

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144502.11 + phase: 0
(675 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
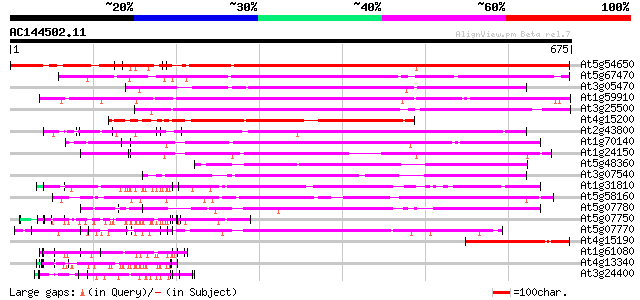
Score E
Sequences producing significant alignments: (bits) Value
At5g54650 unknown protein 689 0.0
At5g67470 formin-like protein 407 e-114
At3g05470 unknown protein 403 e-112
At1g59910 hypothetical protein 398 e-111
At3g25500 formin-like protein AHF1 395 e-110
At4g15200 p140mDia like protein 386 e-107
At2g43800 unknown protein 364 e-100
At1g70140 hypothetical protein 357 2e-98
At1g24150 unknown protein 290 2e-78
At5g48360 unknown protein 253 2e-67
At3g07540 putative protein 223 4e-58
At1g31810 hypothetical protein 215 7e-56
At5g58160 strong similarity to unknown protein (gb|AAD23008.1) 190 3e-48
At5g07780 putative protein 156 4e-38
At5g07750 putative protein 147 2e-35
At5g07770 unknown protein 144 2e-34
At4g15190 hypothetical protein 130 2e-30
At1g61080 hypothetical protein 124 1e-28
At4g13340 extensin-like protein 123 3e-28
At3g24400 protein kinase, putative 115 1e-25
>At5g54650 unknown protein
Length = 900
Score = 689 bits (1777), Expect = 0.0
Identities = 377/683 (55%), Positives = 463/683 (67%), Gaps = 53/683 (7%)
Query: 2 NDYSFGSSSNNLGDNKKTSLQGNQSMGAFAVVGSPFELNPPPGRVGTIHSGMPPLRPPPG 61
+DYS GSS N G + K QG+QS ++ G + G+ L
Sbjct: 260 SDYSVGSSIN-YGGSVKGDKQGHQSFNIYSNQGKMSSFD------GSNSDTSDSLE---- 308
Query: 62 RMNPLPHEPPSFTPFGNTAVSAAAPPPPRQSGVASARPPATPSPPQSLLAGAKPVPPRPP 121
L HE N + PPP R + V S + S +P+PP PP
Sbjct: 309 --ERLSHEGLRNNSITNHGLPPLKPPPGRTASVLSGK---------SFSGKVEPLPPEPP 357
Query: 122 LHPALPGAKPSPPPPPAPLGAKP---GPPPPPPPAPPSAKPGPPPPPPPAPSGAKPGPRP 178
+ K S PPPP P P GPP PPPPAPP GP PPPPP P G +P
Sbjct: 358 KFLKVSSKKASAPPPPVPAPQMPSSAGPPRPPPPAPPPGSGGPKPPPPPGPKGPRP---- 413
Query: 179 PPPPPKSGVAPPRPPIGPKAGGPKATENAEAGAEGGADTSKAKLKPFFWDKVPANSDQSM 238
PPP G PRPP GP A+A + D K KLKPFFWDKV AN + SM
Sbjct: 414 -PPPMSLGPKAPRPPSGP----------ADALDD---DAPKTKLKPFFWDKVQANPEHSM 459
Query: 239 VWNQIKSGSFQFNEEMIETLFGYNAVNKNNGQRQKESSSSQDPSPQYIQIVDKKKAQNLL 298
VWN I+SGSFQFNEEMIE+LFGY A +KN + K+ SS Q PQ++QI++ KK QNL
Sbjct: 460 VWNDIRSGSFQFNEEMIESLFGYAAADKN--KNDKKGSSGQAALPQFVQILEPKKGQNLS 517
Query: 299 ILLRALNVTMEEVCDALYEGNELPSEFLQTLLKMAPTSDEELKLRLFNGDLSQLGPADRF 358
ILLRALN T EEVCDAL EGNELP EF+QTLLKMAPT +EELKLRL+ G+++QLG A+RF
Sbjct: 518 ILLRALNATTEEVCDALREGNELPVEFIQTLLKMAPTPEEELKLRLYCGEIAQLGSAERF 577
Query: 359 LKAMVDIPSAFKRMEVLLFMCTFKEELTTTMESFAVLEVACKELRNSRLFHKLLEAVLKT 418
LKA+VDIP AFKR+E LLFMCT EE+ ESF LEVACKELR SRLF KLLEAVLKT
Sbjct: 578 LKAVVDIPFAFKRLEALLFMCTLHEEMAFVKESFQKLEVACKELRGSRLFLKLLEAVLKT 637
Query: 419 GNRMNDGTYRGGAQAFKLDTLLKLSDVKGTDGKTTLLHFVVQEIIRSEGIKAARAAKDSQ 478
GNRMNDGT+RGGAQAFKLDTLLKL+DVKGTDGKTTLLHFVVQEIIR+EG++AAR ++SQ
Sbjct: 638 GNRMNDGTFRGGAQAFKLDTLLKLADVKGTDGKTTLLHFVVQEIIRTEGVRAARTIRESQ 697
Query: 479 SLSNIKTDEL------HETEDHYRELGLEMVSHLSTELENVKRGSVLDADSLTATTIKLG 532
S S++KT++L E+E++YR LGLE VS LS+ELE+VK+ + +DAD LT T +K+G
Sbjct: 698 SFSSVKTEDLLVEETSEESEENYRNLGLEKVSGLSSELEHVKKSANIDADGLTGTVLKMG 757
Query: 533 HGLVKAKDILNKNLKNVEDDRGFRETVESFVKNAEADVKKLLEDEKKIMALVKSTGDYFH 592
H L KA+D +N +K+ ++ GFRE +E F++NAE + +LE+EK+IMALVKSTGDYFH
Sbjct: 758 HALSKARDFVNSEMKSSGEESGFREALEDFIQNAEGSIMSILEEEKRIMALVKSTGDYFH 817
Query: 593 GNATKDDGLRLFVVVRDFLIMLDKVCKEVRDAQKKPAKPIKQETSRGLSSSDT-RPSPS- 650
G A KD+GLRLFV+VRDFLI+LDK CKEVR+A+ +P + +++ S +SS+T R +PS
Sbjct: 818 GKAGKDEGLRLFVIVRDFLIILDKSCKEVREARGRPVRMARKQGSTASASSETPRQTPSL 877
Query: 651 DFRQRLFPAIAERRIDDDSSDEE 673
D RQ+LFPAI ERR+D SSD +
Sbjct: 878 DPRQKLFPAITERRVDQSSSDSD 900
Score = 48.1 bits (113), Expect = 2e-05
Identities = 25/60 (41%), Positives = 27/60 (44%), Gaps = 7/60 (11%)
Query: 136 PPAPLGAKPGPPPPPPPA-PPSAKPGPPPPP------PPAPSGAKPGPRPPPPPPKSGVA 188
P L KPG P P P+ PP GPP PP PP S P PPPPP K +
Sbjct: 134 PRRNLATKPGSSPSPSPSRPPKRSRGPPRPPTRPKSPPPRKSSFPPSRSPPPPPAKKNAS 193
Score = 47.0 bits (110), Expect = 3e-05
Identities = 24/62 (38%), Positives = 27/62 (42%), Gaps = 5/62 (8%)
Query: 127 PGAKPSPPPPPAPLGAKPGPPPP-----PPPAPPSAKPGPPPPPPPAPSGAKPGPRPPPP 181
PG+ PSP P P ++ P PP PPP S P PPPPPA A P
Sbjct: 142 PGSSPSPSPSRPPKRSRGPPRPPTRPKSPPPRKSSFPPSRSPPPPPAKKNASKNSTSAPV 201
Query: 182 PP 183
P
Sbjct: 202 SP 203
Score = 40.0 bits (92), Expect = 0.004
Identities = 23/73 (31%), Positives = 30/73 (40%), Gaps = 7/73 (9%)
Query: 96 SARPPATPSPPQSLLAGAKPVPPRPPLHPALPGAKPSPPPPPAPLGAKPGPPPPPPPAPP 155
+ +P ++PSP S PPRPP +P PPP PPPPP
Sbjct: 139 ATKPGSSPSPSPSRPPKRSRGPPRPP-------TRPKSPPPRKSSFPPSRSPPPPPAKKN 191
Query: 156 SAKPGPPPPPPPA 168
++K P PA
Sbjct: 192 ASKNSTSAPVSPA 204
Score = 32.7 bits (73), Expect = 0.68
Identities = 21/77 (27%), Positives = 29/77 (37%), Gaps = 15/77 (19%)
Query: 77 GNTAVSAAAPPPPRQSGVASARPPATPSPPQSLLAGAKPVPPRPPLHPALPGAKPSPPPP 136
G++ + + PP R G RPP P P PP + P ++ PPPP
Sbjct: 143 GSSPSPSPSRPPKRSRG--PPRPPTRPKSP-------------PPRKSSFPPSRSPPPPP 187
Query: 137 PAPLGAKPGPPPPPPPA 153
+K P PA
Sbjct: 188 AKKNASKNSTSAPVSPA 204
>At5g67470 formin-like protein
Length = 899
Score = 407 bits (1047), Expect = e-114
Identities = 267/657 (40%), Positives = 363/657 (54%), Gaps = 77/657 (11%)
Query: 59 PPGRMNPLPHEPPSF--TPFGNTAVSAAAPPPPRQS----GVASARPPATPSPPQSLLAG 112
P LPH + + FG+ +AA+ P + + PP PP L
Sbjct: 271 PRSANGSLPHSKRTSPRSKFGSAPTTAASRSPEMKHVIIPSIKQKLPPPVQPPPLRGLES 330
Query: 113 AKPVPPRPPLHPALPGAKPSPPPPPAPLGA---KPGPPPPPPPAPPSAKPGPPPPPPPAP 169
+ P P ++P PPP A A + P PPP +PP + PPPPPPP
Sbjct: 331 DEQELPYSQNKPKF--SQPPPPPNRAAFQAITQEKSPVPPPRRSPPPLQTPPPPPPPP-- 386
Query: 170 SGAKPGPRPPPPPPK--------------------SGVAPPRPPI----GPKAGGPKATE 205
P PPPPP+ S +P R PK +
Sbjct: 387 ------PLAPPPPPQKRPRDFQMLRKVTNSEATTNSTTSPSRKQAFKTPSPKTKAVEEVN 440
Query: 206 NAEAGA-----EGGADTSKAKLKPFFWDKVPANSDQSMVWNQIKSGSFQFNEEMIETLFG 260
+ AG+ +G D SK KLKP WDKV A+SD++ VW+Q+KS SFQ NE+ +E LFG
Sbjct: 441 SVSAGSLEKSGDGDTDPSKPKLKPLHWDKVRASSDRATVWDQLKSSSFQLNEDRMEHLFG 500
Query: 261 YNAVNKNNGQRQKESSSSQDPSPQYIQIVDKKKAQNLLILLRALNVTMEEVCDALYEGN- 319
N+ ++ ++ S + +++D KK+QN+ ILLRALNVT EEV +AL +GN
Sbjct: 501 CNS--GSSAPKEPVRRSVIPLAENENRVLDPKKSQNIAILLRALNVTREEVSEALTDGNP 558
Query: 320 -ELPSEFLQTLLKMAPTSDEELKLRLFNGDLSQLGPADRFLKAMVDIPSAFKRMEVLLFM 378
L +E L+TL+KMAPT +EE+KLR ++GD+S+LG A+RFLK ++DIP AFKR+E +L+
Sbjct: 559 ESLGAELLETLVKMAPTKEEEIKLREYSGDVSKLGTAERFLKTILDIPFAFKRVEAMLYR 618
Query: 379 CTFKEELTTTMESFAVLEVACKELRNSRLFHKLLEAVLKTGNRMNDGTYRGGAQAFKLDT 438
F E+ SF LE A EL+ SRLF KLLEAVL TGNRMN GT RG A AFKLDT
Sbjct: 619 ANFDAEVKYLRNSFQTLEEASLELKASRLFLKLLEAVLMTGNRMNVGTNRGDAIAFKLDT 678
Query: 439 LLKLSDVKGTDGKTTLLHFVVQEIIRSEGIKAARAAKDSQSLSNIKTDELHETEDHYREL 498
LLKL D+KG DGKTTLLHFVVQEI RSEG KD LH D +R+
Sbjct: 679 LLKLVDIKGVDGKTTLLHFVVQEITRSEG---TTTTKDETI--------LHGNNDGFRKQ 727
Query: 499 GLEMVSHLSTELENVKRGSVLDADSLTATTIKLGHGLVKAKDILNKNLKNVEDDRGFRET 558
GL++V+ LS +L NVK+ + +D D L++ KL GL D L LK F ++
Sbjct: 728 GLQVVAGLSRDLVNVKKSAGMDFDVLSSYVTKLEMGL----DKLRSFLKTETTQGRFFDS 783
Query: 559 VESFVKNAEADVKKLLEDEKKIMALVKSTGDYFHGNATKDDG--LRLFVVVRDFLIMLDK 616
+++F+K AE +++K+ E+K +++VK +YFHGNA +++ LR+F+VVRDFL +LD
Sbjct: 784 MKTFLKEAEEEIRKIKGGERKALSMVKEVTEYFHGNAAREEAHPLRIFMVVRDFLGVLDN 843
Query: 617 VCKEVRDAQKKPAKPIKQETSRGLSSSDTRPSPSDFRQRLFPAIAERRIDDDSSDEE 673
VCKEV+ Q+ + + ++R S T P R + R DD SSD E
Sbjct: 844 VCKEVKTMQEM-STSMGSASARSFRISATASLPVLHRYK-------ARQDDTSSDSE 892
Score = 39.3 bits (90), Expect = 0.007
Identities = 23/65 (35%), Positives = 28/65 (42%), Gaps = 5/65 (7%)
Query: 101 ATPSPPQSLLAGAKPVPPRP-----PLHPALPGAKPSPPPPPAPLGAKPGPPPPPPPAPP 155
+TP PP + P+P P P +P+ P PPPPP G P P
Sbjct: 40 STPPPPDFQSTPSPPLPDTPDQPFFPENPSTPQQTLFPPPPPPVSADVNGGLPIPTATTQ 99
Query: 156 SAKPG 160
SAKPG
Sbjct: 100 SAKPG 104
Score = 34.7 bits (78), Expect = 0.18
Identities = 20/51 (39%), Positives = 23/51 (44%), Gaps = 4/51 (7%)
Query: 139 PLGAKPGPPPPP----PPAPPSAKPGPPPPPPPAPSGAKPGPRPPPPPPKS 185
PL + PPPP P+PP P P PS + PPPPPP S
Sbjct: 34 PLFPESSTPPPPDFQSTPSPPLPDTPDQPFFPENPSTPQQTLFPPPPPPVS 84
Score = 34.3 bits (77), Expect = 0.23
Identities = 24/71 (33%), Positives = 28/71 (38%), Gaps = 8/71 (11%)
Query: 124 PALPGAKPSPPP-----PPAPLGAKPGPP-PPPPPAPPSAKPGPPPPPPPAP--SGAKPG 175
P P + PPP P PL P P P P+ P PPPPPP + +G P
Sbjct: 34 PLFPESSTPPPPDFQSTPSPPLPDTPDQPFFPENPSTPQQTLFPPPPPPVSADVNGGLPI 93
Query: 176 PRPPPPPPKSG 186
P K G
Sbjct: 94 PTATTQSAKPG 104
Score = 33.9 bits (76), Expect = 0.30
Identities = 21/65 (32%), Positives = 23/65 (35%), Gaps = 6/65 (9%)
Query: 132 SPPPPPAPLGAKPGPPPPPPPAPPSAKPGPPPP------PPPAPSGAKPGPRPPPPPPKS 185
S PPP + P PP P P P P P PPP P A P P +
Sbjct: 39 SSTPPPPDFQSTPSPPLPDTPDQPFFPENPSTPQQTLFPPPPPPVSADVNGGLPIPTATT 98
Query: 186 GVAPP 190
A P
Sbjct: 99 QSAKP 103
Score = 33.5 bits (75), Expect = 0.40
Identities = 23/70 (32%), Positives = 31/70 (43%), Gaps = 8/70 (11%)
Query: 147 PPPPPPAPPSAKPGPPPPPPPAPSGAKPG-PRPPPPPPKSGVAPPRPPIGPKAGG----P 201
P P PP + P PP P P +P P P P ++ PP PP+ G P
Sbjct: 37 PESSTPPPPDFQSTPSPPLPDTPD--QPFFPENPSTPQQTLFPPPPPPVSADVNGGLPIP 94
Query: 202 KA-TENAEAG 210
A T++A+ G
Sbjct: 95 TATTQSAKPG 104
Score = 33.1 bits (74), Expect = 0.52
Identities = 23/70 (32%), Positives = 26/70 (36%), Gaps = 14/70 (20%)
Query: 117 PPRPPLHPALPGAKPSPPPPPAP----LGAKPGPP-----PPPPPAPPSAKPGPPPPPPP 167
PP P + PSPP P P P P PPPPP + G P P
Sbjct: 42 PPPPDFQ-----STPSPPLPDTPDQPFFPENPSTPQQTLFPPPPPPVSADVNGGLPIPTA 96
Query: 168 APSGAKPGPR 177
AKPG +
Sbjct: 97 TTQSAKPGKK 106
>At3g05470 unknown protein
Length = 884
Score = 403 bits (1036), Expect = e-112
Identities = 231/530 (43%), Positives = 317/530 (59%), Gaps = 72/530 (13%)
Query: 140 LGAKPGPPPPPPPAP------------------------------------------PSA 157
+ A P PPPPPPP P P++
Sbjct: 365 VSAPPPPPPPPPPLPQFSNKRIHTLSSPETANLQTLSSQLCEKLCASSSKTSFPINVPNS 424
Query: 158 KPGPPPPPPPAPSGAKPGPRPPPPPPKSGVAPPRPPIGPKAGGPKATENAEAGAEGGADT 217
+P PPPPPPP G PPPP S R P+G K G P
Sbjct: 425 QPRPPPPPPPPQQLQVAGINKTPPPPLSLDFSERRPLG-KDGAP---------------- 467
Query: 218 SKAKLKPFFWDKVPANSDQSMVWNQIKSGSFQFNEEMIETLFGYNAVNKNNGQRQKESSS 277
KLKP WDKV A D++MVW+++++ SF+ +EEMIE+LFGY + E
Sbjct: 468 -LPKLKPLHWDKVRATPDRTMVWDKLRTSSFELDEEMIESLFGYTM----QSSTKNEEGK 522
Query: 278 SQDPSPQYIQIVDKKKAQNLLILLRALNVTMEEVCDALYEGNELPSEFLQTLLKMAPTSD 337
S+ PSP +++ K+ QN ILL+ALN T +++C AL +G L + L+ L+KM PT +
Sbjct: 523 SKTPSPGK-HLLEPKRLQNFTILLKALNATADQICSALGKGEGLCLQQLEALVKMVPTKE 581
Query: 338 EELKLRLFNGDLSQLGPADRFLKAMVDIPSAFKRMEVLLFMCTFKEELTTTMESFAVLEV 397
EELKLR + G + +LG A++FL+A+V +P AF+R E +L+ TF++E+ SF++LE
Sbjct: 582 EELKLRSYKGAVDELGSAEKFLRALVGVPFAFQRAEAMLYRETFEDEVVHLRNSFSMLEE 641
Query: 398 ACKELRNSRLFHKLLEAVLKTGNRMNDGTYRGGAQAFKLDTLLKLSDVKGTDGKTTLLHF 457
ACKEL++SRLF KLLEAVLKTGNRMN GT RGGA+AFKLD LLKLSDVKGTDGKTTLLHF
Sbjct: 642 ACKELKSSRLFLKLLEAVLKTGNRMNVGTIRGGAKAFKLDALLKLSDVKGTDGKTTLLHF 701
Query: 458 VVQEIIRSEGIKAARAAKD---SQSLSNIKTDELHETEDHYRELGLEMVSHLSTELENVK 514
VVQEI RSEGI+ + + +Q + +T E E E+ YR +GL++VS L+TEL NVK
Sbjct: 702 VVQEISRSEGIRVSDSIMGRIMNQRSNKNRTPE--EKEEDYRRMGLDLVSGLNTELRNVK 759
Query: 515 RGSVLDADSLTATTIKLGHGLVKAKDILNKNLKNVEDDRGFRETVESFVKNAEADVKKLL 574
+ + +D + L + L GL + + ++ LK E++R F ++ SF++ E +++L
Sbjct: 760 KTATIDLEGLVTSVSNLRDGLGQLSCLASEKLKGDEENRAFVSSMSSFLRYGEKSLEELR 819
Query: 575 EDEKKIMALVKSTGDYFHGNATKDD--GLRLFVVVRDFLIMLDKVCKEVR 622
EDEK+IM V +YFHG+ D+ LR+FV+VRDFL MLD VC+E+R
Sbjct: 820 EDEKRIMERVGEIAEYFHGDVRGDEKNPLRIFVIVRDFLGMLDHVCRELR 869
Score = 32.3 bits (72), Expect = 0.89
Identities = 15/41 (36%), Positives = 18/41 (43%)
Query: 127 PGAKPSPPPPPAPLGAKPGPPPPPPPAPPSAKPGPPPPPPP 167
P PSP P P+ + PP PP P PP + P P
Sbjct: 98 PAPSPSPFPNGGPIESPAYPPAPPRPIPPHLRRPLPQRTHP 138
Score = 30.0 bits (66), Expect = 4.4
Identities = 17/37 (45%), Positives = 19/37 (50%), Gaps = 7/37 (18%)
Query: 146 PPPPPPPAP-PSAKPGPPPPPPPAPSGAKPGPRPPPP 181
P P P P+P P+ P P PPAP PRP PP
Sbjct: 96 PWPAPSPSPFPNGGPIESPAYPPAP------PRPIPP 126
Score = 29.6 bits (65), Expect = 5.7
Identities = 19/50 (38%), Positives = 21/50 (42%), Gaps = 5/50 (10%)
Query: 131 PSPPPPPAPLGAKPGPPPPPPPAPPSAKPGPPPP--PPPAPSGAKPGPRP 178
P P P P+P P P PA P A P P PP P P P +P
Sbjct: 96 PWPAPSPSPF---PNGGPIESPAYPPAPPRPIPPHLRRPLPQRTHPLEQP 142
Score = 29.3 bits (64), Expect = 7.5
Identities = 17/37 (45%), Positives = 18/37 (47%), Gaps = 6/37 (16%)
Query: 156 SAKPGPPPPPPPAPSGAKPGPRPPPPPPKSGVAPPRP 192
S P P P P P P+G P P PP APPRP
Sbjct: 93 SFSPWPAPSPSPFPNGG-PIESPAYPP-----APPRP 123
>At1g59910 hypothetical protein
Length = 929
Score = 398 bits (1023), Expect = e-111
Identities = 262/681 (38%), Positives = 363/681 (52%), Gaps = 53/681 (7%)
Query: 36 PFELNPPPGRVGTIHSGMPPLRPPP-----------GRMNPLPHEPPSFTPFGNTAVSAA 84
P PPPG+ ++ P PPP PLP P P N +S +
Sbjct: 260 PSSTQPPPGQYMAGNASFPSSTPPPPGQYMAGNAPFSSSTPLP---PGQYPAVNAQLSTS 316
Query: 85 APPPPRQSGVASA--RPPATPSPPQSL-----LAGAKPVPPRPPLHPALPGAKPSPPPPP 137
AP P G +A P +T + P SL + G + PL P +PP PP
Sbjct: 317 APSVPLPPGQYTAVNAPFSTSTQPVSLPPGQYMPGNAALSASTPLTPGQFTTANAPPAPP 376
Query: 138 APLGAKPGPPPPPP-----PAPPSAKPGPPPPPPPAPSGAKPGPRPPPPPPKSGVAPPRP 192
P PPPPPP P PP K GP PPPP P G K G PPPPPP S PP+P
Sbjct: 377 GPANQTSPPPPPPPSAAAPPPPPPPKKGPAAPPPPPPPGKK-GAGPPPPPPMSKKGPPKP 435
Query: 193 PIGPKAGGPKATENAEAGAEGGADTSKAKLKPFFWDKVPANSDQSMVWNQIKSGSFQFNE 252
P PK GP + D ++ KLKP WDK+ ++ +SMVW++I GSF F+
Sbjct: 436 PGNPK--GPTKSGETSLAVGKTEDPTQPKLKPLHWDKMNPDASRSMVWHKIDGGSFNFDG 493
Query: 253 EMIETLFGYNA--VNKNNGQRQKESSSSQDPSPQYIQIVDKKKAQNLLILLRALNVTMEE 310
+++E LFGY A +++N Q ++ S+ P Q I+D +K+QN I+L++L +T EE
Sbjct: 494 DLMEALFGYVARKPSESNSVPQNQTVSNSVPHNQ-TYILDPRKSQNKAIVLKSLGMTKEE 552
Query: 311 VCDALYEGNELPSEFLQTLLKMAPTSDEELKLRLFNGDLSQLGPADRFLKAMVD-IPSAF 369
+ D L EG++ S+ L+ L +APT +E+ ++ F+G+ L AD L ++ +PSAF
Sbjct: 553 IIDLLTEGHDAESDTLEKLAGIAPTPEEQTEIIDFDGEPMTLAYADSLLFHILKAVPSAF 612
Query: 370 KRMEVLLFMCTFKEELTTTMESFAVLEVACKELRNSRLFHKLLEAVLKTGNRMNDGTYRG 429
R V+LF + E+ S LE AC ELR LF KLLEA+LK GNRMN GT RG
Sbjct: 613 NRFNVMLFKINYGSEVAQQKGSLLTLESACNELRARGLFMKLLEAILKAGNRMNAGTARG 672
Query: 430 GAQAFKLDTLLKLSDVKGTDGKTTLLHFVVQEIIRSEGIKAA----RAAKDSQSLSNIKT 485
AQAF L L KLSDVK D KTTLLHFVV+E++RSEG +AA + D+ S N
Sbjct: 673 NAQAFNLTALRKLSDVKSVDAKTTLLHFVVEEVVRSEGKRAAMNKNMMSSDNGSGENADM 732
Query: 486 DELHETEDHYRELGLEMVSHLSTELENVKRGSVLDADSLTATTIKLGHGLVKAKDILNKN 545
E E + ++GL ++ LS+E NVK+ + +D DS ATT+ LG + + K +L+++
Sbjct: 733 SR-EEQEIEFIKMGLPIIGGLSSEFTNVKKAAGIDYDSFVATTLALGTRVKETKRLLDQS 791
Query: 546 LKNVEDDRGFRETVESFVKNAEADVKKLLEDEKKIMALVKSTGDYFHGNATKDDGL-RLF 604
K ED G + SF ++AE ++K + E++ +IM LVK T +Y+ A K+ L +LF
Sbjct: 792 -KGKED--GCLTKLRSFFESAEEELKVITEEQLRIMELVKKTTNYYQAGALKERNLFQLF 848
Query: 605 VVVRDFLIMLDKVCKEVRDAQKKPAKPIKQETSRGLSSSDTR-PSPSDFRQR---LFPAI 660
V++RDFL M+D C E+ Q+K + T G SSS PS + QR FP +
Sbjct: 849 VIIRDFLGMVDNACSEIARNQRKQQQQRPATTVAGASSSPAETPSVAAAPQRNAVRFPIL 908
Query: 661 -------AERRIDDDSSDEES 674
+ R SD ES
Sbjct: 909 PPNFMSESSRYSSSSDSDSES 929
>At3g25500 formin-like protein AHF1
Length = 1051
Score = 395 bits (1015), Expect = e-110
Identities = 236/542 (43%), Positives = 325/542 (59%), Gaps = 42/542 (7%)
Query: 151 PPAPPSAKPGPPPPPPPAP---------SGAKPGPRPPP-PPPKSGVAPPRPPIGPKAGG 200
P S P PPPPPPP P + A RPP PP P + P
Sbjct: 518 PQILQSRVPPPPPPPPPLPLWGRRSQVTTKADTISRPPSLTPPSHPFVIPSENL-PVTSS 576
Query: 201 PKATENAEAGAEGGADTSKAKLKPFFWDKVPANSDQSMVWNQIKSGSFQFNEEMIETLFG 260
P T +E +T K KLK WDKV A+SD+ MVW+ ++S SF+ +EEMIETLF
Sbjct: 577 PMETPETVCASEAAEETPKPKLKALHWDKVRASSDREMVWDHLRSSSFKLDEEMIETLFV 636
Query: 261 YNAVNKNNGQRQKESSSSQDPSPQYIQIVDKKKAQNLLILLRALNVTMEEVCDALYEGNE 320
++N Q Q Q +++D KKAQN+ ILLRALNVT+EEVC+AL EGN
Sbjct: 637 AKSLNNKPNQSQTTPRCVLPSPNQENRVLDPKKAQNIAILLRALNVTIEEVCEALLEGNA 696
Query: 321 --LPSEFLQTLLKMAPTSDEELKLRLFNGDLS-QLGPADRFLKAMVDIPSAFKRMEVLLF 377
L +E L++LLKMAPT +EE KL+ +N D +LG A++FLKAM+DIP AFKR++ +L+
Sbjct: 697 DTLGTELLESLLKMAPTKEEERKLKAYNDDSPVKLGHAEKFLKAMLDIPFAFKRVDAMLY 756
Query: 378 MCTFKEELTTTMESFAVLEVACKELRNSRLFHKLLEAVLKTGNRMNDGTYRGGAQAFKLD 437
+ F+ E+ +SF LE AC+ELRNSR+F KLLEAVLKTGNRMN GT RG A AFKLD
Sbjct: 757 VANFESEVEYLKKSFETLEAACEELRNSRMFLKLLEAVLKTGNRMNVGTNRGDAHAFKLD 816
Query: 438 TLLKLSDVKGTDGKTTLLHFVVQEIIRSEGIKAARAAKDSQSLSNIKTDELHETEDHYRE 497
TLLKL DVKG DGKTTLLHFVVQEIIR+EG + S +N +TD++ R+
Sbjct: 817 TLLKLVDVKGADGKTTLLHFVVQEIIRAEGTRL--------SGNNTQTDDI-----KCRK 863
Query: 498 LGLEMVSHLSTELENVKRGSVLDADSLTATTIKLGHGLVKAKDIL--NKNLKNVEDDRGF 555
LGL++VS L +EL NVK+ + +D++ L++ KL G+ K + + + + + F
Sbjct: 864 LGLQVVSSLCSELSNVKKAAAMDSEVLSSYVSKLSQGIAKINEAIQVQSTITEESNSQRF 923
Query: 556 RETVESFVKNAEADVKKLLEDEKKIMALVKSTGDYFHGNATKDDG--LRLFVVVRDFLIM 613
E++++F+K AE ++ ++ E ++LVK +YFHGN+ K++ R+F+VVRDFL +
Sbjct: 924 SESMKTFLKRAEEEIIRVQAQESVALSLVKEITEYFHGNSAKEEAHPFRIFLVVRDFLGV 983
Query: 614 LDKVCKEVRDAQKKPAKPIKQETSRGLSSSDTRPSP-SDFRQRLFPAIAERRIDDDSSDE 672
+D+VCKEV ++ +SS+ P P + + P + RR SS
Sbjct: 984 VDRVCKEVGMINERTM----------VSSAHKFPVPVNPMMPQPLPGLVGRRQSSSSSSS 1033
Query: 673 ES 674
S
Sbjct: 1034 SS 1035
Score = 37.4 bits (85), Expect = 0.028
Identities = 29/96 (30%), Positives = 41/96 (42%), Gaps = 21/96 (21%)
Query: 68 HEPPSFTPFGNTAVSAAAPPPPRQSGVASARPPATPSPPQSLLAGAKPVPPRPPLHPALP 127
HEP F P + S +PPP + +S PP++ P S P PL+P+ P
Sbjct: 27 HEP--FFPIDSPPPSPPSPPPLPKLPFSSTTPPSSSDPNAS---------PFFPLYPSSP 75
Query: 128 GAKPSPPPPPAPLGAKPGP-----PPPPPPAPPSAK 158
PPP PA + P P +PP++K
Sbjct: 76 -----PPPSPASFASFPANISSLIVPHATKSPPNSK 106
Score = 37.0 bits (84), Expect = 0.036
Identities = 24/69 (34%), Positives = 27/69 (38%), Gaps = 10/69 (14%)
Query: 127 PGAKPSPPP-PPAPLGAKPGPPPPPPPAPP--SAKPGPPPPPPPAPSGAKPG-------P 176
P + PSPPP P P + P P A P P PPPP PA + P P
Sbjct: 37 PPSPPSPPPLPKLPFSSTTPPSSSDPNASPFFPLYPSSPPPPSPASFASFPANISSLIVP 96
Query: 177 RPPPPPPKS 185
PP S
Sbjct: 97 HATKSPPNS 105
Score = 37.0 bits (84), Expect = 0.036
Identities = 23/62 (37%), Positives = 26/62 (41%), Gaps = 8/62 (12%)
Query: 131 PSPPPPPAPLGAKPGPPPPPPPAPPSAKPGPPPPPPPAPSGAKP--GPRPPPPPPKSGVA 188
P PPP+P P PPP P P + PP P S P PPPP P S +
Sbjct: 32 PIDSPPPSP------PSPPPLPKLPFSSTTPPSSSDPNASPFFPLYPSSPPPPSPASFAS 85
Query: 189 PP 190
P
Sbjct: 86 FP 87
Score = 36.6 bits (83), Expect = 0.047
Identities = 19/50 (38%), Positives = 24/50 (48%), Gaps = 5/50 (10%)
Query: 115 PVPPRPPLHPALPGAKPSPPPPPAPLGAK-----PGPPPPPPPAPPSAKP 159
P PP PP P LP + +PP P + P PPPP PA ++ P
Sbjct: 38 PSPPSPPPLPKLPFSSTTPPSSSDPNASPFFPLYPSSPPPPSPASFASFP 87
Score = 31.6 bits (70), Expect = 1.5
Identities = 18/48 (37%), Positives = 20/48 (41%), Gaps = 3/48 (6%)
Query: 146 PPPPPPPAPPSAKPGPPPPPPPAPSGAKPGPRPPPPPPKSGVAPPRPP 193
P PPP+PPS PP P P S P P P + P PP
Sbjct: 32 PIDSPPPSPPS---PPPLPKLPFSSTTPPSSSDPNASPFFPLYPSSPP 76
>At4g15200 p140mDia like protein
Length = 645
Score = 386 bits (991), Expect = e-107
Identities = 223/369 (60%), Positives = 258/369 (69%), Gaps = 46/369 (12%)
Query: 120 PPLHPALPGAKPSPPPPPAPLGAKPGPPPPPPPAPPSAKPGPPPPPPPAPSGAKPGPRPP 179
PPL LP + +PPPPPA PPP PPP PP P P PPPPP + RPP
Sbjct: 299 PPLK--LPPGRSAPPPPPAA-----APPPQPPPPPP---PKPQPPPPPKIA------RPP 342
Query: 180 PPPPKSGVAPPRPPIGPKAGGPKATENAEAGAEGGADTSKAKLKPFFWDKVPANSDQSMV 239
P PPK G AP R G + G + ++E GA K KLKPFFWDK+ AN DQ MV
Sbjct: 343 PAPPK-GAAPKRQ--GNTSSGDASDVDSETGA------PKTKLKPFFWDKM-ANPDQKMV 392
Query: 240 WNQIKSGSFQFNEEMIETLFGYNAVNKN-NGQRQKESSSSQDPSPQYIQIVDKKKAQNLL 298
W++I +GSFQFNEE +E+LFGYN NKN NGQ+ +SS + P QYIQI+D +KAQNL
Sbjct: 393 WHEISAGSFQFNEEAMESLFGYNDGNKNKNGQKSTDSSLRESPL-QYIQIIDTRKAQNLS 451
Query: 299 ILLRALNVTMEEVCDALYEGNELPSEFLQTLLKMAPTSDEELKLRLFNGDLSQLGPADRF 358
ILLRALNVT EEV DA+ EGNELP E LQTLLKMAPTS+EELKLRL++GDL LGP
Sbjct: 452 ILLRALNVTTEEVVDAIKEGNELPVELLQTLLKMAPTSEEELKLRLYSGDLHLLGP---- 507
Query: 359 LKAMVDIPSAFKRMEVLLFMCTFKEELTTTMESFAVLEVACKELRNSRLFHKLLEAVLKT 418
R E LLFM + +EE++ E+ LEVACK+LRNSRLF KLLEAVLKT
Sbjct: 508 ------------RSESLLFMISLQEEVSGLKEALGTLEVACKKLRNSRLFLKLLEAVLKT 555
Query: 419 GNRMNDGTYRGGAQAFKLDTLLKLSDVKGTDGKTTLLHFVVQEIIRSEGIKAARAAKDSQ 478
GNRMN GT+RG AQAFKLDTLLKLSDVKGTDGKTTLLHFVV EIIRSEG++A R S+
Sbjct: 556 GNRMNVGTFRGDAQAFKLDTLLKLSDVKGTDGKTTLLHFVVLEIIRSEGVRALRL--QSR 613
Query: 479 SLSNIKTDE 487
S S++KTD+
Sbjct: 614 SFSSVKTDD 622
Score = 42.0 bits (97), Expect = 0.001
Identities = 25/64 (39%), Positives = 27/64 (42%), Gaps = 9/64 (14%)
Query: 127 PGAKPSPPPPPAP-------LGAKPGPPPPPPPAPPSAKPGPPPPPPP--APSGAKPGPR 177
PG PS P PAP L P PP P + P P P APS + PGP
Sbjct: 124 PGPGPSFAPGPAPNPRSYDWLAPASSPNEPPAETPDESSPSPSEETPSVVAPSQSVPGPP 183
Query: 178 PPPP 181
PPP
Sbjct: 184 RPPP 187
Score = 38.5 bits (88), Expect = 0.012
Identities = 29/72 (40%), Positives = 31/72 (42%), Gaps = 8/72 (11%)
Query: 128 GAKPSPPPPPAPLGAKPGPPPPPPPAPPSAKPGPPPPPPPA--PSGAKPGPRPPPP---- 181
G PS P P P A PGP P P A P P PPA P + P P P
Sbjct: 117 GPAPSFAPGPGPSFA-PGPAPNPRSYDWLA-PASSPNEPPAETPDESSPSPSEETPSVVA 174
Query: 182 PPKSGVAPPRPP 193
P +S PPRPP
Sbjct: 175 PSQSVPGPPRPP 186
Score = 33.9 bits (76), Expect = 0.30
Identities = 24/77 (31%), Positives = 28/77 (36%), Gaps = 7/77 (9%)
Query: 99 PPATPSPPQSLLAGAKPVPPRPPLHPALPGAKPSPPPPPAPLGAKPGPPPPPPP--APPS 156
P P P S G P P P + P+ PP P + P P P AP
Sbjct: 120 PSFAPGPGPSFAPG--PAPNPRSYDWLAPASSPNEPPAETPDESSPSPSEETPSVVAPSQ 177
Query: 157 AKPGPPPPPPPAPSGAK 173
+ PGPP P P K
Sbjct: 178 SVPGPP---RPPPQREK 191
Score = 33.9 bits (76), Expect = 0.30
Identities = 27/77 (35%), Positives = 33/77 (42%), Gaps = 33/77 (42%)
Query: 51 SGMPPLRPPPGRM------------NPLPHEPPSFTPFGNTAVSAAAPPPPRQSGVASAR 98
+G+PPL+ PPGR P P PP P PPPP+ AR
Sbjct: 296 AGLPPLKLPPGRSAPPPPPAAAPPPQPPPPPPPKPQP----------PPPPK-----IAR 340
Query: 99 PPATPSPPQSLLAGAKP 115
PP P+PP+ GA P
Sbjct: 341 PP--PAPPK----GAAP 351
Score = 31.6 bits (70), Expect = 1.5
Identities = 22/81 (27%), Positives = 30/81 (36%), Gaps = 13/81 (16%)
Query: 71 PSFTPFGNTAVSAAAPPPPRQSGVASARPPATPSPPQSLLAGAKPVPPRPPLHPALPGAK 130
PSF P + + P PR + P ++P+ P P + P
Sbjct: 120 PSFAPGPGPSFAPGPAPNPRSYDWLA--PASSPNEP-----------PAETPDESSPSPS 166
Query: 131 PSPPPPPAPLGAKPGPPPPPP 151
P AP + PGPP PPP
Sbjct: 167 EETPSVVAPSQSVPGPPRPPP 187
>At2g43800 unknown protein
Length = 894
Score = 364 bits (934), Expect = e-100
Identities = 245/606 (40%), Positives = 330/606 (54%), Gaps = 51/606 (8%)
Query: 41 PPPGRVGTIHS-------GMPPLRPPPGRMNPLPHEPPSFTPFGNTAVSAAAPPPPRQSG 93
PPP + S G P LRP P PLP + SFTP S P RQ
Sbjct: 266 PPPASSSSSSSYSQYHKLGSPELRPLP----PLP-KLQSFTPVYK---STEQLNPKRQDF 317
Query: 94 VASARPPATPSPPQSLLAGAKPVPPRPPLHPALP-------GAKPSPPPPPAP-LGAKPG 145
P+ +G K P R + G+ P AP L A PG
Sbjct: 318 DGDDNENDEFFSPRGS-SGRKQSPTRVSDVDQIDNRSINGSGSNSCSPTNFAPSLNASPG 376
Query: 146 PPPPP----PPAPPSAKPGPPPPPPPAPSGAKPGPRPPPPPPKSGVAPPRPPIGPKAGGP 201
P PP ++ P A+P P PPPPP S V A
Sbjct: 377 TSLKPKSISPPVSLHSQISSNNGIPKRLCPARPPPPPPPPPQVSEVP---------ATMS 427
Query: 202 KATENAEAGAEGGADTSKAKLKPFFWDKVPANSDQSMVWNQIKSGSFQFNEEMIETLFGY 261
+ ++ E +T K KLK WDKV A+S + MVW+QIKS SFQ NEEMIETLF
Sbjct: 428 HSLPGDDSDPEKKVETMKPKLKTLHWDKVRASSSRVMVWDQIKSNSFQVNEEMIETLFKV 487
Query: 262 NAVNKNNGQRQKESSSSQDPSPQYIQIVDKKKAQNLLILLRALNVTMEEVCDALYEGNE- 320
N +S S ++ + +D +K+ N+ ILLRALNVT +EVC+AL EGN
Sbjct: 488 NDPTSRTRDGVVQSVSQEN------RFLDPRKSHNIAILLRALNVTADEVCEALIEGNSD 541
Query: 321 -LPSEFLQTLLKMAPTSDEELKLRLF----NGDLSQLGPADRFLKAMVDIPSAFKRMEVL 375
L E L+ LLKMAPT +EE KL+ +G S++GPA++FLKA+++IP AFKR++ +
Sbjct: 542 TLGPELLECLLKMAPTKEEEDKLKELKDDDDGSPSKIGPAEKFLKALLNIPFAFKRIDAM 601
Query: 376 LFMCTFKEELTTTMESFAVLEVACKELRNSRLFHKLLEAVLKTGNRMNDGTYRGGAQAFK 435
L++ F+ E+ SF LE A EL+N+R+F KLLEAVLKTGNRMN GT RG A AFK
Sbjct: 602 LYIVKFESEIEYLNRSFDTLEAATGELKNTRMFLKLLEAVLKTGNRMNIGTNRGDAHAFK 661
Query: 436 LDTLLKLSDVKGTDGKTTLLHFVVQEIIRSEGIKAARAAKDSQSLSNIKTDELHETEDHY 495
LDTLLKL D+KG DGKTTLLHFVVQEII+ EG + S N+ + +
Sbjct: 662 LDTLLKLVDIKGADGKTTLLHFVVQEIIKFEGARVPFTPSQSHIGDNMAEQSAFQDDLEL 721
Query: 496 RELGLEMVSHLSTELENVKRGSVLDADSLTATTIKLGHGLVKAKDILNKNLKNVEDDRGF 555
++LGL++VS LS++L NVK+ + +D++SL T ++ G+ K K+++ + LK F
Sbjct: 722 KKLGLQVVSGLSSQLINVKKAAAMDSNSLINETAEIARGIAKVKEVITE-LKQETGVERF 780
Query: 556 RETVESFVKNAEADVKKLLEDEKKIMALVKSTGDYFHGNATKDDGLRLFVVVRDFLIMLD 615
E++ SF+ E ++ +L +M +VK +YFHGN ++ R+F VVRDFL +LD
Sbjct: 781 LESMNSFLNKGEKEITELQSHGDNVMKMVKEVTEYFHGN-SETHPFRIFAVVRDFLTILD 839
Query: 616 KVCKEV 621
+VCKEV
Sbjct: 840 QVCKEV 845
Score = 51.6 bits (122), Expect = 1e-06
Identities = 33/101 (32%), Positives = 41/101 (39%), Gaps = 17/101 (16%)
Query: 81 VSAAAPPPPRQSGVASARPPATPSPPQSLLAGAKPVPPRPPLHPALPGAKPSPPPPPAPL 140
V AAPPP +PP + PP P H +PPP L
Sbjct: 37 VVTAAPPP--------YQPPVSSQPPSP--------SPHTHHHHKKHLTTTTPPPHEKHL 80
Query: 141 GAKPGPPPPPPPAPPSAKPGPPPPPPPAPSGAKPGPRPPPP 181
+ PPPPPP+PP P P P + + + P P PPPP
Sbjct: 81 FSSVANPPPPPPSPPHPNPFFPSSDPTS-TASHPPPAPPPP 120
Score = 48.5 bits (114), Expect = 1e-05
Identities = 29/92 (31%), Positives = 36/92 (38%), Gaps = 4/92 (4%)
Query: 85 APPPPRQSGVASARPPATPSPPQSLLAGAKPVPPRPPLHPALPGAKPSPPPPPAPLGAKP 144
A PPP Q V+S P +P P P PPPPP+P P
Sbjct: 40 AAPPPYQPPVSSQPPSPSPHTHHHHKKHLTTTTPPPHEKHLFSSVANPPPPPPSP----P 95
Query: 145 GPPPPPPPAPPSAKPGPPPPPPPAPSGAKPGP 176
P P P + P++ PPP PP P+ P
Sbjct: 96 HPNPFFPSSDPTSTASHPPPAPPPPASLPTFP 127
Score = 44.7 bits (104), Expect = 2e-04
Identities = 29/97 (29%), Positives = 34/97 (34%), Gaps = 14/97 (14%)
Query: 124 PALPGAKPSPPPPPAPLGAKPGPPPP--------------PPPAPPSAKPGPPPPPPPAP 169
P P +PPP P+ ++P P P PPP PPPP P
Sbjct: 33 PFFPVVTAAPPPYQPPVSSQPPSPSPHTHHHHKKHLTTTTPPPHEKHLFSSVANPPPPPP 92
Query: 170 SGAKPGPRPPPPPPKSGVAPPRPPIGPKAGGPKATEN 206
S P P P P S + P P P A P N
Sbjct: 93 SPPHPNPFFPSSDPTSTASHPPPAPPPPASLPTFPAN 129
Score = 39.3 bits (90), Expect = 0.007
Identities = 31/104 (29%), Positives = 42/104 (39%), Gaps = 23/104 (22%)
Query: 59 PPGRMNPLPHEPPSFTPFGN----TAVSAAAPPPPRQSGVAS-ARPPATPSPPQSLLAGA 113
PP P+ +PPS +P + ++ PPP + +S A PP P
Sbjct: 42 PPPYQPPVSSQPPSPSPHTHHHHKKHLTTTTPPPHEKHLFSSVANPPPPP---------- 91
Query: 114 KPVPPRP-PLHPALPGAKPSPPPPPAPLGAKPGPPPPPPPAPPS 156
P PP P P P+ + PPPAP PPP P P+
Sbjct: 92 -PSPPHPNPFFPSSDPTSTASHPPPAP------PPPASLPTFPA 128
Score = 33.1 bits (74), Expect = 0.52
Identities = 26/81 (32%), Positives = 30/81 (36%), Gaps = 8/81 (9%)
Query: 41 PPPGRVGTIHSGMPPLRPPPGRMNPLPHEPPSFTPFGNTAVSAAAPPPPRQSGVASARPP 100
PPP S P PPP +P P P S P + APPPP A P
Sbjct: 73 PPPHEKHLFSSVANPPPPPPSPPHPNPFFPSS-DPTSTASHPPPAPPPP-------ASLP 124
Query: 101 ATPSPPQSLLAGAKPVPPRPP 121
P+ SLL +PP
Sbjct: 125 TFPANISSLLFPTHNKQSKPP 145
>At1g70140 hypothetical protein
Length = 760
Score = 357 bits (915), Expect = 2e-98
Identities = 213/526 (40%), Positives = 303/526 (57%), Gaps = 42/526 (7%)
Query: 135 PPPAPLGAKPGPPPPPPPAPPSAKPGPPPPPPPAPSGAKPGPRPPPPPPKSGV---APPR 191
PPP ++P PPPPPP P PPPP G+ P P PPPP K G + +
Sbjct: 227 PPPQVKQSEPTPPPPPPSIAVKQSAPTPSPPPPIKKGSSPSPPPPPPVKKVGALSSSASK 286
Query: 192 PPIGPKAGGPKATENAEAGAEGGADTSKAKLKPFFWDKVPANSDQSMVWNQIKSGSFQFN 251
PP P G A GG + + KLKP WDKV +SD SMVW++I GSF F+
Sbjct: 287 PPPAPVRG-----------ASGGETSKQVKLKPLHWDKVNPDSDHSMVWDKIDRGSFSFD 335
Query: 252 EEMIETLFGYNAVNKNNGQRQKESSSSQDPSPQYIQIVDKKKAQNLLILLRALNVTMEEV 311
+++E LFGY AV G++ E ++P I I+D +K+QN I+L++L +T EE+
Sbjct: 336 GDLMEALFGYVAV----GKKSPEQGDEKNPKSTQIFILDPRKSQNTAIVLKSLGMTREEL 391
Query: 312 CDALYEGNELPSEFLQTLLKMAPTSDEELKLRLFNGDLSQLGPADRFLKAMV-DIPSAFK 370
++L EGN+ + L+ L ++APT +E+ + F+GD ++L A+ FL ++ +P+AF
Sbjct: 392 VESLIEGNDFVPDTLERLARIAPTKEEQSAILEFDGDTAKLADAETFLFHLLKSVPTAFT 451
Query: 371 RMEVLLFMCTFKEELTTTMESFAVLEVACKELRNSRLFHKLLEAVLKTGNRMNDGTYRGG 430
R+ LF + E+ + L++ACKELR+ LF KLLEA+LK GNRMN GT RG
Sbjct: 452 RLNAFLFRANYYPEMAHHSKCLQTLDLACKELRSRGLFVKLLEAILKAGNRMNAGTARGN 511
Query: 431 AQAFKLDTLLKLSDVKGTDGKTTLLHFVVQEIIRSEGIKAARAAKDSQSL---------- 480
AQAF L LLKLSDVK DGKT+LL+FVV+E++RSEG K + S SL
Sbjct: 512 AQAFNLTALLKLSDVKSVDGKTSLLNFVVEEVVRSEG-KRCVMNRRSHSLTRSGSSNYNG 570
Query: 481 --SNIKTDELHETEDHYRELGLEMVSHLSTELENVKRGSVLDADSLTATTIKLGHGLVKA 538
S+++ E E Y +LGL +V LS+E NVK+ + +D +++ AT L V+A
Sbjct: 571 GNSSLQVMSKEEQEKEYLKLGLPVVGGLSSEFSNVKKAACVDYETVVATCSALA---VRA 627
Query: 539 KDILNKNLKNVEDDRG--FRETVESFVKNAEADVKKLLEDEKKIMALVKSTGDYFHGNAT 596
KD + ED G F +T+ +F+ + E +VK +E+K+M LVK T DY+ A
Sbjct: 628 KD-AKTVIGECEDGEGGRFVKTMMTFLDSVEEEVKIAKGEERKVMELVKRTTDYYQAGAV 686
Query: 597 K--DDGLRLFVVVRDFLIMLDKVCKEV-RDAQ-KKPAKPIKQETSR 638
+ L LFV+VRDFL M+DKVC ++ R+ Q +K PI + R
Sbjct: 687 TKGKNPLHLFVIVRDFLAMVDKVCLDIMRNMQRRKVGSPISPSSQR 732
Score = 43.1 bits (100), Expect = 5e-04
Identities = 28/80 (35%), Positives = 35/80 (43%), Gaps = 8/80 (10%)
Query: 68 HEPPSFTPFGNTAVSAAAPPPPRQSGVASARPPATPSPPQSLLAGAKPVPPRPPLHPALP 127
H+PP S PPPP S +A + TPSPP + G+ P PP PP +
Sbjct: 225 HQPPP-----QVKQSEPTPPPPPPS-IAVKQSAPTPSPPPPIKKGSSPSPPPPPPVKKVG 278
Query: 128 G--AKPSPPPPPAPLGAKPG 145
+ S PPP GA G
Sbjct: 279 ALSSSASKPPPAPVRGASGG 298
Score = 37.4 bits (85), Expect = 0.028
Identities = 27/89 (30%), Positives = 39/89 (43%), Gaps = 5/89 (5%)
Query: 44 GRVGTIHSGM--PPLRPPPG--RMNPLPHEPPSFTPFGNTAVSAAAPPPPRQSGVASARP 99
GR T HS + +PPP + P P PP +A + + PPPP + G + + P
Sbjct: 211 GRSSTSHSVIHNEDHQPPPQVKQSEPTPPPPPPSIAVKQSAPTPS-PPPPIKKGSSPSPP 269
Query: 100 PATPSPPQSLLAGAKPVPPRPPLHPALPG 128
P P L+ + PP P+ A G
Sbjct: 270 PPPPVKKVGALSSSASKPPPAPVRGASGG 298
Score = 34.7 bits (78), Expect = 0.18
Identities = 19/57 (33%), Positives = 25/57 (43%), Gaps = 2/57 (3%)
Query: 41 PPPGRVGTIHSGMPPLRPPPGR--MNPLPHEPPSFTPFGNTAVSAAAPPPPRQSGVA 95
PPP + S P PPP + +P P PP G + SA+ PPP G +
Sbjct: 240 PPPPSIAVKQSAPTPSPPPPIKKGSSPSPPPPPPVKKVGALSSSASKPPPAPVRGAS 296
>At1g24150 unknown protein
Length = 725
Score = 290 bits (741), Expect = 2e-78
Identities = 187/542 (34%), Positives = 279/542 (50%), Gaps = 88/542 (16%)
Query: 143 KPGPPPPPPPAPP-SAKPGPPPPPPPAPSGAKPGPRPPPPPPK------SGVAPPRPPIG 195
KP P PPPPP PP K PPPPP P GP PPPPPP S A +PP
Sbjct: 238 KPDPTPPPPPPPPIPVKQSATPPPPPPPKLKNNGPSPPPPPPLKKTAALSSSASKKPPPA 297
Query: 196 PKAGGPKATENAEAGAEGGADTSKAKLKPFFWDKVPANSDQSMVWNQIKSGSFQFNEEMI 255
P+ + G + + KLKP WDKV +SD SMVW++I GSF F+ +++
Sbjct: 298 PRGS-----------SSGESSNGQVKLKPLHWDKVNPDSDHSMVWDKIDRGSFSFDGDLM 346
Query: 256 ETLFGYNAVNK---NNGQRQKESSSSQDPSPQYIQIVDKKKAQNLLILLRALNVTMEEVC 312
E LFGY AV K ++G +K SS+ SP I I+D +K+QN I+L++L +T +E+
Sbjct: 347 EALFGYVAVGKKSPDDGGDKKPSSA----SPAQIFILDPRKSQNTAIVLKSLGMTRDELV 402
Query: 313 DALYEGNELPSEFLQTLLKMAPTSDEELKLRLFNGDLSQLGPADRFLKAMVD-IPSAFKR 371
++L EG++ + L+ L ++APT +E+ + F+GD L A+ FL ++ +P AF R
Sbjct: 403 ESLMEGHDFHPDTLERLSRIAPTKEEQSAILQFDGDTKMLADAESFLFHLLKAVPCAFTR 462
Query: 372 MEVLLFMCTFKEELTTTMESFAVLEVACKELRNSRLFHKLLEAVLKTGNRMNDGTYRGGA 431
+ LLF + E++ ++ L++AC ELR+ LF
Sbjct: 463 LNALLFRANYYPEISNHNKNLQTLDLACTELRSRGLF----------------------- 499
Query: 432 QAFKLDTLLKLSDVKGTDGKTTLLHFVVQEIIRSEGIKAARAAKDSQSLSNIKTDEL--- 488
DGKTTLL+FVV+E++RSEG + + ++S S + +
Sbjct: 500 ---------------SVDGKTTLLNFVVEEVVRSEGKRCVLNRRTNRSFSRSSSSSISEV 544
Query: 489 ---HETEDHYRELGLEMVSHLSTELENVKRGSVLDADSLTATTIKLGHGLVKAKDILNKN 545
E E Y LGL +V LS+E NVK+ + +D D++ AT + L A+ +L ++
Sbjct: 545 ISKEEQEKEYLRLGLPVVGGLSSEFTNVKKAAAVDYDTVAATCLALTSRAKDARRVLAQS 604
Query: 546 LKNVEDDRGFRETVESFVKNAEADVKKLLEDEKKIMALVKSTGDYFHGNATK-DDGLRLF 604
+ ++ F + + F+ + E +VK E+EKK++ LVK T +Y+ A K + L LF
Sbjct: 605 EGDNKEGVRFVKKMNEFLDSVEEEVKLAKEEEKKVLELVKRTTEYYQAGAVKGKNPLHLF 664
Query: 605 VVVRDFLIMLDKVCKEV---------------RDAQKKPAKPIKQETSRGLSSSDTRPSP 649
V+VRDFL M+DKVC E+ R+A K P P + R S SD+ S
Sbjct: 665 VIVRDFLAMVDKVCVEIARNLQRRSSMGSTQQRNAVKFPVLPPNFMSDR--SRSDSGGSD 722
Query: 650 SD 651
SD
Sbjct: 723 SD 724
Score = 45.4 bits (106), Expect = 1e-04
Identities = 23/60 (38%), Positives = 27/60 (44%)
Query: 86 PPPPRQSGVASARPPATPSPPQSLLAGAKPVPPRPPLHPALPGAKPSPPPPPAPLGAKPG 145
PPPP V + P P PP+ G P PP P A + S PPPAP G+ G
Sbjct: 245 PPPPPPIPVKQSATPPPPPPPKLKNNGPSPPPPPPLKKTAALSSSASKKPPPAPRGSSSG 304
Score = 37.0 bits (84), Expect = 0.036
Identities = 21/58 (36%), Positives = 25/58 (42%), Gaps = 2/58 (3%)
Query: 41 PPPGRVGTIHSGMPPLRPPPGRMN--PLPHEPPSFTPFGNTAVSAAAPPPPRQSGVAS 96
PPP + S PP PPP N P P PP + SA+ PPP G +S
Sbjct: 246 PPPPPIPVKQSATPPPPPPPKLKNNGPSPPPPPPLKKTAALSSSASKKPPPAPRGSSS 303
>At5g48360 unknown protein
Length = 782
Score = 253 bits (646), Expect = 2e-67
Identities = 155/408 (37%), Positives = 248/408 (59%), Gaps = 53/408 (12%)
Query: 223 KPFFWDKVPANSDQSMVWNQIKSGSFQFNEEMIETLFGYNAVNKNNGQRQKESSSSQDPS 282
K F D+ P + + W +++S S + ++EM+ET+F N+ N +
Sbjct: 421 KQSFSDQPP----KQLHWERLRSSSSKLSKEMVETMFIANSSNPRD-------------L 463
Query: 283 PQYIQIVDKKKAQNLLILLRALNVTMEEVCDALYEGN--ELPSEFLQTLLKMAPTSDEEL 340
P Q++D +KAQN+ LL+ LN++ ++VC AL +G+ L +E L+ L ++AP+ +EE
Sbjct: 464 PIQNQVLDPRKAQNIATLLQLLNLSTKDVCQALLDGDCDVLGAELLECLSRLAPSKEEER 523
Query: 341 KLRLFNGDLSQLGPADRFLKAMVDIPSAFKRMEVLLFMCTFKEELTTTMESFAVLEVACK 400
KL+ F+ D S++GPA+RFLK ++ +P FKR++ LLF+ F E+ +SF+V++VAC+
Sbjct: 524 KLKSFS-DGSEIGPAERFLKELLHVPFVFKRVDALLFVANFHSEIKRLRKSFSVVQVACE 582
Query: 401 ELRNSRLFHKLLEAVLKTGNRMNDGTYR-GGAQAFKLDTLLKLSDVKGTDGKTTLLHFVV 459
ELRNSR+F LLEA+LKTGN M+ T R G A AFKLDTLLKL DVKG DG+++LLHFVV
Sbjct: 583 ELRNSRMFSILLEAILKTGNMMSVRTNRCGDADAFKLDTLLKLVDVKGLDGRSSLLHFVV 642
Query: 460 QEIIRSEGIKAARAAKDSQSLSNIKTDELHETEDHYRELGLEMVSHLSTELENVKRGSVL 519
QE+++SEG A LE + +L+TEL NVK+ + +
Sbjct: 643 QEMMKSEGSVRA----------------------------LEGIRNLNTELSNVKKSADI 674
Query: 520 DADSLTATTIKLGHGL--VKAKDILNKNLKNVEDD-RGFRETVESFVKNAEADVKKLLED 576
+ L + ++ GL ++A +L++ + D F+E + F+K A ++ K+
Sbjct: 675 EYGVLRSNVSRICQGLKNIEALLLLSEESGSYGDQWLKFKERMTRFLKTAAEEIVKIKIR 734
Query: 577 EKKIMALVKSTGDYFHGNATKD-DGLRLFVVVRDFLIMLDKVCKEVRD 623
E ++ ++ + FHG+A+K+ +R+F++VRDFL +LD+VCKE+ D
Sbjct: 735 ESSTLSALEEVTEQFHGDASKEGHTMRIFMIVRDFLSVLDQVCKEMGD 782
Score = 31.6 bits (70), Expect = 1.5
Identities = 15/31 (48%), Positives = 17/31 (54%), Gaps = 5/31 (16%)
Query: 144 PGPPPPPPPAPP-----SAKPGPPPPPPPAP 169
P P P P +PP S+ P PPPP PP P
Sbjct: 40 PLPLPLSPISPPFFPLESSPPSPPPPLPPTP 70
Score = 30.8 bits (68), Expect = 2.6
Identities = 13/23 (56%), Positives = 15/23 (64%), Gaps = 1/23 (4%)
Query: 135 PPPAPL-GAKPGPPPPPPPAPPS 156
PP PL + P PPPP PP PP+
Sbjct: 50 PPFFPLESSPPSPPPPLPPTPPT 72
Score = 30.0 bits (66), Expect = 4.4
Identities = 16/37 (43%), Positives = 19/37 (51%), Gaps = 6/37 (16%)
Query: 115 PVP-PRPPLHPALPGAKPSPPPPPAPLGAKPGPPPPP 150
P+P P P+ P + SPP PP PL PP PP
Sbjct: 40 PLPLPLSPISPPFFPLESSPPSPPPPL-----PPTPP 71
Score = 29.6 bits (65), Expect = 5.7
Identities = 15/32 (46%), Positives = 16/32 (49%), Gaps = 2/32 (6%)
Query: 137 PAPLGAKPGPPP--PPPPAPPSAKPGPPPPPP 166
P PL P PP P +PPS P PP PP
Sbjct: 40 PLPLPLSPISPPFFPLESSPPSPPPPLPPTPP 71
>At3g07540 putative protein
Length = 841
Score = 223 bits (567), Expect = 4e-58
Identities = 151/472 (31%), Positives = 249/472 (51%), Gaps = 75/472 (15%)
Query: 161 PPPPPPPAPSGAKPGPRPPPPPPKSGVAPPRPPIGPKAGGPKAT--ENAEAGAEGGADTS 218
PPP PP P P PPP + PP + G K + E ++ EG D
Sbjct: 427 PPPQRPP--------PAMPEPPP---LVPPSQSFMVQKSGKKLSFSELPQSCGEGTTDRP 475
Query: 219 KAKLKPFFWDKVPANSDQSMVWNQIKSGSFQFNEEMIETLFGYNAVNKNNGQRQKESSSS 278
K KLKP WDKV +S ++ W+++ YN+ N N+ +Q+ S
Sbjct: 476 KPKLKPLPWDKVRPSSRRTNTWDRLP----------------YNSSNANS--KQRSLSCD 517
Query: 279 QDPSPQYIQIVDKKKAQNLLILLRALNVTMEEVCDALYEGN--ELPSEFLQTLLKMAPTS 336
Q +++D +K+QN+ +LL L +T +VC AL +G+ L E L++L ++AP+
Sbjct: 518 LPMLNQESKVLDPRKSQNVAVLLTTLKLTTNDVCQALRDGHYDALGVELLESLARVAPSE 577
Query: 337 DEELKLRLFNGD-LSQLGPADRFLKAMVDIPSAFKRMEVLLFMCTFKEELTTTMESFAVL 395
+EE KL ++ D + +L P++RFLK ++++P FKR++ LL + +F ++ SF+V+
Sbjct: 578 EEEKKLISYSDDSVIKLAPSERFLKELLNVPFVFKRVDALLSVASFDSKVKHLKRSFSVI 637
Query: 396 EVACKELRNSRLFHKLLEAVLKTGNRMNDGTYRGGAQAFKLDTLLKLSDVKGTDGKTTLL 455
+ AC+ LRNSR+ +L+ A L+ G + G A FKL+ LL L D+K +DG+T++L
Sbjct: 638 QAACEALRNSRMLLRLVGATLEAGMK------SGNAHDFKLEALLGLVDIKSSDGRTSIL 691
Query: 456 HFVVQEIIRSEGIKAARAAKDSQSLSNIKTDELHETEDHYRELGLEMVSHLSTELENVKR 515
VVQ+I SEGIK GL++V +LS+ L + K+
Sbjct: 692 DSVVQKITESEGIK-----------------------------GLQVVRNLSSVLNDAKK 722
Query: 516 GSVLDADSLTATTIKLGHGLVKAKDILN--KNLKNVEDDR--GFRETVESFVKNAEADVK 571
+ LD + KL + K ++L + + E+ + FRE+V F++ A ++K
Sbjct: 723 SAELDYGVVRMNVSKLYEEVQKISEVLRLCEETGHSEEHQWWKFRESVTRFLETAAEEIK 782
Query: 572 KLLEDEKKIMALVKSTGDYFHGNATKDDG--LRLFVVVRDFLIMLDKVCKEV 621
K+ +E + VK +YFH + K++ L++FV+VRDFL +L+ VCK++
Sbjct: 783 KIEREEGSTLFAVKKITEYFHVDPAKEEAQLLKVFVIVRDFLKILEGVCKKM 834
Score = 38.9 bits (89), Expect = 0.009
Identities = 21/51 (41%), Positives = 22/51 (42%)
Query: 135 PPPAPLGAKPGPPPPPPPAPPSAKPGPPPPPPPAPSGAKPGPRPPPPPPKS 185
PP PL + PPPPP P P P P PA A PR P P S
Sbjct: 48 PPFFPLYSSTSPPPPPSPPQPLPPPAPTFATFPANISALVLPRSPKPQTPS 98
Score = 37.0 bits (84), Expect = 0.036
Identities = 24/56 (42%), Positives = 24/56 (42%), Gaps = 10/56 (17%)
Query: 120 PPLHPALPGAKPSPPP-PPAPLGAKPGPPPPPP----PAPPSAKPGPPPPPPPAPS 170
PP P P PPP PP PL PPP P PA SA P P P PS
Sbjct: 48 PPFFPLYSSTSPPPPPSPPQPL-----PPPAPTFATFPANISALVLPRSPKPQTPS 98
Score = 30.8 bits (68), Expect = 2.6
Identities = 22/76 (28%), Positives = 28/76 (35%), Gaps = 25/76 (32%)
Query: 69 EPPSFTPFGNTAVSAAAPPPPRQSGVASARPPATPSPPQSLLAGAKPVPPRPPLHPALPG 128
+P +F+P S+ +PPPP PSPPQ P+PP P P
Sbjct: 42 DPSTFSPPFFPLYSSTSPPPP-------------PSPPQ-------PLPPPAPTFATFPA 81
Query: 129 -----AKPSPPPPPAP 139
P P P P
Sbjct: 82 NISALVLPRSPKPQTP 97
>At1g31810 hypothetical protein
Length = 1201
Score = 215 bits (547), Expect = 7e-56
Identities = 183/655 (27%), Positives = 274/655 (40%), Gaps = 115/655 (17%)
Query: 41 PPPGRVGTIHSGMPPL-RPPPGRMNPLPHEPPSFTPFGNTAVSAAAPPPPRQSGVASARP 99
PPP S PPL +PPP R P P PPS + + PPPP G +
Sbjct: 576 PPPPPPLPSRSIPPPLAQPPPPRPPPPPPPPPSSRSIPSPSAPPPPPPPPPSFGSTGNKR 635
Query: 100 PATPSPPQSLLAGAKPVPPRPPLHPALPGAKPSPPPPPA---------PLGAKPGPPPPP 150
A P PP P PP P PA A P PPPPP P P PPPPP
Sbjct: 636 QAQPPPP--------PPPPPPTRIPAAKCAPPPPPPPPTSHSGSIRVGPPSTPPPPPPPP 687
Query: 151 ----------PPAPPSAKP-----GPPPPPPPAPSGAKPGPRPPP------PPPKSGV-- 187
PPAPP P G PPPPPP P P P PPP PPP G+
Sbjct: 688 PKANISNAPKPPAPPPLPPSSTRLGAPPPPPPPPLSKTPAPPPPPLSKTPVPPPPPGLGR 747
Query: 188 ----APP--------RPPIGPKAGGPKATENAEAGAEGGADTS---KAKLKPFFWDKVPA 232
PP PP P AG +A+ G T+ K LKP W KV
Sbjct: 748 GTSSGPPPLGAKGSNAPPPPPPAGRGRASLGLGRGRGVSVPTAAPKKTALKPLHWSKV-T 806
Query: 233 NSDQSMVW-------NQIKSGSFQFNEEMIETLFGYNAVNKNNGQRQKESSSSQDPSPQY 285
+ + +W NQ ++ +E +E+LF +AV+ ++ S P+
Sbjct: 807 RAAKGSLWADTQKQENQPRAPEIDISE--LESLF--SAVSDTTAKKSTGRRGSSISKPEK 862
Query: 286 IQIVDKKKAQNLLILLRALNVTMEEVCDALYEGNELPSEF--LQTLLKMAPTSDEELKLR 343
+Q+VD ++A N I+L + + + ++ A+ + L + ++ L+K PT +E LR
Sbjct: 863 VQLVDLRRANNCEIMLTKIKIPLPDMLSAVLALDSLALDIDQVENLIKFCPTKEEMELLR 922
Query: 344 LFNGDLSQLGPADRFLKAMVDIPSAFKRMEVLLFMCTFKEELTTTMESFAVLEVACKELR 403
+ GD LG ++ +P ++ V F TF ++ + A KE++
Sbjct: 923 NYTGDKEMLGKCEQ-------VPRIEAKLRVFGFKITFASQVEELKSCLNTINAATKEVK 975
Query: 404 NSRLFHKLLEAVLKTGNRMNDGTYRGGAQAFKLDTLLKLSDVKGTDGKTTLLHFVVQEII 463
S ++++ +L GN +N GT RG A FKLD+LLKLSD + + K TL+H++ + +
Sbjct: 976 ESAKLRQIMQTILTLGNALNQGTARGSAVGFKLDSLLKLSDTRARNNKMTLMHYLCKLV- 1034
Query: 464 RSEGIKAARAAKDSQSLSNIKTDELHETEDHYRELGLEMVSHLSTELENVKRGSVLDADS 523
+ L + D +H LE S + EL+ + A+
Sbjct: 1035 ----------GEKMPELLDFANDLVH----------LEAASKI--ELKTL-------AEE 1065
Query: 524 LTATTIKLGHGLVKAKDILNKNLKNVEDDRGFRETVESFVKNAEADVKKLLEDEKKIMAL 583
+ A T GL K + L + + GFR+ ++ F+ A+ +VK L ++
Sbjct: 1066 MQAAT----KGLEKVEQELMASENDGAISLGFRKVLKEFLDMADEEVKTLASLYSEVGRN 1121
Query: 584 VKSTGDYFHGNATKDDGLRLFVVVRDFLIMLDKVCKEVRDAQKKPAKPIKQETSR 638
S YF +D F V L + K + R+ +K A+ K++ +
Sbjct: 1122 ADSLSHYF----GEDPARCPFEQVTKILTLFMKTFIKSREENEKQAEAEKKKLEK 1172
Score = 105 bits (263), Expect = 6e-23
Identities = 69/186 (37%), Positives = 81/186 (43%), Gaps = 37/186 (19%)
Query: 41 PPPGRVGTIHSGMPPLRPPPGRMNPLPHEPPSF---TPFGNTAVSAAAPPPPRQSGVASA 97
PPP + T + P +PPP P PP F T F + PPPP + S
Sbjct: 487 PPPPPLFTSTTSFSPSQPPPP-----PPPPPLFMSTTSFSPSQPPPPPPPPPLFTSTTSF 541
Query: 98 RPPATPSPPQSLLAGAKPVPPRPPLHPALPGAKPSPPPPPAPLGAK----------PGPP 147
P P PP L P LH + P PPPPP PL ++ P P
Sbjct: 542 SPSQPPPPPP--LPSFSNRDPLTTLHQPINKTPPPPPPPPPPLPSRSIPPPLAQPPPPRP 599
Query: 148 PPPPPAPPSAK-----PGPPPPPPPAPS--------GAKPGPRPPPPP----PKSGVAPP 190
PPPPP PPS++ PPPPPPP PS A+P P PPPPP P + APP
Sbjct: 600 PPPPPPPPSSRSIPSPSAPPPPPPPPPSFGSTGNKRQAQPPPPPPPPPPTRIPAAKCAPP 659
Query: 191 RPPIGP 196
PP P
Sbjct: 660 PPPPPP 665
Score = 94.0 bits (232), Expect = 2e-19
Identities = 58/161 (36%), Positives = 68/161 (42%), Gaps = 33/161 (20%)
Query: 66 LPHEPPSFTPFGNTAVSAAAPPPPRQSGVASARPPATPSPPQSLLAGAKPVPPRPPLHPA 125
LP +PPS G+ PPPP + ++ +PS P P PP PPL +
Sbjct: 467 LPSDPPSS---GDHVTLLPPPPPPPPPPLFTSTTSFSPSQPP-------PPPPPPPLFMS 516
Query: 126 LPGAKPS---PPPPPAPL-----GAKPGPPPPPPPAPPSAK---------------PGPP 162
PS PPPPP PL P PPPPPP P + P PP
Sbjct: 517 TTSFSPSQPPPPPPPPPLFTSTTSFSPSQPPPPPPLPSFSNRDPLTTLHQPINKTPPPPP 576
Query: 163 PPPPPAPSGAKPGPRPPPPPPKSGVAPPRPPIGPKAGGPKA 203
PPPPP PS + P P PPPP+ PP PP P A
Sbjct: 577 PPPPPLPSRSIPPPLAQPPPPRPPPPPPPPPSSRSIPSPSA 617
Score = 93.2 bits (230), Expect = 4e-19
Identities = 69/210 (32%), Positives = 78/210 (36%), Gaps = 55/210 (26%)
Query: 33 VGSPFELNPPPGRVGTIHSGMPPLRPPPGRMNPLPHEPPSFTPFGNTAVSAAAPPPPRQS 92
V +P L P G + +PP PPP PP FT + + S PPPP
Sbjct: 461 VDNPLNLPSDPPSSGDHVTLLPPPPPPP--------PPPLFTSTTSFSPSQPPPPPPPPP 512
Query: 93 GVASARPPATPSPPQSLLAGAKPVPPRPPLHPALPGAKPSPPPPPAPLGA---------- 142
S + PP P PP PPL + PS PPPP PL +
Sbjct: 513 LFMSTTSFSPSQPP--------PPPPPPPLFTSTTSFSPSQPPPPPPLPSFSNRDPLTTL 564
Query: 143 -------KPGPPPPPPPAPPSAKPGP----PPPPPPAPSGAKPG----------PRPPPP 181
P PPPPPPP P + P P PPP PP P P P PPPP
Sbjct: 565 HQPINKTPPPPPPPPPPLPSRSIPPPLAQPPPPRPPPPPPPPPSSRSIPSPSAPPPPPPP 624
Query: 182 PPKSGVA--------PPRPPIGPKAGGPKA 203
PP G PP PP P P A
Sbjct: 625 PPSFGSTGNKRQAQPPPPPPPPPPTRIPAA 654
>At5g58160 strong similarity to unknown protein (gb|AAD23008.1)
Length = 1307
Score = 190 bits (482), Expect = 3e-48
Identities = 168/670 (25%), Positives = 283/670 (42%), Gaps = 119/670 (17%)
Query: 52 GMPPLRPPP----GRMNPLPHEPPSFTPFGNTAVSAAAPPPPRQSGVASARPPATPSPPQ 107
G P PPP + LP PP PPPP Q + PP P PP
Sbjct: 668 GQPARSPPPISNSDKKPALPRPPPP------------PPPPPMQHSTVTKVPP--PPPP- 712
Query: 108 SLLAGAKPVPPRPPLHPALPGAKPSPPPPPAPLGAKPG------PPPPPPPAPP-----S 156
A P PP P +H + P P PPPPPAP + PP PPAPP S
Sbjct: 713 -----APPAPPTPIVHTSSPPPPPPPPPPPAPPTPQSNGISAMKSSPPAPPAPPRLPTHS 767
Query: 157 AKPGPPPPPPPAPSGAKPGPR-PPPPPPKSGV-----APPRPPIGPKAGGPKATENAEAG 210
A P PP PPP P G P PPPPPPK G P PP GP ++
Sbjct: 768 ASPPPPTAPPPPPLGQTRAPSAPPPPPPKLGTKLSPSGPNVPPTPALPTGPLSSGKGRML 827
Query: 211 AEGGADTSKAKLKPFFWDKVPAN-------------------------------SDQSMV 239
++ KLKP+ W K+ D M+
Sbjct: 828 RVNLKNSPAKKLKPYHWLKLTRAVNGSLWAETQMSSEASKYALFILLSLISLMPPDSCMI 887
Query: 240 WNQ------IKSGSFQFNEEMIETLFGYNAVNKNNGQRQKESSSSQDPSPQYIQIVDKKK 293
N +++ E +E+LF +A + R SS+ P P+ +Q+++ ++
Sbjct: 888 SNSLILYLLVRAPDIDMTE--LESLFSASAPEQAGKSRL---DSSRGPKPEKVQLIEHRR 942
Query: 294 AQNLLILLRALNVTMEEVCDALY--EGNELPSEFLQTLLKMAPTSDEELKLRLFNGDLSQ 351
A N I+L + V ++++ +++ E + L ++ ++ L+K PT +E L+ + GD +
Sbjct: 943 AYNCEIMLSKVKVPLQDLTNSVLNLEESALDADQVENLIKFCPTREEMELLKGYTGDKDK 1002
Query: 352 LGPADRFLKAMVDIPSAFKRMEVLLFMCTFKEELTTTMESFAVLEVACKELRNSRLFHKL 411
LG + F M+ +P R+E L + +FK + T+ +++NS F ++
Sbjct: 1003 LGKCELFFLEMMKVP----RVETKLRVFSFKMQFTS-------------QVKNSEKFKRI 1045
Query: 412 LEAVLKTGNRMNDGTYRGGAQAFKLDTLLKLSDVKGTDGKTTLLHFVVQEIIRSEGIKAA 471
++ +L GN +N GT RG A FKLD+L KLS+ + + + TL+H++ K +
Sbjct: 1046 MQTILSLGNALNQGTARGAAVGFKLDSLPKLSETRARNNRMTLMHYL---------CKVS 1096
Query: 472 RAAKDSQSLSNIKTDELHETEDHYRELG--LEMVSHLSTELENVKRGSVLDADSLTATTI 529
+ S ++ +E + D + L + V + EL +++ + + L
Sbjct: 1097 FYSLRFCSFVDVLEEERYSLMDSLQILAEKIPEVLDFTKELSSLEPATKIQLKFLAEEMQ 1156
Query: 530 KLGHGLVKAKDILNKNLKNVEDDRGFRETVESFVKNAEADVKKLLEDEKKIMALVKSTGD 589
+ GL K L+ + + F + ++ F+ AEA+V+ L + V
Sbjct: 1157 AINKGLEKVVQELSLSENDGPISHNFNKILKEFLHYAEAEVRSLASLYSGVGRNVDGLIL 1216
Query: 590 YFHGNATKDDGLRLFVVVRDFLIMLDKVCKE-----VRDAQKKPAKPIKQETSRGLSSSD 644
YF + K ++ + +F+ + ++ +E +A+K A+ K +T GL +
Sbjct: 1217 YFGEDPAKCPFEQVVSTLLNFVRLFNRAHEENGKQLEAEAKKNAAEKEKPKTG-GLDTEI 1275
Query: 645 TRPSPSDFRQ 654
+P + ++
Sbjct: 1276 KKPLNEEVKE 1285
Score = 40.8 bits (94), Expect = 0.002
Identities = 25/75 (33%), Positives = 33/75 (43%), Gaps = 6/75 (8%)
Query: 104 SPPQSLLAGAKPVPPRPPLHPALPGAKPSPPPPPAPLGAK------PGPPPPPPPAPPSA 157
SP L+ + V P + A+ + P P+ K P P PP P P A
Sbjct: 484 SPSLKLVHHSATVKPLVDDSKSPENAEENFPKSPSAHDGKAISFSPPTPSPPHPVRPQLA 543
Query: 158 KPGPPPPPPPAPSGA 172
+ G PPPPPP P+ A
Sbjct: 544 QAGAPPPPPPLPAAA 558
Score = 34.7 bits (78), Expect = 0.18
Identities = 21/65 (32%), Positives = 26/65 (39%), Gaps = 4/65 (6%)
Query: 130 KPSPPPPPAPLGAKPGPPPPPPPAPPSA----KPGPPPPPPPAPSGAKPGPRPPPPPPKS 185
KP +P A+ P P A P P PP P P A+ G PPPPP +
Sbjct: 497 KPLVDDSKSPENAEENFPKSPSAHDGKAISFSPPTPSPPHPVRPQLAQAGAPPPPPPLPA 556
Query: 186 GVAPP 190
+ P
Sbjct: 557 AASKP 561
Score = 33.5 bits (75), Expect = 0.40
Identities = 25/83 (30%), Positives = 30/83 (36%), Gaps = 2/83 (2%)
Query: 66 LPHEPPSFTPFGNTAVSAAAPPPPRQSGVASARPPATPSPPQSLLAGAKPVPPRPPLHPA 125
L E PS ++A + A P +PS P P PP HP
Sbjct: 480 LHQESPSLKLVHHSATVKPLVDDSKSPENAEENFPKSPSAHDGKAISFSPPTPSPP-HPV 538
Query: 126 LPG-AKPSPPPPPAPLGAKPGPP 147
P A+ PPPP PL A P
Sbjct: 539 RPQLAQAGAPPPPPPLPAAASKP 561
>At5g07780 putative protein
Length = 464
Score = 156 bits (394), Expect = 4e-38
Identities = 128/525 (24%), Positives = 219/525 (41%), Gaps = 91/525 (17%)
Query: 131 PSPPPPPAPLGAKPGPPPPPPPAPPSAKPGPPPPPPPAPSGAKPGPRPPPPPPKSGVAPP 190
P PPPPP PL + P PPPPP P + PPPPPPP P P PPPP P+ PP
Sbjct: 14 PLPPPPP-PLMRRRAPLPPPPPPPLMRRRAPPPPPPPLMRRRAPPPPPPPPLPRPCSRPP 72
Query: 191 RPPIGPKAGGPKATENAEAGAEGGADTSKAKLKPFFWDKVPANSDQSMVWNQIKSG---- 246
+ +K LKP W K S+ W++++
Sbjct: 73 K--------------------------TKCSLKPLHWVKKTRALPGSL-WDELQRRQECR 105
Query: 247 ---------SFQFNEEMIETLFGYNAVNKNNGQRQKESSSSQDPSPQYIQIVDKKKAQNL 297
+ + + IET+F A K P P+ + ++D ++A N
Sbjct: 106 DIEDEQILCAIELSVSEIETIFSLGAKPKPK------------PEPEKVPLIDLRRATNT 153
Query: 298 LILLRALNVTMEEVCDALYEGNEL---PSEFLQTLLKMAPTSDEELKLRLFNGDLSQLGP 354
I L LN+ + ++ A +E + ++ L+ + PT ++ L + GD
Sbjct: 154 EIRLMLLNIRLPDMIAAAMAMDESRLDDFDQIENLINLFPTKEDMKFLLTYTGDKGNCEQ 213
Query: 355 ADRFLKAMVDIPSAFKRMEVLLFMCTFKEELTTTMESFAVLEVACKELRNSRLFHKLLEA 414
++L+ +V +P ++ V F F ++T + + AC+E+R S+ ++E
Sbjct: 214 LFQYLQEVVKVPRVESKLRVFSFKIQFGTQITKLTKGLNAVNSACEEIRTSQKLKDIMEN 273
Query: 415 VLKTGNRMNDGTYRGGAQAFKLDTLLKLSDVKGTDGKTTLLHFVVQEIIRSEGIKAARAA 474
+L GN +N GT RG A F+LD+LL LS+ + + K TL+H++ + + A+
Sbjct: 274 ILCLGNILNQGTGRGRAVGFRLDSLLILSETRADNSKMTLMHYLCKVL----------AS 323
Query: 475 KDSQSLSNIKTDELHETEDHYRELGLEMVSHLSTELENVKRGSVLDADSLTATTIKLGHG 534
K S L + H+ +LE+++ S + SL + G
Sbjct: 324 KASDLL------DFHK------------------DLESLESASKIQLKSLAEEIQAITKG 359
Query: 535 LVKAKDILNKNLKNVEDDRGFRETVESFVKNAEADVKKLLEDEKKIMALVKSTGDYF-HG 593
L K K L + + + FR+ ++ F+ NAE V ++ ++ YF +
Sbjct: 360 LEKLKQELTASETDGPVSQVFRKLLKEFISNAETQVATVMALYYPARGNAEALAHYFGYH 419
Query: 594 NATKDDGLRLFVVVRDFLIMLDKVCKEVRDAQKKPAKPIKQETSR 638
+ L +R F ++ K+ +KK AK + E ++
Sbjct: 420 YPFEQVTATLLSFIRLFKKAHEENVKQAELEKKKAAKEAEMEKTK 464
Score = 44.7 bits (104), Expect = 2e-04
Identities = 29/74 (39%), Positives = 32/74 (43%), Gaps = 10/74 (13%)
Query: 86 PPPPRQSGVASARPPATPSPPQSLLAGAKPVPPRPPLHPALPGAKPSPPPPPAPLGAKPG 145
PPPP + PP P PP + A P PP P + P PPPPP PL P
Sbjct: 17 PPPPPLMRRRAPLPP--PPPPPLMRRRAPPPPPPPLMRRRAP-----PPPPPPPL---PR 66
Query: 146 PPPPPPPAPPSAKP 159
P PP S KP
Sbjct: 67 PCSRPPKTKCSLKP 80
Score = 38.1 bits (87), Expect = 0.016
Identities = 27/80 (33%), Positives = 30/80 (36%), Gaps = 13/80 (16%)
Query: 65 PLPHEPPSFTPFGNTAVSAAAPPPPRQSGVASARPPATPSPPQSLLAGAKPVPPRPPLHP 124
PLP PP A PPPP + PP P PP + A P PP PPL
Sbjct: 14 PLPPPPPPLM-----RRRAPLPPPPPPPLMRRRAPP--PPPPPLMRRRAPPPPPPPPL-- 64
Query: 125 ALPGAKPSPPPPPAPLGAKP 144
+P PP KP
Sbjct: 65 ----PRPCSRPPKTKCSLKP 80
Score = 34.3 bits (77), Expect = 0.23
Identities = 26/77 (33%), Positives = 33/77 (42%), Gaps = 12/77 (15%)
Query: 28 GAFAVVGSPFELNPPPGRVGTIHSGMPPLRPPP--GRMNPLPHEPPSFTPFGNTAVSAAA 85
GA+++V P PPP + + +PP PPP R P P PP A
Sbjct: 8 GAYSLVPLP----PPPPPLMRRRAPLPPPPPPPLMRRRAPPPPPPPLMRR------RAPP 57
Query: 86 PPPPRQSGVASARPPAT 102
PPPP +RPP T
Sbjct: 58 PPPPPPLPRPCSRPPKT 74
Score = 32.3 bits (72), Expect = 0.89
Identities = 38/137 (27%), Positives = 44/137 (31%), Gaps = 44/137 (32%)
Query: 55 PLRPPPG----RMNPLPHEPPSFTPFGNTAVSAAAPPPPRQSGVASARPPATPSPPQSLL 110
PL PPP R PLP PP + APPPP + R P P PP
Sbjct: 14 PLPPPPPPLMRRRAPLPPPPPP------PLMRRRAPPPPPPP-LMRRRAPPPPPPPPLPR 66
Query: 111 AGAKPVPPRPPLHP--------ALPGA-------------------------KPSPPPPP 137
++P + L P ALPG+ S
Sbjct: 67 PCSRPPKTKCSLKPLHWVKKTRALPGSLWDELQRRQECRDIEDEQILCAIELSVSEIETI 126
Query: 138 APLGAKPGPPPPPPPAP 154
LGAKP P P P P
Sbjct: 127 FSLGAKPKPKPEPEKVP 143
>At5g07750 putative protein
Length = 1289
Score = 147 bits (371), Expect = 2e-35
Identities = 102/284 (35%), Positives = 123/284 (42%), Gaps = 46/284 (16%)
Query: 41 PPPGRVGTIHSGMPPLRPPPGRMNPLPHEPPSFTPFGNTAVSAAAPPPPRQSGVASARPP 100
PPP +H G PP PPP P PP P + PPPP G PP
Sbjct: 1010 PPPPPPPPMHGGAPPPPPPPPMHGGAPPPPPP--PPMHGGAPPPPPPPPMHGGAPPPPPP 1067
Query: 101 -----ATPSPPQSLLAGAKPVPPRPPLHPALPGAKPSPPPPPAPLGAKPGPPPP-----P 150
A P PP + GA P PP P + G P PPPPP GA P PPPP P
Sbjct: 1068 PMFGGAQPPPPPPMRGGAPPPPP-----PPMRGGAPPPPPPPMRGGAPPPPPPPMHGGAP 1122
Query: 151 PPAPPSAKPGPPPPPPPAPSGAKPGPRPPPPPPKSGVAPPRPPIGPK--AGGP------- 201
PP PP + G PPPPPP P G PG PPPPPP P PP GP+ GGP
Sbjct: 1123 PPPPPPMRGGAPPPPPP-PGGRGPGAPPPPPPPGGRAPGPPPPPGPRPPGGGPPPPPMLG 1181
Query: 202 ---KATENAEAG--------AEGGADTSKAKLKPFFWDKVPANSDQSMVWNQI-KSGSFQ 249
A + AG G A K+ LKP W KV + Q +W+++ + G Q
Sbjct: 1182 ARGAAVDPRGAGRGRGLPRPGFGSAAQKKSSLKPLHWVKV-TRALQGSLWDELQRHGESQ 1240
Query: 250 ----FNEEMIETLFGYNAVNKNNGQRQKESSSSQDPSPQYIQIV 289
F+ IETLF +A + + S P+ +Q+V
Sbjct: 1241 TPSEFDVSEIETLF--SATVQKPADKSGSRRKSVGAKPEKVQLV 1282
Score = 133 bits (335), Expect = 3e-31
Identities = 80/184 (43%), Positives = 87/184 (46%), Gaps = 30/184 (16%)
Query: 34 GSPFELNPPPGRVGTIHSGMPPLRPPPGRMNPLPHEPPSFTPFGNTAVSAAAPPPPRQSG 93
GSP PPP G S PP PPP +P P PP P G + PPPP
Sbjct: 941 GSPPPPPPPPPSYG---SPPPPPPPPPSYGSPPPPPPP---PPGYGSPPPPPPPPPSYGS 994
Query: 94 VASARPPATPSPPQSLLAGAKPVPPRPPLHPALPGAKPSPPPPPAPLGAKPGPPPPP--- 150
PP P PP S ++ P PP PP+H GA P PPPPP GA P PPPPP
Sbjct: 995 -----PPPPPPPPFSHVSSIPPPPPPPPMHG---GAPPPPPPPPMHGGAPPPPPPPPMHG 1046
Query: 151 -----PPAPPSAKPGPPPPPPPAPSGAKPGPRPP--------PPPPKSGVAPPRPPIGPK 197
PP PP PPPPPPP GA+P P PP PPPP G APP PP +
Sbjct: 1047 GAPPPPPPPPMHGGAPPPPPPPMFGGAQPPPPPPMRGGAPPPPPPPMRGGAPPPPPPPMR 1106
Query: 198 AGGP 201
G P
Sbjct: 1107 GGAP 1110
Score = 119 bits (299), Expect = 4e-27
Identities = 75/170 (44%), Positives = 85/170 (49%), Gaps = 29/170 (17%)
Query: 42 PPGRVGTIHSGMPPLRPPPGRMNPLPHEPPSFTPFGNTAVSAAAPPPPRQSGVASARPPA 101
P ++ +H G+ PP + P P PP PF N A S +PPPP PP
Sbjct: 896 PSVKILPLH-GISSAPSPPVKTAPPPPPPP---PFSN-AHSVLSPPPPSYGS-----PPP 945
Query: 102 TPSPPQSLLAGAKPVPPRPPLHPALPGAKPSPPPPPAPLGAKPGPPPPPP----PAPPSA 157
P PP S G+ P PP PP P+ G+ P PPPPP G+ P PPPPPP P PP
Sbjct: 946 PPPPPPSY--GSPPPPPPPP--PSY-GSPPPPPPPPPGYGSPPPPPPPPPSYGSPPPPPP 1000
Query: 158 KP-------GPPPPPPPAPSGAKPGPRPPPPPPKSGVAPPRPPIGPKAGG 200
P PPPPPPP GA P PPPPPP G APP PP P GG
Sbjct: 1001 PPFSHVSSIPPPPPPPPMHGGAPP---PPPPPPMHGGAPPPPPPPPMHGG 1047
Score = 111 bits (278), Expect = 1e-24
Identities = 70/182 (38%), Positives = 81/182 (44%), Gaps = 36/182 (19%)
Query: 51 SGMPPLRPPPGRMNPLPHE-------PPSFTPFGNTAVS-----------AAAPPPPRQS 92
S PP PPP +PL PP P+ + A S ++AP PP
Sbjct: 857 SSSPPPPPPPPPFSPLNTTKANDYILPPPPLPYTSIAPSPSVKILPLHGISSAPSPP--- 913
Query: 93 GVASARPPATPSPPQSLLAGAKPVPPR---PPLHPALPGAKPSPPPPPAPLGAKPGPPPP 149
V +A PP P P + + P PP PP P P + SPPPPP P + PPPP
Sbjct: 914 -VKTAPPPPPPPPFSNAHSVLSPPPPSYGSPPPPPPPPPSYGSPPPPPPPPPSYGSPPPP 972
Query: 150 PPPAPPSAKPGPPPPPPPAPSGAKPG-----------PRPPPPPPKSGVAPPRPPIGPKA 198
PPP P P PPPPPPP+ P P PPPPPP G APP PP P
Sbjct: 973 PPPPPGYGSPPPPPPPPPSYGSPPPPPPPPFSHVSSIPPPPPPPPMHGGAPPPPPPPPMH 1032
Query: 199 GG 200
GG
Sbjct: 1033 GG 1034
Score = 107 bits (268), Expect = 2e-23
Identities = 74/209 (35%), Positives = 82/209 (38%), Gaps = 54/209 (25%)
Query: 41 PPPGRVGTIHSGMPPLRPPPGRMNPLPHEPPSFTPFGNTAVSAAAPPPP----------- 89
PPP ++ L PPP P P PP + N+ PPPP
Sbjct: 793 PPPPPFASVRRNSETLLPPP----PPPPPPPFASVRRNSETLLPPPPPPPPWKSLYASTF 848
Query: 90 ---RQSGVASARPPATPSPPQSLLAGAK------PVPPRP-------------PLH---- 123
+S+ PP P PP S L K P PP P PLH
Sbjct: 849 ETHEACSTSSSPPPPPPPPPFSPLNTTKANDYILPPPPLPYTSIAPSPSVKILPLHGISS 908
Query: 124 ---PALPGAKPSPPPPP--------APLGAKPGPPPPPPPAPPSAKPGPPPPPPPAPSGA 172
P + A P PPPPP +P G PPPPPP PPS G PPPPPP P
Sbjct: 909 APSPPVKTAPPPPPPPPFSNAHSVLSPPPPSYGSPPPPPPPPPSY--GSPPPPPPPPPSY 966
Query: 173 KPGPRPPPPPPKSGVAPPRPPIGPKAGGP 201
P PPPPPP G PP PP P G P
Sbjct: 967 GSPPPPPPPPPGYGSPPPPPPPPPSYGSP 995
Score = 85.5 bits (210), Expect = 9e-17
Identities = 80/250 (32%), Positives = 98/250 (39%), Gaps = 72/250 (28%)
Query: 12 NLGDNKKTSLQGNQSMGAFAVVGSPFELNPPPGRVGTIH---SGMPPLRPPPGRMN---- 64
+L KT L +Q+ V SP PPP H S +PP PPP +
Sbjct: 629 SLTSEAKTVLHSSQA------VASPPPPPPPPPLPTYSHYQTSQLPPPPPPPPPFSSERP 682
Query: 65 -------PLPHEPPSFTPFGNTAVSAAAPPPPRQSGVASARP------PATPSPP-QSLL 110
P P PP F+ + + PPPP +S RP P PSPP +S+
Sbjct: 683 NSGTVLPPPPPPPPPFSSERPNSGTVLPPPPPPPLPFSSERPNSGTVLPPPPSPPWKSVY 742
Query: 111 AGAKPVPPRPPLHPALPGAKPSPPPPPAP---LGAK---------PGPPPPPPPAP---- 154
A A +P A P + P+PPPPP +G K P PPPPPPP P
Sbjct: 743 ASALAIPAICSTSQA-PTSSPTPPPPPPAYYSVGQKSSDLQTSQLPSPPPPPPPPPFASV 801
Query: 155 -PSAKPGPPPPPPPAP---------SGAKPGPRPPPPPPKSGVA---------------- 188
+++ PPPPPP P S P PPPPP KS A
Sbjct: 802 RRNSETLLPPPPPPPPPPFASVRRNSETLLPPPPPPPPWKSLYASTFETHEACSTSSSPP 861
Query: 189 --PPRPPIGP 196
PP PP P
Sbjct: 862 PPPPPPPFSP 871
Score = 71.6 bits (174), Expect = 1e-12
Identities = 56/180 (31%), Positives = 67/180 (37%), Gaps = 44/180 (24%)
Query: 67 PHEPPSFTPFGNTAVSAAAPPPPRQSGVASARPPATPSPPQSLLAGAKPVPPRPPL---- 122
P P TP S PPP S + A+ T +A P PP PPL
Sbjct: 604 PDSSPKETPSSLPPASPHQAPPPLPSLTSEAK---TVLHSSQAVASPPPPPPPPPLPTYS 660
Query: 123 HPALPGAKPSPPPPPAPLGAKPG-----PPPPPPPAPPSAK--------PGPPPPPPP-- 167
H P PPPPP +P PPPPPPP P S++ P PPPPP P
Sbjct: 661 HYQTSQLPPPPPPPPPFSSERPNSGTVLPPPPPPPPPFSSERPNSGTVLPPPPPPPLPFS 720
Query: 168 --APSGAKPGPRPPPPP--------------------PKSGVAPPRPPIGPKAGGPKATE 205
P+ P PP PP P S PP PP + G K+++
Sbjct: 721 SERPNSGTVLPPPPSPPWKSVYASALAIPAICSTSQAPTSSPTPPPPPPAYYSVGQKSSD 780
Score = 69.7 bits (169), Expect = 5e-12
Identities = 64/215 (29%), Positives = 80/215 (36%), Gaps = 38/215 (17%)
Query: 14 GDNKKTSLQGNQSMGAFAVVGSPFELNP-PPGRVGTIHSGMPPLRPPPGRMNPLPHEPPS 72
GD+ K + ++ G + P ++ P G+ M + PP R+N P +
Sbjct: 503 GDSLKPKSKQQETQGPNVRMAKPNAVSRWIPSNKGSYKDSMH-VAYPPTRINSAPASITT 561
Query: 73 FTPFGNTAVS----------------AAAPPPPRQSGVASARPPATPSPPQSLLAGAKPV 116
G A S + + P R + P ++P S L A P
Sbjct: 562 SLKDGKRATSPDGVIPKDAKTKYLRASVSSPDMRSRAPICSSPDSSPKETPSSLPPASPH 621
Query: 117 PPRPPLHPALPGAKPSPPPPPAPLGAKPGPPPPPPPAPP------SAKPGPPPPPPPAPS 170
PPL AK A A P PPPPPPP P S P PPPPPPP S
Sbjct: 622 QAPPPLPSLTSEAKTVLHSSQAV--ASPPPPPPPPPLPTYSHYQTSQLPPPPPPPPPFSS 679
Query: 171 -----GAKPGPRPPPPPPKSG-------VAPPRPP 193
G P PPPPPP S V PP PP
Sbjct: 680 ERPNSGTVLPPPPPPPPPFSSERPNSGTVLPPPPP 714
>At5g07770 unknown protein
Length = 721
Score = 144 bits (362), Expect = 2e-34
Identities = 158/620 (25%), Positives = 259/620 (41%), Gaps = 103/620 (16%)
Query: 27 MGAFAVVGSPFELNPPP-GRVGTIHSGMPPLR-----PPP--GRMNPLPHEPPSFTPFGN 78
M + G+ + PP GRV PP+R PPP GR+ P P PP F P G
Sbjct: 1 MSPVEITGADAVVTPPMRGRVPLPPPPPPPMRRSAPSPPPMSGRVPPPPPPPPMFDPKGA 60
Query: 79 TAVSAAAPPPPRQS--------------------GVASARPPATPSPPQSLLAGAKPVPP 118
V P +S G A P S ++L + A V
Sbjct: 61 GRVICCLRPGQNKSSLKRFQCGKLTNAWEELQRHGEAQTAPEFDLSEIEALFSAA--VQN 118
Query: 119 RPPLHPALPGAKPSPPPPPAPLGAKPGPPPPPPPAPPSAKPGPPPPPPPAPSGAKPGPRP 178
+ + A + P + P PPP PPP P S P
Sbjct: 119 QADKSGSRREAFEANPDKLQLISGADALVPLPPP----------PPPMPRRS-------P 161
Query: 179 PPPPPKSGVAPPRPPIGPKAGGPKATENAEAGAEGGADTSKAKLKPFFWDKVPANSDQSM 238
PPPPP+ + G + T ++ LKP W K+ + Q
Sbjct: 162 PPPPPRFDAFDHK--------GARMVCGFRCPV-----TKRSSLKPLHWVKI-TRALQGS 207
Query: 239 VWN--QIKSGSFQFNEEM----IETLFGYNAVNKNNGQRQKESSSSQDPSPQYIQIVDKK 292
+W+ QI+ G Q E+ IETLF A + P P+ + ++D K
Sbjct: 208 LWDELQIQYGESQTAIELDVPEIETLFSVGAKPRPK------------PKPEKVPLIDLK 255
Query: 293 KAQNLLILLRALNVTMEEVCDALYEGNE--LPSEFLQTLLKMAPTSDEELKLRLFNGDLS 350
+A N ++ L+ L + + ++ A+ +E L + ++ L+++ PT +E L+ + GD +
Sbjct: 256 RANNTIVNLKILKMPLPDMMAAVMAMDESVLDVDQIENLIQLCPTKEEMELLKNYTGDKA 315
Query: 351 QLGPADRFLKAMVDIPSAFKRMEVLLFMCTFKEELTTTMESFAVLEVACKELRNSRLFHK 410
LG +++ L ++ +P ++ VL F F ++T + V+ AC+E+R+S++ +
Sbjct: 316 TLGKSEQCLLELMKVPRFEAKLRVLSFKIPFGTKITKFRKMLNVVNSACEEVRSSQMLKE 375
Query: 411 LLEAVLKTGNRMNDGTYRGGAQAFKLDTLLKLSDVKGTDGKTTLLHFVVQEIIRSEGIKA 470
+++ +L GN +N GT RG A F+LD+LL LS+ + + K TL+H++ +++ S+
Sbjct: 376 IMKIILFLGNTLNQGTARGSAVGFRLDSLLILSETRADNNKMTLMHYLC-KVLASKAADL 434
Query: 471 ARAAKDSQSLS-----NIKT--DELHETEDHYRELGLEMVSH-----LSTELENVKRGSV 518
KD QSL N+K+ +E+H +L E+ + +S + + +
Sbjct: 435 LDFHKDLQSLESTLEINLKSLAEEIHAITKGLEKLKQELTASETDGPVSQVFRKLLKDFI 494
Query: 519 LDADSLTATTIKLGHGLVKAKDILNKNLKNVEDDR--GFRETVESFV---KNAEADVKKL 573
A++ AT L K KN DR T+ SF+ K A + K
Sbjct: 495 SSAETQVATVSTLYSSAKKVAHCYVVANKNTCGDRLPAVSATLLSFIRLFKKAHQENVKQ 554
Query: 574 LEDEKKIMALVKSTGDYFHG 593
+ EKK A T D F G
Sbjct: 555 EDLEKKKAA----TEDVFGG 570
Score = 66.2 bits (160), Expect = 6e-11
Identities = 43/120 (35%), Positives = 51/120 (41%), Gaps = 26/120 (21%)
Query: 86 PPPPRQSGVASARPPATPSPPQSLLAGAKPVP-------PRPPLHPALPGAKPSPPPPPA 138
P PP + A R PA +PP L+ G P+P P PP P++ G P PPPPP
Sbjct: 599 PRPPSRPRYACCRIPAV-NPPPRLVCGPYPLPRLVRVGSPSPP-PPSMSGGAPPPPPPPP 656
Query: 139 PLGAKPGPPPP-----------------PPPAPPSAKPGPPPPPPPAPSGAKPGPRPPPP 181
L A PPP P P + G PPP P+ SG P P PP P
Sbjct: 657 MLVASRTAPPPHLSHVRSIPFQTRLVMGTSPLPLLVREGAPPPTLPSMSGGAPPPPPPLP 716
Score = 53.1 bits (126), Expect = 5e-07
Identities = 35/98 (35%), Positives = 44/98 (44%), Gaps = 17/98 (17%)
Query: 95 ASARPPATPSPPQSLLAGAKPVPPRPPLHPALPGAK-PSPPPPPAPLGAKPGPPPPPPPA 153
+ A+ P+ PP S+ PRPP P + P+ PPP + P P
Sbjct: 584 SEAKSPSRIRPPPSI--------PRPPSRPRYACCRIPAVNPPPRLVCG-------PYPL 628
Query: 154 PPSAKPGPPPPPPPAPSGAKPGPRPPPPP-PKSGVAPP 190
P + G P PPPP+ SG P P PPPP S APP
Sbjct: 629 PRLVRVGSPSPPPPSMSGGAPPPPPPPPMLVASRTAPP 666
Score = 50.4 bits (119), Expect = 3e-06
Identities = 45/160 (28%), Positives = 65/160 (40%), Gaps = 35/160 (21%)
Query: 6 FGSSSNNLGDNKKTSLQGNQSMGAFAVVGSPFELNPPPGRVGTIHSGMPPLRPPPGRMNP 65
FG +N+ + TSL +++ + P + PP R +P + PPP R+
Sbjct: 568 FGGPDHNIDSD--TSLDDSEAKSPSRIRPPP-SIPRPPSRPRYACCRIPAVNPPP-RLVC 623
Query: 66 LPHEPPSFTPFGNTAVSAAAPPPPRQSGVASARPPATP-------SPPQSL--------- 109
P+ P G+ + PPPP SG A PP P +PP L
Sbjct: 624 GPYPLPRLVRVGSPS-----PPPPSMSGGAPPPPPPPPMLVASRTAPPPHLSHVRSIPFQ 678
Query: 110 ---LAGAKPVP------PRPPLHPALPGAKPSPPPPPAPL 140
+ G P+P PP P++ G P PPPPP P+
Sbjct: 679 TRLVMGTSPLPLLVREGAPPPTLPSMSGGAP-PPPPPLPM 717
Score = 43.5 bits (101), Expect = 4e-04
Identities = 26/63 (41%), Positives = 27/63 (42%), Gaps = 13/63 (20%)
Query: 147 PPPPPPAPPSAK-------PGPPPPPP------PAPSGAKPGPRPPPPPPKSGVAPPRPP 193
PPP P PPS P PPP P P + G PPPP SG APP PP
Sbjct: 594 PPPSIPRPPSRPRYACCRIPAVNPPPRLVCGPYPLPRLVRVGSPSPPPPSMSGGAPPPPP 653
Query: 194 IGP 196
P
Sbjct: 654 PPP 656
>At4g15190 hypothetical protein
Length = 130
Score = 130 bits (328), Expect = 2e-30
Identities = 68/127 (53%), Positives = 93/127 (72%), Gaps = 4/127 (3%)
Query: 549 VEDDRGFRETVESFVKNAEADVKKLLEDEKKIMALVKSTGDYFHGNATKDDGLRLFVVVR 608
++++ F + F++ A+AD K L E+E++IM LVKS+ DYFHG + K++GLRLF +VR
Sbjct: 1 MDEESDFERALAGFIERADADFKWLKEEEERIMVLVKSSADYFHGKSAKNEGLRLFAIVR 60
Query: 609 DFLIMLDKVCKEVRDAQKKPAKPIKQETSRGLSSSDTRPSPSDFRQRLFPAIAERRID-- 666
DFLIML+KVC+EV++ K K+E+ S S+ +PSP DFRQRLFPAIAERR+D
Sbjct: 61 DFLIMLEKVCREVKETTKTTNHSGKKESEMTTSDSN-QPSP-DFRQRLFPAIAERRMDSS 118
Query: 667 DDSSDEE 673
DDS DEE
Sbjct: 119 DDSDDEE 125
>At1g61080 hypothetical protein
Length = 907
Score = 124 bits (312), Expect = 1e-28
Identities = 81/213 (38%), Positives = 96/213 (45%), Gaps = 54/213 (25%)
Query: 41 PPPGRVGTIHSGMPPLRPPPGRMNPLPH------EPPSFTPFGNTAVSAAAPPPPRQSGV 94
PPP + H PP P P + PL H PP+F P SA PPPP
Sbjct: 462 PPPAVMPLKHFAPPPPPPLPPAVMPLKHFAPPPPTPPAFKPLKG---SAPPPPPPPPLPT 518
Query: 95 ASARPPATPSPPQSLLAGAKPVPPRPPLHPALPGAKPSPPPPPAPLGAKPGPPPPPP--- 151
A PP P PP++ +A P PP PP PG +PPPPP P G + PPPPPP
Sbjct: 519 TIAAPPPPPPPPRAAVA---PPPPPPP-----PGTAAAPPPPPPPPGTQAAPPPPPPPPM 570
Query: 152 ----PAPP------SAKPGPPPPPPPAP--SGAKPGP------------RPPPPPPKSGV 187
P+PP S GPPPPPPP P +GA P P PPPPPP+ G+
Sbjct: 571 QNRAPSPPPMPMGNSGSGGPPPPPPPMPLANGATPPPPPPPMAMANGAAGPPPPPPRMGM 630
Query: 188 A-----PPRPP-----IGPKAGGPKATENAEAG 210
A PP PP + PK K + + G
Sbjct: 631 ANGAAGPPPPPGAARSLRPKKAATKLKRSTQLG 663
Score = 120 bits (302), Expect = 2e-27
Identities = 73/177 (41%), Positives = 86/177 (48%), Gaps = 22/177 (12%)
Query: 54 PPLRPPPGRMN-------PLPHEPPSFTPFGNTAVSAAAPPPPRQSGVAS------ARPP 100
PP PPP ++ PLP PP+ P + A+S PPPP A A PP
Sbjct: 418 PPPPPPPPPLSFIKTASLPLPSPPPT-PPIADIAISMPPPPPPPPPPPAVMPLKHFAPPP 476
Query: 101 ATPSPPQSL-LAGAKPVPPRPPLHPALPGAKPSPPPPPAPLGAKPGPPPPPPPAPPSAKP 159
P PP + L P PP PP L G+ P PPPPP PL PPPPPP P +A
Sbjct: 477 PPPLPPAVMPLKHFAPPPPTPPAFKPLKGSAPPPPPPP-PLPTTIAAPPPPPPPPRAAVA 535
Query: 160 GPPPPPPPAPSGAKPGPRPPPPPPKSGVAPPRPPIGP---KAGGPKATENAEAGAEG 213
PPPPPPP + A P PPPPPP + APP PP P +A P +G+ G
Sbjct: 536 PPPPPPPPGTAAAPP---PPPPPPGTQAAPPPPPPPPMQNRAPSPPPMPMGNSGSGG 589
Score = 114 bits (284), Expect = 2e-25
Identities = 66/169 (39%), Positives = 76/169 (44%), Gaps = 17/169 (10%)
Query: 36 PFELNPPPGRVGTIHSGMPPLRPPPGRMNPLPHEPPSFTPFGNTAVSAAAPPPPRQSGVA 95
P PP + I MPP PPP PP+ P + A P PP +
Sbjct: 436 PLPSPPPTPPIADIAISMPPPPPPPP-------PPPAVMPLKHFAPPPPPPLPPAVMPLK 488
Query: 96 SARPPATPSPPQSLLAGAKPVPPRPPLHPALPGAKPSPPPPPAPLGAKPGPPPPPPPAPP 155
PP P L G+ P PP PP LP +PPPPP P A PPPPPPP
Sbjct: 489 HFAPPPPTPPAFKPLKGSAPPPPPPP---PLPTTIAAPPPPPPPPRAAVAPPPPPPPPGT 545
Query: 156 SAKPGPPPPPPPAPSGAKPGPRPPPPPPKSGVAPPRPPI---GPKAGGP 201
+A P PPPPPP G + P PPPPPP AP PP+ +GGP
Sbjct: 546 AAAPPPPPPPP----GTQAAPPPPPPPPMQNRAPSPPPMPMGNSGSGGP 590
Score = 77.8 bits (190), Expect = 2e-14
Identities = 52/145 (35%), Positives = 57/145 (38%), Gaps = 51/145 (35%)
Query: 86 PPPPRQSGVASARPPATPSPPQSLLAGAKPVPPRPPLHPALPGAKPSPPPPPAPLGAKPG 145
PPPP PP P P + + P+P PP P A PPPPP
Sbjct: 416 PPPP---------PPPPPPPLSFIKTASLPLPSPPPTPPIADIAISMPPPPP-------- 458
Query: 146 PPPPPPPAPPS---AKPGPPP-P-----------PPPAPSGAKP---------------- 174
PPPPPP P A P PPP P PPP P KP
Sbjct: 459 PPPPPPAVMPLKHFAPPPPPPLPPAVMPLKHFAPPPPTPPAFKPLKGSAPPPPPPPPLPT 518
Query: 175 ---GPRPPPPPPKSGVAPPRPPIGP 196
P PPPPPP++ VAPP PP P
Sbjct: 519 TIAAPPPPPPPPRAAVAPPPPPPPP 543
Score = 45.8 bits (107), Expect = 8e-05
Identities = 27/72 (37%), Positives = 35/72 (48%), Gaps = 2/72 (2%)
Query: 40 NPPPGRVGTIHSGMPPLRPPPGRMNPLPHEPPSFTPFGNTAVSAAAPPPPRQSGVA--SA 97
+PPP +G SG PP PPP + PP P +A PPPP + G+A +A
Sbjct: 576 SPPPMPMGNSGSGGPPPPPPPMPLANGATPPPPPPPMAMANGAAGPPPPPPRMGMANGAA 635
Query: 98 RPPATPSPPQSL 109
PP P +SL
Sbjct: 636 GPPPPPGAARSL 647
>At4g13340 extensin-like protein
Length = 760
Score = 123 bits (309), Expect = 3e-28
Identities = 65/156 (41%), Positives = 80/156 (50%), Gaps = 14/156 (8%)
Query: 39 LNPPPGRVGTIHSGMPPLRPPPGRMNPLPHEPPSFTPFGNTAVSAAAPPPPRQSGVASAR 98
L+ PP G+ G + P P + PLP PPS + + PP P S +
Sbjct: 376 LSRPPVNCGSFSCGRS-VSPRPPVVTPLP--PPS--------LPSPPPPAPIFSTPPTLT 424
Query: 99 PPATPSPPQSLLAGAKPVPPRPPLHPALPGAKPSPPPPPAPLGAKPGPPPPPPPAPPSAK 158
P PSPP + + P PP PP++ P P PPPPP P+ + P PPPPPPP PP
Sbjct: 425 SPPPPSPPPPVYSPPPPPPPPPPVYSPPP---PPPPPPPPPVYSPPPPPPPPPPPPPVYS 481
Query: 159 PGPPPPPPPAPSGAKPGPRPPPPPPKSGVAPPRPPI 194
P PP PPPP P P P PPPPPP +PP PP+
Sbjct: 482 PPPPSPPPPPPPVYSPPPPPPPPPPPPVYSPPPPPV 517
Score = 109 bits (272), Expect = 6e-24
Identities = 65/170 (38%), Positives = 78/170 (45%), Gaps = 18/170 (10%)
Query: 41 PPPGRVGTIHSGMPPLRPPPGRMNPLPHEPPSFTPFGNTAVSAAAPPPPRQSGVASARPP 100
PPP ++S PP PPP +P P PP P + PPPP V S PP
Sbjct: 427 PPPSPPPPVYSPPPPPPPPPPVYSPPPPPPPPPPP-PVYSPPPPPPPPPPPPPVYSPPPP 485
Query: 101 ATPSPPQSLLAGAKPVPPRPP---LHPALPGAKPSPPPPPAPLG-----AKPGPPPPPPP 152
+ P PP + + P PP PP P P SPPPPP+P +P PPPP P
Sbjct: 486 SPPPPPPPVYSPPPPPPPPPPPPVYSPPPPPVYSSPPPPPSPAPTPVYCTRPPPPPPHSP 545
Query: 153 APPSAKPGPP-------PPPPPA--PSGAKPGPRPPPPPPKSGVAPPRPP 193
PP P PP PPPP + P + P P PPPP ++PP PP
Sbjct: 546 PPPQFSPPPPEPYYYSSPPPPHSSPPPHSPPPPHSPPPPIYPYLSPPPPP 595
Score = 107 bits (267), Expect = 2e-23
Identities = 64/173 (36%), Positives = 74/173 (41%), Gaps = 33/173 (19%)
Query: 54 PPLRPPPGRMNPLPHEPPSFT----PFGNTAVSAAAPPPPRQSGVASARPPATPSPPQSL 109
PP P P P+ PP+ T P V + PPPP V S PP P PP +
Sbjct: 404 PPSLPSPPPPAPIFSTPPTLTSPPPPSPPPPVYSPPPPPPPPPPVYSPPPPPPPPPPPPV 463
Query: 110 LAGAKPVPPRPPLHPALPGAKPSPPPPPAPLGAKPGPPPPPPPAP--------------- 154
+ P PP PP P PSPPPPP P+ + P PPPPPPP P
Sbjct: 464 YSPPPPPPPPPPPPPVYSPPPPSPPPPPPPVYSPPPPPPPPPPPPVYSPPPPPVYSSPPP 523
Query: 155 ---PSAKP----GPPPPPPPAPSGAKPGPRPP-------PPPPKSGVAPPRPP 193
P+ P PPPPPP +P + P PP PPPP S P PP
Sbjct: 524 PPSPAPTPVYCTRPPPPPPHSPPPPQFSPPPPEPYYYSSPPPPHSSPPPHSPP 576
Score = 100 bits (250), Expect = 2e-21
Identities = 65/168 (38%), Positives = 78/168 (45%), Gaps = 40/168 (23%)
Query: 41 PPPGRVGT-IHSGMPPLRPPPGRMNPLPHEPPSFTPFGNTAVSAAAPPPPRQSGVASARP 99
PPP T ++ PP PPP P PP F+P ++PPPP S+ P
Sbjct: 523 PPPSPAPTPVYCTRPP--PPPPHSPP----PPQFSPPPPEPYYYSSPPPPH-----SSPP 571
Query: 100 PATPSPPQSLLAGAKPVPPRPPLHPALPGAKPSPPPPPAPLGAKP-----GPPPPPP--- 151
P +P PP S P PP++P L SPPPPP P+ + P PPPPPP
Sbjct: 572 PHSPPPPHS---------PPPPIYPYL-----SPPPPPTPVSSPPPTPVYSPPPPPPCIE 617
Query: 152 -PAPPSAKPGPPPPPPPAPSGAKPGPRP-----PPPPPKSGVAPPRPP 193
P PP PPPPPP + P P P PPPPP +PP PP
Sbjct: 618 PPPPPPCIEYSPPPPPPVVHYSSPPPPPVYYSSPPPPPVYYSSPPPPP 665
Score = 100 bits (249), Expect = 3e-21
Identities = 69/205 (33%), Positives = 82/205 (39%), Gaps = 43/205 (20%)
Query: 33 VGSPFELNPPPGRVGTIHSGMPPLRPPPGRM-------NPLPHEPPSFTPFGNTAVSAAA 85
V SP PPP ++S PP PPP P P PP ++P S+
Sbjct: 463 VYSPPPPPPPPPPPPPVYSPPPPSPPPPPPPVYSPPPPPPPPPPPPVYSPPPPPVYSSPP 522
Query: 86 PPP-PRQSGVASARPP----------------------ATPSPPQSLLAGAKPVPPRPPL 122
PPP P + V RPP ++P PP S P PP P
Sbjct: 523 PPPSPAPTPVYCTRPPPPPPHSPPPPQFSPPPPEPYYYSSPPPPHSSPPPHSPPPPHSPP 582
Query: 123 HPALPGAKPSPP------PPPAPLGAKPGPPP--PPPPAPPSAKPGPPPPPPPAPSGAKP 174
P P P PP PPP P+ + P PPP PPP PP + PPPPPP + P
Sbjct: 583 PPIYPYLSPPPPPTPVSSPPPTPVYSPPPPPPCIEPPPPPPCIEYSPPPPPPVVHYSSPP 642
Query: 175 GP-----RPPPPPPKSGVAPPRPPI 194
P PPPPP PP PP+
Sbjct: 643 PPPVYYSSPPPPPVYYSSPPPPPPV 667
Score = 91.7 bits (226), Expect = 1e-18
Identities = 51/132 (38%), Positives = 59/132 (44%), Gaps = 5/132 (3%)
Query: 71 PSFTPFGNTAVSAAAPPPPRQSGVASARPPATPSPPQSLLAGAKPVPPRPPLHPALPGAK 130
P F N A P Q +RPP + + V PRPP+ LP
Sbjct: 351 PEFDDRRNCLPGRPAQRSPGQCKAFLSRPPVNCGS----FSCGRSVSPRPPVVTPLPPPS 406
Query: 131 PSPPPPPAPLGAKPGP-PPPPPPAPPSAKPGPPPPPPPAPSGAKPGPRPPPPPPKSGVAP 189
PPPPAP+ + P PPPP+PP PPPPPPP P P P PPPPPP +P
Sbjct: 407 LPSPPPPAPIFSTPPTLTSPPPPSPPPPVYSPPPPPPPPPPVYSPPPPPPPPPPPPVYSP 466
Query: 190 PRPPIGPKAGGP 201
P PP P P
Sbjct: 467 PPPPPPPPPPPP 478
Score = 87.4 bits (215), Expect = 2e-17
Identities = 68/202 (33%), Positives = 84/202 (40%), Gaps = 54/202 (26%)
Query: 54 PPLRPPPGRMNPLPHEPPSFT--PFGNTAVSAAAPPPPRQSG-----VASARPPATP--S 104
PP PPP + +P P EP ++ P +++ +PPPP S PP TP S
Sbjct: 541 PPHSPPPPQFSPPPPEPYYYSSPPPPHSSPPPHSPPPPHSPPPPIYPYLSPPPPPTPVSS 600
Query: 105 PPQSLLAGAKPVPP---RPPLHPALPGAKPSPPP-------PPAPLGAKPGPPPP----- 149
PP + + P PP PP P + + P PPP PP P+ PPPP
Sbjct: 601 PPPTPVYSPPPPPPCIEPPPPPPCIEYSPPPPPPVVHYSSPPPPPVYYSSPPPPPVYYSS 660
Query: 150 PPPAPP---SAKPGP----------------PPPPPPAPSGAKPGPRP----PPPPPKSG 186
PPP PP S+ P P PPPPP AP P P P PPPP
Sbjct: 661 PPPPPPVHYSSPPPPEVHYHSPPPSPVHYSSPPPPPSAPCEESPPPAPVVHHSPPPPMVH 720
Query: 187 VAPP-------RPPIGPKAGGP 201
+PP PP P+ GP
Sbjct: 721 HSPPPPVIHQSPPPPSPEYEGP 742
Score = 86.7 bits (213), Expect = 4e-17
Identities = 65/203 (32%), Positives = 72/203 (35%), Gaps = 47/203 (23%)
Query: 36 PFELNPPPGRVGTIHSGMPPLRPPPGRMNPLPHEPPS------FTPFGNTAVSAAAPPPP 89
P + +PPP S PP PP P PH PP P T VS+ PPP
Sbjct: 547 PPQFSPPPPEPYYYSSPPPPHSSPPPHSPPPPHSPPPPIYPYLSPPPPPTPVSS---PPP 603
Query: 90 RQSGVASARPPATPSPPQSLLAGAKPVPPRPPLHPALPGAKP----SPPPPPAPLGAKPG 145
PP PP P PP P +H + P P SPPPPP + P
Sbjct: 604 TPVYSPPPPPPCIEPPPPPPCIEYSPPPPPPVVHYSSPPPPPVYYSSPPPPPVYYSSPPP 663
Query: 146 PPP-----PPPPAPPSAKPGP-------PPPPPPAPSGAKPGPRP--------------- 178
PPP PPPP P P PPPPP AP P P P
Sbjct: 664 PPPVHYSSPPPPEVHYHSPPPSPVHYSSPPPPPSAPCEESPPPAPVVHHSPPPPMVHHSP 723
Query: 179 -------PPPPPKSGVAPPRPPI 194
PPPP P PP+
Sbjct: 724 PPPVIHQSPPPPSPEYEGPLPPV 746
Score = 69.7 bits (169), Expect = 5e-12
Identities = 62/181 (34%), Positives = 76/181 (41%), Gaps = 49/181 (27%)
Query: 40 NPPPGRVGTIHSGMPPLRPPPGRMNPLP---HEPPSFTPFGNTAVSAAAPPPPRQSGVAS 96
+PPP V + P + PPP P P + PP P + + + PPPP V
Sbjct: 600 SPPPTPVYSPPPPPPCIEPPP----PPPCIEYSPPPPPPVVHYS---SPPPPP----VYY 648
Query: 97 ARPPATPSPPQSLLAGAKPVPPRPPLHPALPGAKPSPP------PPPAPLGAKPGPPPP- 149
+ PP P S PP PP+H + P PP PPP+P+ PPPP
Sbjct: 649 SSPPPPPVYYSS-------PPPPPPVH----YSSPPPPEVHYHSPPPSPVHYSSPPPPPS 697
Query: 150 ------PPPAPPSAKPGPPP-----PPPPAPSGAKPGPRPP---PPPPKSGV---APPRP 192
PPPAP PPP PPPP + P P P P PP GV +PP P
Sbjct: 698 APCEESPPPAPVVHHSPPPPMVHHSPPPPVIHQSPPPPSPEYEGPLPPVIGVSYASPPPP 757
Query: 193 P 193
P
Sbjct: 758 P 758
Score = 29.6 bits (65), Expect = 5.7
Identities = 30/96 (31%), Positives = 35/96 (36%), Gaps = 28/96 (29%)
Query: 40 NPPPGRVGTIHSGMPPLRPPPGRMNPLPHEPPSFTPFGNTAVSAAAPPPPRQSGVASARP 99
+PPP V H PP PPP P PP V +PPPP V + P
Sbjct: 681 SPPPSPV---HYSSPP--PPPSA--PCEESPPP------APVVHHSPPPPM---VHHSPP 724
Query: 100 PA----TPSPPQSLLAGAKPV--------PPRPPLH 123
P +P PP G P PP PP +
Sbjct: 725 PPVIHQSPPPPSPEYEGPLPPVIGVSYASPPPPPFY 760
>At3g24400 protein kinase, putative
Length = 694
Score = 115 bits (287), Expect = 1e-25
Identities = 80/218 (36%), Positives = 96/218 (43%), Gaps = 47/218 (21%)
Query: 36 PFELNPPPGRVGTIHSGMPPLRPPPGRMNP-LPHEPPSFTPFGNTAVSAAAPPPPRQSGV 94
P + PPP + PP PPP + P LP PP TA+ A PPPP + V
Sbjct: 17 PLPIPPPPQPLPV----TPP--PPPTALPPALPPPPPP------TALPPALPPPPPPTTV 64
Query: 95 ASARPPATPSPPQSLLAG--------------AKPVPPRPPLHPALPGA-KPSPPPPPAP 139
PP+TPSPP L P P PPL P+ P A PSPP P+P
Sbjct: 65 PPI-PPSTPSPPPPLTPSPLPPSPTTPSPPLTPSPTTPSPPLTPSPPPAITPSPPLTPSP 123
Query: 140 LGAKPGPPPPPPPAP----PSAKPGPPP--------PPPPAPSGAKPGPRPPP------P 181
L P P PPPP+P P P PPP PPPP+P+ PR PP P
Sbjct: 124 LPPSPTTPSPPPPSPSIPSPPLTPSPPPSSPLRPSSPPPPSPATPSTPPRSPPPPSTPTP 183
Query: 182 PPKSGVAPPRPPIGPKAGGPKATENAEAGAEGGADTSK 219
PP+ G P PP P G +T + G+ A +SK
Sbjct: 184 PPRVGSLSPPPPASPSGGRSPSTPSTTPGSSPPAQSSK 221
Score = 81.6 bits (200), Expect = 1e-15
Identities = 53/134 (39%), Positives = 61/134 (44%), Gaps = 19/134 (14%)
Query: 84 AAPPPPRQSGVASARPPATPSPPQSLLAGAKPVPPRPPLHPALPGAKPSPPPPPAPLGAK 143
++ PPP + +P P PPQ L PV P PP ALP A P PPPP A A
Sbjct: 2 SSAPPPGGTPSPPPQPLPIPPPPQPL-----PVTPPPP-PTALPPALPPPPPPTALPPAL 55
Query: 144 PGPPPPP--PPAPPSAKPGPPP----PPPPAPSGAKPGPRPPPPPPKSGVAP-------P 190
P PPPP PP PPS PPP P PP+P+ P P P P + P P
Sbjct: 56 PPPPPPTTVPPIPPSTPSPPPPLTPSPLPPSPTTPSPPLTPSPTTPSPPLTPSPPPAITP 115
Query: 191 RPPIGPKAGGPKAT 204
PP+ P P T
Sbjct: 116 SPPLTPSPLPPSPT 129
Score = 80.1 bits (196), Expect = 4e-15
Identities = 47/108 (43%), Positives = 51/108 (46%), Gaps = 6/108 (5%)
Query: 94 VASARPPA-TPSPPQSLLAGAKPVPPRPPLHPALPGAKPSPPPPPAPLGAKPGPPPPPPP 152
++SA PP TPSPP L P P P P P A P PPP P A P P PPPP
Sbjct: 1 MSSAPPPGGTPSPPPQPLPIPPPPQPLPVTPPPPPTALPPALPPPPPPTALP-PALPPPP 59
Query: 153 APPSAKPGPP----PPPPPAPSGAKPGPRPPPPPPKSGVAPPRPPIGP 196
P + P PP PPPP PS P P P PP P PP+ P
Sbjct: 60 PPTTVPPIPPSTPSPPPPLTPSPLPPSPTTPSPPLTPSPTTPSPPLTP 107
Score = 58.2 bits (139), Expect = 2e-08
Identities = 61/195 (31%), Positives = 72/195 (36%), Gaps = 77/195 (39%)
Query: 35 SPFELNPPPGRVGTIHSGMPPLRPPPGRMNPLPHEPPSFTPFGNTAVSAAAPPPPRQSGV 94
SP +PPP PPL P P PPS + + ++PPPP
Sbjct: 127 SPTTPSPPPPSPSI---PSPPLTPSP---------PPS------SPLRPSSPPPP----- 163
Query: 95 ASARPPATPS-PPQSLLAGAKPVPPRPPLHPALPGAKPSPPPPPAPLGAKPGPPPPPPPA 153
PATPS PP+S P PP P P G+ SPPPP +P G + P+
Sbjct: 164 ----SPATPSTPPRS------PPPPSTPTPPPRVGSL-SPPPPASPSGGR-------SPS 205
Query: 154 PPSAKPGPPPPPPPA---PSGAKPG-------------------------------PRPP 179
PS PG PP + GA G P P
Sbjct: 206 TPSTTPGSSPPAQSSKELSKGAMVGIAIGGGFVLLVALALIFFLCKKKRRRDNEAPPAPI 265
Query: 180 PPPPKS-GVAPPRPP 193
PPPKS APPRPP
Sbjct: 266 VPPPKSPSSAPPRPP 280
Score = 47.4 bits (111), Expect = 3e-05
Identities = 55/213 (25%), Positives = 75/213 (34%), Gaps = 36/213 (16%)
Query: 31 AVVGSPFELNPPPGRVGTIHSGMPPLRPPPGRMNPLPHEPPSFTPFGNTAVSAAAPPPPR 90
++ P +PPP + PP P P PP TP V + +PPPP
Sbjct: 139 SIPSPPLTPSPPPS--SPLRPSSPPPPSPATPSTPPRSPPPPSTPTPPPRVGSLSPPPP- 195
Query: 91 QSGVASARPPATP------SPPQSLLAGAKPVPPRPPLHPALPGA--------------- 129
+ + R P+TP SPP +K + + A+ G
Sbjct: 196 -ASPSGGRSPSTPSTTPGSSPPAQ---SSKELSKGAMVGIAIGGGFVLLVALALIFFLCK 251
Query: 130 KPSPPPPPAPLGAKPGPPPPPPPAPPSAKPGPPPPPPPAPSGAKPGPRPPPPPPKSGVAP 189
K AP P P PPP +P SA PP PP S G +S + P
Sbjct: 252 KKRRRDNEAP----PAPIVPPPKSPSSA----PPRPPHFMSSGSSGDYDSNYSDQSVLPP 303
Query: 190 PRPPIGPKAGGPKATENAEAGAEGGADTSKAKL 222
P P + G + T N E + S+A L
Sbjct: 304 PSPGLALGLGIYQGTFNYEELSRATNGFSEANL 336
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.133 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,586,062
Number of Sequences: 26719
Number of extensions: 1110617
Number of successful extensions: 44482
Number of sequences better than 10.0: 702
Number of HSP's better than 10.0 without gapping: 480
Number of HSP's successfully gapped in prelim test: 238
Number of HSP's that attempted gapping in prelim test: 6104
Number of HSP's gapped (non-prelim): 7077
length of query: 675
length of database: 11,318,596
effective HSP length: 106
effective length of query: 569
effective length of database: 8,486,382
effective search space: 4828751358
effective search space used: 4828751358
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC144502.11


