSSRSKAVKS------KMLSSTVVSAVDKEKDAFFVVRKGDVIGIYKDLSDCQAQVGSSV 110
Query: 121 CDPPVSVYKGYSMSNETEEYLLSHGLKNALYTIRASDLKEDLFGTLVPCPFQDPS----- 175
D PVSVYKGYS+ +TEEYL S GLK LY++RASDLK+D+FG L PC FQ+P+
Sbjct: 111 FDLPVSVYKGYSLPKDTEEYLSSVGLKKPLYSLRASDLKDDMFGALTPCLFQEPAPCTVK 170
Query: 176 ----STQGTTSNADSSKKRA---------LEVLEQNNV------KTCIVEFDGASKGNPG 216
T T + D K + LE L + +TC +EFDGASKGNPG
Sbjct: 171 VSEDETTSETKSKDDKKDQLPSASISYDPLEKLSKVEPSAYISDETCFIEFDGASKGNPG 230
Query: 217 KAGAGAILRSKDGNLIYRVREGVGIATNNVAEYRAMILGMRHALKKGFTSICIQGDSKLV 276
+GA A+L+++DG+LI RVR+G+GIATNN AEY A+ILG+++A++KG+ +I ++GDSKLV
Sbjct: 231 LSGAAAVLKTEDGSLICRVRQGLGIATNNAAEYHALILGLKYAIEKGYKNIKVKGDSKLV 290
Query: 277 CMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNSEADAQANLAVNL 334
CMQ +QI G WKV +E L+ L+K AK L +K VSF+ISHVLR+LN++AD QANLAV L
Sbjct: 291 CMQKQQIKGQWKVNHEVLAKLHKEAKLLCNKCVSFEISHVLRNLNADADEQANLAVRL 348
>At3g01410 putative RNase H
Length = 290
Score = 259 bits (661), Expect = 2e-69
Identities = 134/292 (45%), Positives = 185/292 (62%), Gaps = 41/292 (14%)
Query: 88 QNESDAFYVVRKGDVVGIYNTLTDSQAQVGSSVCDPPVSVYKGYSMSNETEEYLLSHGLK 147
++E DAFY+VRKGD++G+Y +L++ Q Q GSSV P +SVYKGY E+ L S G+K
Sbjct: 2 EDEKDAFYIVRKGDIIGVYRSLSECQGQAGSSVSHPAMSVYKGYGWPKGAEDLLSSFGIK 61
Query: 148 NALYTIRASDLKEDLFGTLVPCPFQDPSSTQGTTSNADSSKKRALEV------------- 194
NAL+++ AS +K+D FG L+PCP Q PSS+QG + N S KR ++
Sbjct: 62 NALFSVNASHVKDDAFGKLIPCPVQQPSSSQGESLNKSSPSKRLQDMGSGESGSFSPSPP 121
Query: 195 -----------------------LEQNNVKTCIVEFDGASKGNPGKAGAGAILRSKDGNL 231
+ QN+ +C +EFDGASKGNPGKAGAGA+LR+ D ++
Sbjct: 122 QKQLKIENDMLRRIPSSLLTRTPIRQND--SCTIEFDGASKGNPGKAGAGAVLRASDNSV 179
Query: 232 IYRVREGVGIATNNVAEYRAMILGMRHALKKGFTSICIQGDSKLVCMQMKQIDGLWKVKN 291
++ +REGVG ATNNVAEYRA++LG+R AL KGF ++ + GDS LVCM Q+ G WK +
Sbjct: 180 LFYLREGVGNATNNVAEYRALLLGLRSALDKGFKNVHVLGDSMLVCM---QVQGAWKTNH 236
Query: 292 ENLSTLYKVAKELKDKFVSFKISHVLRDLNSEADAQANLAVNLAGEAFKIIN 343
++ L K AKEL + F +F I H+ R+ NSEAD QAN A+ LA ++I+
Sbjct: 237 PKMAELCKQAKELMNSFKTFDIKHIAREKNSEADKQANSAIFLADGQTQVIS 288
>At2g13330 F14O4.9
Length = 889
Score = 65.9 bits (159), Expect = 3e-11
Identities = 45/134 (33%), Positives = 71/134 (52%), Gaps = 5/134 (3%)
Query: 198 NNVKTCIVEFDGASKGNPGKAGAGAILRSKDGNLIYRVREGVGIATNNVAEYRAMILGMR 257
N+ K ++ DG+S N +G G L S G +I + + A+NN +EY A+I G++
Sbjct: 638 NSNKKWLLHVDGSS--NRQGSGVGIQLTSPTGEVIEQSLQLGFNASNNESEYEALIAGIK 695
Query: 258 HALKKGFTSICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVL 317
A +KG I DS+LV Q G ++ K+E + ++ K L +F SFK++ +
Sbjct: 696 LAQEKGIREIHAYSDSQLVT---SQFHGEYEAKDERMEAYLELVKTLAQQFESFKLTRIP 752
Query: 318 RDLNSEADAQANLA 331
R N+ AD A LA
Sbjct: 753 RGENTSADTLAALA 766
>At1g17390 hypothetical protein
Length = 322
Score = 65.1 bits (157), Expect = 6e-11
Identities = 45/125 (36%), Positives = 66/125 (52%), Gaps = 6/125 (4%)
Query: 208 DGASKGNPGKAGAGAILRSKDGNLIYRVREGVGIATNNVAEYRAMILGMRHALKKGFTSI 267
DGAS+GNP A AG ++R DGN Y +GI + +AE G+ A ++G T +
Sbjct: 117 DGASRGNPRLATAGGVVRDGDGNWCYGFSLNIGICSAPLAELWGAYYGLNIAWERGVTQL 176
Query: 268 CIQGDSKLVCMQMKQ-IDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNSEADA 326
++ DS++V ++ ID + LS L ++ L K S +ISHV R+ N AD
Sbjct: 177 EMEIDSEMVVGFLRTGID-----DSHPLSFLVRLCHGLLSKDWSVRISHVYREANRLADG 231
Query: 327 QANLA 331
AN A
Sbjct: 232 LANYA 236
>At1g20390 hypothetical protein
Length = 1791
Score = 62.4 bits (150), Expect = 4e-10
Identities = 38/104 (36%), Positives = 58/104 (55%), Gaps = 3/104 (2%)
Query: 241 IATNNVAEYRAMILGMRHALKKGFTSICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKV 300
+ATNNVAEY +I G+R A T+I DS+L+ Q+ G ++ KNE + K+
Sbjct: 1246 VATNNVAEYEVLIAGLRLAAGMQITTIHAFTDSQLIA---GQLSGEYEAKNEKMDAYLKI 1302
Query: 301 AKELKDKFVSFKISHVLRDLNSEADAQANLAVNLAGEAFKIINV 344
+ + F +FK+S + R N+ ADA A LA+ + +II V
Sbjct: 1303 VQLMTKDFENFKLSKIPRGDNAPADALAALALTSDSDLRRIIPV 1346
>At3g32110 non-LTR reverse transcriptase, putative
Length = 1911
Score = 60.8 bits (146), Expect = 1e-09
Identities = 43/131 (32%), Positives = 66/131 (49%), Gaps = 4/131 (3%)
Query: 205 VEFDGASKGNPGKAGAGAILRSKDGNLIYRVREGVGIATNNVAEYRAMILGMRHALKKGF 264
+ DGAS+GNPG A AG +LR+ G +G + +AE + G+ A K
Sbjct: 1753 INTDGASRGNPGLASAGGVLRNSAGAWCGGFAVNIGRCSAPLAELWGVYYGLYMAWAKQL 1812
Query: 265 TSICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNSEA 324
T + ++ DS++V +K G + LS L ++ K + +ISHV R+ NS A
Sbjct: 1813 THLELEVDSEVVVGFLKTGIG----ETHPLSFLVRLCHNFLSKDWTVRISHVYREANSLA 1868
Query: 325 DAQANLAVNLA 335
D AN A +L+
Sbjct: 1869 DGLANHAFSLS 1879
>At4g21420 hypothetical protein
Length = 229
Score = 60.1 bits (144), Expect = 2e-09
Identities = 37/126 (29%), Positives = 65/126 (51%), Gaps = 4/126 (3%)
Query: 205 VEFDGASKGNPGKAGAGAILRSKDGNLIYRVREGVGIATNNVAEYRAMILGMRHALKKGF 264
+ FDG S+G G+A G + R+ + E +G AT+ +AE+ A+ G+ AL+ G
Sbjct: 73 LNFDG-SRGREGQASIGGVFRNHKAEFLLGYSESIGEATSTMAEFAALKRGLELALENGL 131
Query: 265 TSICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNSEA 324
T + ++GD+K++ M I +++ E + K + + + +SHV R+ N A
Sbjct: 132 TDLWLEGDAKII---MDIISRRGRLRCEKTNKHVNYIKVVMPELNNCVLSHVYREGNRVA 188
Query: 325 DAQANL 330
D A L
Sbjct: 189 DKLAKL 194
>At2g27870 putative non-LTR retroelement reverse transcriptase
Length = 314
Score = 59.3 bits (142), Expect = 3e-09
Identities = 46/146 (31%), Positives = 75/146 (50%), Gaps = 4/146 (2%)
Query: 208 DGASKGNPGKAGAGAILRSKDGNLIYRVREGVGIATNNVAEYRAMILGMRHALKKGFTSI 267
DGAS+GNPG A AG +LR ++G +G+ + +AE + G+ A ++ T +
Sbjct: 159 DGASRGNPGLASAGGVLRDEEGAWRGGFALNIGVCSAPLAELWGVYYGLYIAWERRVTRL 218
Query: 268 CIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNSEADAQ 327
I+ DS++V +K G+ +V LS L ++ + + +ISHV R+ N AD
Sbjct: 219 EIEVDSEIVVGFLK--IGINEV--HPLSFLVRLCHDFISRDWRVRISHVYREANRLADGL 274
Query: 328 ANLAVNLAGEAFKIINVCSSLMYMYL 353
AN A +L + V SL ++ L
Sbjct: 275 ANYAFSLPLGFHSLSLVPDSLRFILL 300
>At2g22350 putative non-LTR retroelement reverse transcriptase
Length = 321
Score = 58.5 bits (140), Expect = 5e-09
Identities = 42/128 (32%), Positives = 66/128 (50%), Gaps = 4/128 (3%)
Query: 208 DGASKGNPGKAGAGAILRSKDGNLIYRVREGVGIATNNVAEYRAMILGMRHALKKGFTSI 267
DGAS+GNPG A AG +LR +G I +G+ + +AE + G+ A +G +
Sbjct: 166 DGASRGNPGFATAGGVLRDHNGAWIGGFAVNIGVCSAPLAELWGVYYGLFIAWGRGARRV 225
Query: 268 CIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNSEADAQ 327
++ DSK+V + G+ + LS L ++ + K +ISHV R+ N AD
Sbjct: 226 ELEVDSKMVVGFL--TTGI--ADSHPLSFLLRLCYDFLSKGWIVRISHVYREANRLADGL 281
Query: 328 ANLAVNLA 335
AN A +L+
Sbjct: 282 ANYAFSLS 289
>At1g37200 hypothetical protein
Length = 1564
Score = 57.0 bits (136), Expect = 2e-08
Identities = 46/147 (31%), Positives = 72/147 (48%), Gaps = 5/147 (3%)
Query: 198 NNVKTCIVEFDGASKGNPGKAGAGAILRSKDGNLIYRVREGVGIATNNVAEYRAMILGMR 257
N+ K ++ DG+S N +G G L S G +I + A+NN +EY A+I G++
Sbjct: 1036 NSNKKWLLHVDGSS--NRQGSGEGIQLTSPTGEVIEQSFRLGFNASNNESEYEALIDGIK 1093
Query: 258 HALKKGFTSICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVL 317
A I DS+LV Q G ++ K+E + ++ K L +F SF+++ +
Sbjct: 1094 LAQGMRIRDIHAHSDSQLVT---SQFHGEYEAKDERMEAYLELVKTLTQQFESFELTRIP 1150
Query: 318 RDLNSEADAQANLAVNLAGEAFKIINV 344
R N+ DA A LA L +II V
Sbjct: 1151 RGENTSTDALAALASTLDPFVKRIIPV 1177
>At5g33360 putative protein
Length = 306
Score = 56.6 bits (135), Expect = 2e-08
Identities = 44/156 (28%), Positives = 73/156 (46%), Gaps = 4/156 (2%)
Query: 179 GTTSNADSSKKRALEVLEQNNVKTCIVEFDGASKGNPGKAGAGAILRSKDGNLIYRVREG 238
G+ SN S+ + ++ C + DGAS GNPG A AG LR++ G +
Sbjct: 122 GSGSNNSRSRMERQVRWSKPSLGWCKLNTDGASHGNPGLAIAGGALRNEYGEWCFGFALN 181
Query: 239 VGIATNNVAEYRAMILGMRHALKKGFTSICIQGDSKLVCMQMKQIDGLWKVKNENLSTLY 298
+G + +AE + G+ A +G T + ++ DS++V ++ G + LS L
Sbjct: 182 IGRCSAPLAELWGVYYGLFMAWDRGITRLELEVDSEMVVGFLRTGIG----SSHPLSFLV 237
Query: 299 KVAKELKDKFVSFKISHVLRDLNSEADAQANLAVNL 334
++ + +I HV R+ N AD AN A +L
Sbjct: 238 RMCHGFLSRDWIVRIGHVYREANRLADELANYAFDL 273
>At2g15410 putative retroelement pol polyprotein
Length = 1787
Score = 53.5 bits (127), Expect = 2e-07
Identities = 37/103 (35%), Positives = 53/103 (50%), Gaps = 3/103 (2%)
Query: 242 ATNNVAEYRAMILGMRHALKKGFTSICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVA 301
A+NN AEY A+I G+R A I DS+LV Q G ++ KNE + KV
Sbjct: 1192 ASNNEAEYEALIAGLRLAHGIEVKKIQAYCDSQLVA---SQFSGNYEAKNERMDAYLKVV 1248
Query: 302 KELKDKFVSFKISHVLRDLNSEADAQANLAVNLAGEAFKIINV 344
+EL F F+++ + R N+ ADA A LA + ++I V
Sbjct: 1249 RELSYNFEVFELTKIPRSDNAPADALAVLASTSDPDLRRVIPV 1291
>At2g31080 putative non-LTR retroelement reverse transcriptase
Length = 1231
Score = 49.7 bits (117), Expect = 2e-06
Identities = 40/128 (31%), Positives = 58/128 (45%), Gaps = 6/128 (4%)
Query: 208 DGASKGNPGKAGAGAILRSKDGNLIYRVREGVGIATNNVAEYRAMILGMRHALKKGFTSI 267
DGAS+GN G A AG +R+ G + +G +AE G+ A KGF +
Sbjct: 1076 DGASRGNHGLAAAGGAIRNGQGEWLGGFALNIGSCAAPLAELWGAYYGLLIAWDKGFRRV 1135
Query: 268 CIQGDSKLVCMQMKQIDGLWKVKNEN-LSTLYKVAKELKDKFVSFKISHVLRDLNSEADA 326
+ D KLV + V N + LS L ++ + + ++SHV R+ N AD
Sbjct: 1136 ELDLDCKLVVGFLST-----GVSNAHPLSFLVRLCQGFFTRDWLVRVSHVYREANRLADG 1190
Query: 327 QANLAVNL 334
AN A L
Sbjct: 1191 LANYAFTL 1198
>At1g47320 hypothetical protein
Length = 259
Score = 48.9 bits (115), Expect = 4e-06
Identities = 40/138 (28%), Positives = 66/138 (46%), Gaps = 5/138 (3%)
Query: 198 NNVKTCI-VEFDGASKGNPGKAGAGAILRSKDGNLIYRVREGVGIATNNVAEYRAMILGM 256
N C+ + DGAS+GNPG A AG +L+ +G +G ++ +AE G+
Sbjct: 95 NGESGCLKINTDGASRGNPGLATAGGVLQDNEGRWCGGFSLNIGRSSAPMAELWGAYYGL 154
Query: 257 RHALKKGFTSICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHV 316
A ++ + I ++ DS++V +K G+ + LS L ++ K +I HV
Sbjct: 155 YLAWERKSSHIELEVDSEIVVGFLK--TGI--SDHHPLSFLVRLCHGFISKDWRVRIFHV 210
Query: 317 LRDLNSEADAQANLAVNL 334
R+ N AD AN +L
Sbjct: 211 YREANRFADGLANYVFSL 228
>At3g31530 hypothetical protein
Length = 831
Score = 46.2 bits (108), Expect = 3e-05
Identities = 37/110 (33%), Positives = 58/110 (52%), Gaps = 10/110 (9%)
Query: 226 SKDGNLI-YRVREGVGIATNNVAEYRAMILGMRHA--LKKGFT-SICIQGDSKLVCMQMK 281
SK G+ + R+ G ATNNVAEY A++ G+ A LK G T + C DS+L+
Sbjct: 423 SKQGSGVGIRLTSPTGEATNNVAEYEALVAGLNLAWGLKIGKTRAFC---DSQLIA---N 476
Query: 282 QIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNSEADAQANLA 331
Q +G + +++ + + L F F+++ + R N+ ADA A LA
Sbjct: 477 QFNGEYTTQDKKMEAYLIHVQNLAKNFDEFELTRIPRGENTSADALAALA 526
>At2g07730 putative non-LTR retroelement reverse transcriptase
Length = 970
Score = 44.3 bits (103), Expect = 1e-04
Identities = 40/146 (27%), Positives = 64/146 (43%), Gaps = 5/146 (3%)
Query: 208 DGASKGNPGKAGAGAILRSKDGNLIYRVREGVGIATNNVAEYRAMILGMRHALKKGFTSI 267
DGAS+G+ G A A + + G + +G +AE G+ A KGF +
Sbjct: 815 DGASRGHQGLAAASGAILNLQGEWLGGFALNIGSCDAPLAELWGAYYGLLIAWDKGFRRV 874
Query: 268 CIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNSEADAQ 327
+ DS+LV + G+ K LS L ++ + + ++SHV R+ N AD
Sbjct: 875 ELNLDSELVVGFLS--TGISKA--HPLSFLVRLCQGFFTRDWLVRVSHVYREANRLADGL 930
Query: 328 ANLAVNLAGEAFKIINVCSSLMYMYL 353
AN A L F +C + ++L
Sbjct: 931 ANYAFFLP-LGFHCFEICPEDVLLFL 955
>At1g27870 hypothetical protein
Length = 213
Score = 42.0 bits (97), Expect = 5e-04
Identities = 20/72 (27%), Positives = 41/72 (56%), Gaps = 3/72 (4%)
Query: 207 FDGASKGNPGKAGAGAILRSKDGNLIYRVREGVGIATNNV--AEYRAMILGMRHALKKGF 264
FDG+ K+ AG ++R +G+ + + +G +N +E +A+I+ M+H G+
Sbjct: 64 FDGSFVNGDVKSKAGWVVRDSNGSYLL-AGQAIGRKVDNALESEIQALIISMQHCWSHGY 122
Query: 265 TSICIQGDSKLV 276
+C +GD+K++
Sbjct: 123 KRVCFEGDNKML 134
>At3g25270 hypothetical protein
Length = 343
Score = 40.8 bits (94), Expect = 0.001
Identities = 22/72 (30%), Positives = 42/72 (57%), Gaps = 3/72 (4%)
Query: 207 FDGASKGNPGKAGAGAILRSKDGNLIYRVREGVGIATNNV--AEYRAMILGMRHALKKGF 264
+DGA A AG ++R ++G + + +G T++ +E++A+I+ M+HA +G+
Sbjct: 198 YDGAFNHQTRNAKAGWLMRDENG-VYMGSGQAIGSTTSDSLESEFQALIIAMQHAWSQGY 256
Query: 265 TSICIQGDSKLV 276
+ +GDSK V
Sbjct: 257 RKVIFEGDSKQV 268
>At4g08830 putative protein
Length = 947
Score = 40.4 bits (93), Expect = 0.001
Identities = 23/69 (33%), Positives = 38/69 (54%)
Query: 208 DGASKGNPGKAGAGAILRSKDGNLIYRVREGVGIATNNVAEYRAMILGMRHALKKGFTSI 267
DGAS+GN G A G +LR G+ +G+ + +AE + G+ A ++ FT +
Sbjct: 854 DGASRGNLGLATTGGVLRDGIGHWCGGFALDIGVCSAPLAELWGVYYGLYMAWERRFTRV 913
Query: 268 CIQGDSKLV 276
++ DS+LV
Sbjct: 914 ELEVDSELV 922
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.131 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,471,460
Number of Sequences: 26719
Number of extensions: 305747
Number of successful extensions: 925
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 34
Number of HSP's successfully gapped in prelim test: 20
Number of HSP's that attempted gapping in prelim test: 879
Number of HSP's gapped (non-prelim): 58
length of query: 357
length of database: 11,318,596
effective HSP length: 100
effective length of query: 257
effective length of database: 8,646,696
effective search space: 2222200872
effective search space used: 2222200872
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146308.4


