

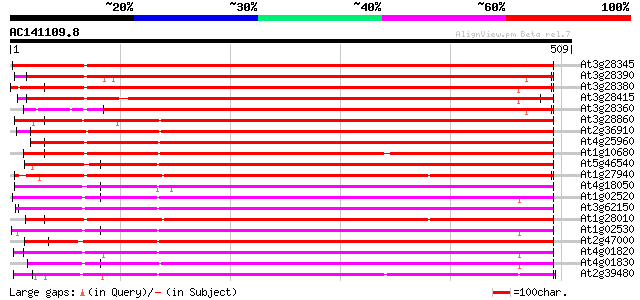
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141109.8 + phase: 0
(509 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g28345 P-glycoprotein, putative, 3'partial 678 0.0
At3g28390 P-glycoprotein, putative 664 0.0
At3g28380 P-glycoprotein, putative 658 0.0
At3g28415 putative protein 617 e-177
At3g28360 P-glycoprotein like protein 595 e-170
At3g28860 P-glycoprotein, putative 418 e-117
At2g36910 putative ABC transporter 402 e-112
At4g25960 P-glycoprotein-2 (pgp2) 374 e-104
At1g10680 putative P-glycoprotein-2 emb|CAA71277 369 e-102
At5g46540 multidrug resistance p-glycoprotein 367 e-102
At1g27940 hypothetical protein 363 e-100
At4g18050 multidrug resistance protein/P-glycoprotein - like 361 e-100
At1g02520 P-glycoprotein, putative 356 1e-98
At3g62150 P-glycoprotein-like proetin 353 1e-97
At1g28010 hypothetical protein 352 4e-97
At1g02530 hypothetical protein 346 2e-95
At2g47000 putative ABC transporter 344 8e-95
At4g01820 P-glycoprotein-like protein pgp3 341 6e-94
At4g01830 putative P-glycoprotein-like protein 333 2e-91
At2g39480 putative ABC transporter 327 1e-89
>At3g28345 P-glycoprotein, putative, 3'partial
Length = 610
Score = 678 bits (1750), Expect = 0.0
Identities = 332/491 (67%), Positives = 407/491 (82%), Gaps = 1/491 (0%)
Query: 3 GGDQKNVSINDKKKKNGSFKSIFMHADVLDWFFMAFGFFGAIGDGMMVPFVLFITSKIMN 62
G +++ S +K GS +SIFMHAD +DW M G GA+GDG P VL ITSK+MN
Sbjct: 2 GKEEEKESGRNKMNCFGSVRSIFMHADGVDWLLMGLGLIGAVGDGFTTPLVLLITSKLMN 61
Query: 63 SVGSASGTSSSNFVHDVNKNAVVVLYMACASFFVCFLEGYCWTRTGERQAARMRVRYLKA 122
++G +S ++ F+ ++KN+V +LY+AC S+ VCFLEGYCWTRTGERQ ARMR +YL+A
Sbjct: 62 NIGGSS-FNTDTFMQSISKNSVALLYVACGSWVVCFLEGYCWTRTGERQTARMREKYLRA 120
Query: 123 VLRQEVSYFDLHVTSTTDVITSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFALLW 182
VLRQ+V YFDLHVTST+DVITSVSSDS VIQDVLS+K+PNFL++AS F+ S IV F LLW
Sbjct: 121 VLRQDVGYFDLHVTSTSDVITSVSSDSFVIQDVLSEKLPNFLMSASTFVGSYIVGFILLW 180
Query: 183 RLAIVGFPFMVLLVIPGYMYKRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFVGESK 242
RLAIVG PF+VLLVIPG MY R + ++RKIREEYN+AG +AEQAISS+RTVY+F GE K
Sbjct: 181 RLAIVGLPFIVLLVIPGLMYGRALISISRKIREEYNEAGFVAEQAISSVRTVYAFSGERK 240
Query: 243 TLAAFSNALEGSVKLGLKQGLAKGLAIGSNGVVYAIWSLIFYYGSIMVMYHGAKGGTVFV 302
T++ FS AL+GSVKLG+KQGLAKG+ IGSNG+ +A+W + +YGS MVMYHGA+GGTVF
Sbjct: 241 TISKFSTALQGSVKLGIKQGLAKGITIGSNGITFAMWGFMSWYGSRMVMYHGAQGGTVFA 300
Query: 303 VGVTLAIGGLAFGTCFSNVRYFAEASVAGERIMEVIKRVPTIDSENMEGEIIEKVLGEVE 362
V +AIGG++ G SN++YF EA+ GERIMEVI RVP IDS+N +G +EK+ GEVE
Sbjct: 301 VAAAIAIGGVSLGGGLSNLKYFFEAASVGERIMEVINRVPKIDSDNPDGHKLEKIRGEVE 360
Query: 363 FNNVEFVYPSRPESVILNDFCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGGEILL 422
F NV+FVYPSR E+ I +DFCL+VPSGKTVALVGGSGSGKSTV+SLLQRFYDP+ GEIL+
Sbjct: 361 FKNVKFVYPSRLETSIFDDFCLRVPSGKTVALVGGSGSGKSTVISLLQRFYDPLAGEILI 420
Query: 423 DGVAIHKLQLKWLRSQMGLVSQEPALFATSIKENILFGREDATYEEIVDAAKASNAHNFI 482
DGV+I KLQ+KWLRSQMGLVSQEPALFAT+IKENILFG+EDA+ +++V+AAKASNAHNFI
Sbjct: 421 DGVSIDKLQVKWLRSQMGLVSQEPALFATTIKENILFGKEDASMDDVVEAAKASNAHNFI 480
Query: 483 SMLPQGYDTQV 493
S LP GY+TQV
Sbjct: 481 SQLPNGYETQV 491
>At3g28390 P-glycoprotein, putative
Length = 1225
Score = 664 bits (1714), Expect = 0.0
Identities = 325/478 (67%), Positives = 394/478 (81%), Gaps = 1/478 (0%)
Query: 16 KKNGSFKSIFMHADVLDWFFMAFGFFGAIGDGMMVPFVLFITSKIMNSVGSASGTSSSNF 75
K GS +SIFMHAD +DW MA G GA+GDG + P + FI SK++N+VG +S F
Sbjct: 3 KSFGSIRSIFMHADGVDWMLMALGLIGAVGDGFITPIIFFICSKLLNNVGGSS-FDDETF 61
Query: 76 VHDVNKNAVVVLYMACASFFVCFLEGYCWTRTGERQAARMRVRYLKAVLRQEVSYFDLHV 135
+ V KNAV ++Y+ACAS+ +CF+EGYCWTRTGERQAA+MR +YLKAVLRQ+V YFDLHV
Sbjct: 62 MQTVAKNAVALVYVACASWVICFIEGYCWTRTGERQAAKMREKYLKAVLRQDVGYFDLHV 121
Query: 136 TSTTDVITSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFALLWRLAIVGFPFMVLL 195
TST+DVITSVSSDSLVIQD LS+K+PNFL+N S F++S IV F LLWRL IVGFPF++LL
Sbjct: 122 TSTSDVITSVSSDSLVIQDFLSEKLPNFLMNTSAFVASYIVGFLLLWRLTIVGFPFIILL 181
Query: 196 VIPGYMYKRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFVGESKTLAAFSNALEGSV 255
+IPG MY R +R++ KIREEYN+AG+IAEQ ISS+RTVY+F E K + FS AL+GSV
Sbjct: 182 LIPGLMYGRALIRISMKIREEYNEAGSIAEQVISSVRTVYAFGSEKKMIEKFSTALQGSV 241
Query: 256 KLGLKQGLAKGLAIGSNGVVYAIWSLIFYYGSIMVMYHGAKGGTVFVVGVTLAIGGLAFG 315
KLGL+QGLAKG+AIGSNG+ YAIW + +YGS MVM HG+KGGTV V V + GG + G
Sbjct: 242 KLGLRQGLAKGIAIGSNGITYAIWGFLTWYGSRMVMNHGSKGGTVSSVIVCVTFGGTSLG 301
Query: 316 TCFSNVRYFAEASVAGERIMEVIKRVPTIDSENMEGEIIEKVLGEVEFNNVEFVYPSRPE 375
SN++YF+EA V GERIM+VI RVP IDS+N+EG+I+EK GEVEFN+V+F YPSRPE
Sbjct: 302 QSLSNLKYFSEAFVVGERIMKVINRVPGIDSDNLEGQILEKTRGEVEFNHVKFTYPSRPE 361
Query: 376 SVILNDFCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGGEILLDGVAIHKLQLKWL 435
+ I +D CL+VPSGKTVALVGGSGSGKSTV+SLLQRFYDPI GEIL+DG+ I+KLQ+KWL
Sbjct: 362 TPIFDDLCLRVPSGKTVALVGGSGSGKSTVISLLQRFYDPIAGEILIDGLPINKLQVKWL 421
Query: 436 RSQMGLVSQEPALFATSIKENILFGREDATYEEIVDAAKASNAHNFISMLPQGYDTQV 493
RSQMGLVSQEP LFATSIKENILFG+EDA+ +E+V+AAKASNAH+FIS P Y TQV
Sbjct: 422 RSQMGLVSQEPVLFATSIKENILFGKEDASMDEVVEAAKASNAHSFISQFPNSYQTQV 479
Score = 228 bits (582), Expect = 5e-60
Identities = 145/495 (29%), Positives = 256/495 (51%), Gaps = 17/495 (3%)
Query: 5 DQKNVSINDKKKKNGSFKSIFMHADVLDWFFMAFGFFGAIGDGMMVPFVLFITSKIMNSV 64
D N+S D K SFK + M + +W +G GA G + P + + +++
Sbjct: 627 DFPNLSPKDGKSLVPSFKRL-MSMNRPEWKHALYGCLGAALFGAVQPIYSYSSGSMVSVY 685
Query: 65 GSASGTSSSNFVHDVNKNAV---VVLYMACA--SFFVCFLEGYCWTRTGERQAARMRVRY 119
AS HD K V+L++ A +F + Y + GE R+R R
Sbjct: 686 FLAS--------HDQIKEKTRIYVLLFVGLALFTFLSNISQHYGFAYMGEYLTKRIRERM 737
Query: 120 LKAVLRQEVSYFDLHVTSTTDVITSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFA 179
L +L EV++FD S+ + + ++ D+ +++ ++ D++ + S + +
Sbjct: 738 LGKILTFEVNWFDKDENSSGAICSRLAKDANMVRSLVGDRMSLLVQTISAVSITCAIGLV 797
Query: 180 LLWRLAIVGFPFMVLLVIPGYMYKRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFVG 239
+ WR +IV ++V+ Y + + ++R + +++ +A +A+S+IRT+ +F
Sbjct: 798 ISWRFSIVMMSVQPVIVVCFYTQRVLLKSMSRNAIKGQDESSKLAAEAVSNIRTITAFSS 857
Query: 240 ESKTLAAFSNALEGSVKLGLKQGLAKGLAIG-SNGVVYAIWSLIFYYGSIMVMYHGAKGG 298
+ + + EG K +Q G+ +G S ++ + +L F+YG ++
Sbjct: 858 QERIINLLKMVQEGPRKDSARQSWLAGIMLGTSQSLITCVSALNFWYGGKLIADGKMMSK 917
Query: 299 TVFVVGVTLAIGGLAFGTCFSNVRYFAEASVAGERIMEVIKRVPTIDSENMEGEIIEKVL 358
+ + A G + + + S A + V+ R TI+ EN +G + +KV
Sbjct: 918 EFLEIFLIFASTGRVIAEAGTMTKDLVKGSDAVASVFAVLDRNTTIEPENPDGYVPKKVK 977
Query: 359 GEVEFNNVEFVYPSRPESVILNDFCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGG 418
G++ F+NV+F YP+RP+ +I +F + + GK+ A+VG SGSGKST++SL++RFYDP+ G
Sbjct: 978 GQISFSNVDFAYPTRPDVIIFQNFSIDIEDGKSTAIVGPSGSGKSTIISLIERFYDPLKG 1037
Query: 419 EILLDGVAIHKLQLKWLRSQMGLVSQEPALFATSIKENILFGREDATYE--EIVDAAKAS 476
+ +DG I L+ LR + LVSQEP LFA +I+ENI++G + EI++AAKA+
Sbjct: 1038 IVKIDGRDIRSCHLRSLRQHIALVSQEPTLFAGTIRENIMYGGASNKIDESEIIEAAKAA 1097
Query: 477 NAHNFISMLPQGYDT 491
NAH+FI+ L GYDT
Sbjct: 1098 NAHDFITSLSNGYDT 1112
>At3g28380 P-glycoprotein, putative
Length = 1240
Score = 658 bits (1697), Expect = 0.0
Identities = 322/493 (65%), Positives = 405/493 (81%), Gaps = 2/493 (0%)
Query: 1 MGGGDQKNVSINDKKKKNGSFKSIFMHADVLDWFFMAFGFFGAIGDGMMVPFVLFITSKI 60
MG D+K S DK K GS +SIFMHAD +DW MA G GA+GDG + P V+FI + +
Sbjct: 1 MGKEDEKE-SGRDKMKSFGSIRSIFMHADGVDWILMALGLIGAVGDGFITPVVVFIFNTL 59
Query: 61 MNSVGSASGTSSSNFVHDVNKNAVVVLYMACASFFVCFLEGYCWTRTGERQAARMRVRYL 120
+N++G++S +++ F+ ++KN V +LY+AC S+ +CFLEGYCWTRTGERQAARMR +YL
Sbjct: 60 LNNLGTSS-SNNKTFMQTISKNVVALLYVACGSWVICFLEGYCWTRTGERQAARMREKYL 118
Query: 121 KAVLRQEVSYFDLHVTSTTDVITSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFAL 180
+AVLRQ+V YFDLHVTST+DVITS+SSDSLVIQD LS+K+PNFL+NAS F++S IV+F L
Sbjct: 119 RAVLRQDVGYFDLHVTSTSDVITSISSDSLVIQDFLSEKLPNFLMNASAFVASYIVSFIL 178
Query: 181 LWRLAIVGFPFMVLLVIPGYMYKRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFVGE 240
+WRL IVGFPF++LL++PG MY R + ++RKI E+YN+AG+IAEQAISS+RTVY+F E
Sbjct: 179 MWRLTIVGFPFIILLLVPGLMYGRALVSISRKIHEQYNEAGSIAEQAISSVRTVYAFGSE 238
Query: 241 SKTLAAFSNALEGSVKLGLKQGLAKGLAIGSNGVVYAIWSLIFYYGSIMVMYHGAKGGTV 300
+K + FS AL GSVKLGL+QGLAKG+ IGSNGV +AIW+ + +YGS +VM HG+KGGTV
Sbjct: 239 NKMIGKFSTALRGSVKLGLRQGLAKGITIGSNGVTHAIWAFLTWYGSRLVMNHGSKGGTV 298
Query: 301 FVVGVTLAIGGLAFGTCFSNVRYFAEASVAGERIMEVIKRVPTIDSENMEGEIIEKVLGE 360
FVV + GG++ G SN++YF+EA VA ERI+EVIKRVP IDS EG+I+E++ GE
Sbjct: 299 FVVISCITYGGVSLGQSLSNLKYFSEAFVAWERILEVIKRVPDIDSNKKEGQILERMKGE 358
Query: 361 VEFNNVEFVYPSRPESVILNDFCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGGEI 420
VEFN+V+F Y SRPE+ I +D CLK+P+GKTVALVGGSGSGKSTV+SLLQRFYDPI GEI
Sbjct: 359 VEFNHVKFTYLSRPETTIFDDLCLKIPAGKTVALVGGSGSGKSTVISLLQRFYDPIAGEI 418
Query: 421 LLDGVAIHKLQLKWLRSQMGLVSQEPALFATSIKENILFGREDATYEEIVDAAKASNAHN 480
L+DGV+I KLQ+ WLRSQMGLVSQEP LFATSI ENILFG+EDA+ +E+V+AAKASNAH
Sbjct: 419 LIDGVSIDKLQVNWLRSQMGLVSQEPVLFATSITENILFGKEDASLDEVVEAAKASNAHT 478
Query: 481 FISMLPQGYDTQV 493
FIS P GY TQV
Sbjct: 479 FISQFPLGYKTQV 491
Score = 231 bits (589), Expect = 7e-61
Identities = 137/464 (29%), Positives = 246/464 (52%), Gaps = 8/464 (1%)
Query: 32 DWFFMAFGFFGAIGDGMMVPFVLFITSKIMNSVGSASGTSSSNFVHDVNKNAVVVLY-MA 90
+W +G A G++ P +++ SV S +S + + + + V++ +A
Sbjct: 668 EWKHALYGCLSAALVGVLQP----VSAYSAGSVISVFFLTSHDQIKEKTRIYVLLFVGLA 723
Query: 91 CASFFVCFLEGYCWTRTGERQAARMRVRYLKAVLRQEVSYFDLHVTSTTDVITSVSSDSL 150
SF V + Y + GE R+R + L +L EV++FD+ S+ + + ++ D+
Sbjct: 724 IFSFLVNISQHYGFAYMGEYLTKRIREQMLSKILTFEVNWFDIDDNSSGAICSRLAKDAN 783
Query: 151 VIQDVLSDKVPNFLVNASRFLSSNIVAFALLWRLAIVGFPFMVLLVIPGYMYKRISMRLA 210
V++ ++ D++ + S + + I+ + WRLAIV L+V+ Y + + L+
Sbjct: 784 VVRSMVGDRMSLLVQTISAVIIACIIGLVIAWRLAIVMISVQPLIVVCFYTQRVLLKSLS 843
Query: 211 RKIREEYNQAGTIAEQAISSIRTVYSFVGESKTLAAFSNALEGSVKLGLKQGLAKGLAIG 270
K + +++ +A +A+S+IRT+ +F + + + EG + + + G+ +G
Sbjct: 844 EKASKAQDESSKLAAEAVSNIRTITAFSSQERIIKLLKKVQEGPRRESVHRSWLAGIVLG 903
Query: 271 -SNGVVYAIWSLIFYYGSIMVMYHGAKGGTVFVVGVTLAIGGLAFGTCFSNVRYFAEASV 329
S ++ +L F+YG ++ F + + G + A
Sbjct: 904 TSRSLITCTSALNFWYGGRLIADGKIVSKAFFEIFLIFVTTGRVIADAGTMTTDLARGLD 963
Query: 330 AGERIMEVIKRVPTIDSENMEGEIIEKVLGEVEFNNVEFVYPSRPESVILNDFCLKVPSG 389
A + V+ R TI+ +N +G + EK+ G++ F NV+F YP+RP+ VI +F +++ G
Sbjct: 964 AVGSVFAVLDRCTTIEPKNPDGYVAEKIKGQITFLNVDFAYPTRPDVVIFENFSIEIDEG 1023
Query: 390 KTVALVGGSGSGKSTVVSLLQRFYDPIGGEILLDGVAIHKLQLKWLRSQMGLVSQEPALF 449
K+ A+VG SGSGKST++ L++RFYDP+ G + +DG I L+ LR + LVSQEP LF
Sbjct: 1024 KSTAIVGTSGSGKSTIIGLIERFYDPLKGTVKIDGRDIRSYHLRSLRKYISLVSQEPMLF 1083
Query: 450 ATSIKENILFG--REDATYEEIVDAAKASNAHNFISMLPQGYDT 491
A +I+ENI++G + EI++AAKA+NAH+FI+ L GYDT
Sbjct: 1084 AGTIRENIMYGGTSDKIDESEIIEAAKAANAHDFITSLSNGYDT 1127
>At3g28415 putative protein
Length = 1098
Score = 617 bits (1591), Expect = e-177
Identities = 306/478 (64%), Positives = 386/478 (80%), Gaps = 9/478 (1%)
Query: 16 KKNGSFKSIFMHADVLDWFFMAFGFFGAIGDGMMVPFVLFITSKIMNSVGSASGTSSSNF 75
K GS +SIFMHA+ +D M G GA+GDG + P + FIT ++N +G +S F
Sbjct: 2 KSFGSVRSIFMHANSVDLVLMGLGLIGAVGDGFITPIIFFITGLLLNDIGDSS-FGDKTF 60
Query: 76 VHDVNKNAVVVLYMACASFFVCFLEGYCWTRTGERQAARMRVRYLKAVLRQEVSYFDLHV 135
+H + KNAV +LY+A AS +CF+ GERQA+RMR +YL+AVLRQ+V YFDLHV
Sbjct: 61 MHAIMKNAVALLYVAGASLVICFV--------GERQASRMREKYLRAVLRQDVGYFDLHV 112
Query: 136 TSTTDVITSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFALLWRLAIVGFPFMVLL 195
TST+DVITSVSSD+LVIQDVLS+K+PNFL++AS F++S IV F +LWRL IVGFPF +LL
Sbjct: 113 TSTSDVITSVSSDTLVIQDVLSEKLPNFLMSASAFVASYIVGFIMLWRLTIVGFPFFILL 172
Query: 196 VIPGYMYKRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFVGESKTLAAFSNALEGSV 255
+IPG M R + ++RKIREEYN+AG+IAEQAIS +RTVY+F E K ++ FS ALEGSV
Sbjct: 173 LIPGLMCGRALINISRKIREEYNEAGSIAEQAISLVRTVYAFGSERKMISKFSAALEGSV 232
Query: 256 KLGLKQGLAKGLAIGSNGVVYAIWSLIFYYGSIMVMYHGAKGGTVFVVGVTLAIGGLAFG 315
KLGL+QG+AKG+AIGSNGV YAIW + +YGS MVMYHGAKGGT+F V + + GG + G
Sbjct: 233 KLGLRQGIAKGIAIGSNGVTYAIWGFMTWYGSRMVMYHGAKGGTIFAVIICITYGGTSLG 292
Query: 316 TCFSNVRYFAEASVAGERIMEVIKRVPTIDSENMEGEIIEKVLGEVEFNNVEFVYPSRPE 375
SN++YF+EA VAGERI+EVIKRVP IDS+N G+++E + GEV+F +V+F+Y SRPE
Sbjct: 293 RGLSNLKYFSEAVVAGERIIEVIKRVPDIDSDNPRGQVLENIKGEVQFKHVKFMYSSRPE 352
Query: 376 SVILNDFCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGGEILLDGVAIHKLQLKWL 435
+ I +D CL++PSGK+VALVGGSGSGKSTV+SLLQRFYDPI GEIL+DGV+I KLQ+KWL
Sbjct: 353 TPIFDDLCLRIPSGKSVALVGGSGSGKSTVISLLQRFYDPIVGEILIDGVSIKKLQVKWL 412
Query: 436 RSQMGLVSQEPALFATSIKENILFGREDATYEEIVDAAKASNAHNFISMLPQGYDTQV 493
RSQMGLVSQEPALFATSI+ENILFG+EDA+++E+V+AAK+SNAH+FIS P GY TQV
Sbjct: 413 RSQMGLVSQEPALFATSIEENILFGKEDASFDEVVEAAKSSNAHDFISQFPLGYKTQV 470
Score = 211 bits (538), Expect = 6e-55
Identities = 128/477 (26%), Positives = 245/477 (50%), Gaps = 7/477 (1%)
Query: 8 NVSINDKKKKNGSFKSIFMHADVLDWFFMAFGFFGAIGDGMMVPFVLFITSKIMNSVGSA 67
N++ + K K SFK + M + +W +G A+ G + P + + +++
Sbjct: 626 NLAGSIPKDKKPSFKRL-MAMNKPEWKHALYGCLSAVLYGALHPIYAYASGSMVSVYFL- 683
Query: 68 SGTSSSNFVHDVNKNAVVVLYMACASFFVCFLEGYCWTRTGERQAARMRVRYLKAVLRQE 127
TS ++ + +A F + ++ Y + GE R+R L +L E
Sbjct: 684 --TSHDEMKEKTRIYVLLFVGLAVLCFLISIIQQYSFAYMGEYLTKRIRENILSKLLTFE 741
Query: 128 VSYFDLHVTSTTDVITSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFALLWRLAIV 187
VS+FD S+ + + ++ D+ V++ ++ ++V + S + + A+ W+L+IV
Sbjct: 742 VSWFDEDENSSGSICSRLAKDANVVRSLVGERVSLLVQTISAVSVACTLGLAISWKLSIV 801
Query: 188 GFPFMVLLVIPGYMYKRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFVGESKTLAAF 247
++V Y + + +++K + +++ +A +A+S+IRT+ +F + + L
Sbjct: 802 MIAIQPVVVGCFYTQRIVLKSISKKAIKAQDESSKLAAEAVSNIRTITAFSSQERILKLL 861
Query: 248 SNALEGSVKLGLKQGLAKGLAIG-SNGVVYAIWSLIFYYGSIMVMYHGAKGGTVFVVGVT 306
EG + ++Q G+ + S ++ +L ++YG+ +++ F + +
Sbjct: 862 KMVQEGPQRENIRQSWLAGIVLATSRSLMTCTSALNYWYGARLIIDGKITSKAFFELFIL 921
Query: 307 LAIGGLAFGTCFSNVRYFAEASVAGERIMEVIKRVPTIDSENMEGEIIEKVLGEVEFNNV 366
G + A+ S A + V+ R I+ E +G + + + G+++F NV
Sbjct: 922 FVSTGRVIADAGAMTMDLAKGSDAVGSVFAVLDRYTNIEPEKPDGFVPQNIKGQIKFVNV 981
Query: 367 EFVYPSRPESVILNDFCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGGEILLDGVA 426
+F YP+RP+ +I +F + + GK+ A+VG SGSGKST++ L++RFYDP+ G + +DG
Sbjct: 982 DFAYPTRPDVIIFKNFSIDIDEGKSTAIVGPSGSGKSTIIGLIERFYDPLKGIVKIDGRD 1041
Query: 427 IHKLQLKWLRSQMGLVSQEPALFATSIKENILFG--REDATYEEIVDAAKASNAHNF 481
I L+ LR +GLVSQEP LFA +I+ENI++G + EI++AAKA+NAH+F
Sbjct: 1042 IRSYHLRSLRQHIGLVSQEPILFAGTIRENIMYGGASDKIDESEIIEAAKAANAHDF 1098
>At3g28360 P-glycoprotein like protein
Length = 1158
Score = 595 bits (1535), Expect = e-170
Identities = 289/408 (70%), Positives = 351/408 (85%)
Query: 86 VLYMACASFFVCFLEGYCWTRTGERQAARMRVRYLKAVLRQEVSYFDLHVTSTTDVITSV 145
+LY+ACAS+ +CFLEGYCWTRTGERQAA+MR RYL+AVLRQ+V YFDLHVTST+D+ITSV
Sbjct: 1 MLYVACASWVICFLEGYCWTRTGERQAAKMRERYLRAVLRQDVGYFDLHVTSTSDIITSV 60
Query: 146 SSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFALLWRLAIVGFPFMVLLVIPGYMYKRI 205
SSDSLVIQD LS+K+PN L+NAS F+ S IV F LLWRL IVGFPF++LL+IPG MY R
Sbjct: 61 SSDSLVIQDFLSEKLPNILMNASAFVGSYIVGFMLLWRLTIVGFPFIILLLIPGLMYGRA 120
Query: 206 SMRLARKIREEYNQAGTIAEQAISSIRTVYSFVGESKTLAAFSNALEGSVKLGLKQGLAK 265
+ ++RKIREEYN+AG+IAEQAISS+RTVY+FV E K + FS+AL+GSVKLGL+QGLAK
Sbjct: 121 LIGISRKIREEYNEAGSIAEQAISSVRTVYAFVSEKKMIEKFSDALQGSVKLGLRQGLAK 180
Query: 266 GLAIGSNGVVYAIWSLIFYYGSIMVMYHGAKGGTVFVVGVTLAIGGLAFGTCFSNVRYFA 325
G+AIGSNG+VYAIW + +YGS MVM +G KGGTV V V + GG A G SN++YF+
Sbjct: 181 GIAIGSNGIVYAIWGFLTWYGSRMVMNYGYKGGTVSTVTVCVTFGGTALGQALSNLKYFS 240
Query: 326 EASVAGERIMEVIKRVPTIDSENMEGEIIEKVLGEVEFNNVEFVYPSRPESVILNDFCLK 385
EA VAGERI ++IKRVP IDS+N+ G I+E + GEVEFNNV+ YPSRPE++I +D CLK
Sbjct: 241 EAFVAGERIQKMIKRVPDIDSDNLNGHILETIRGEVEFNNVKCKYPSRPETLIFDDLCLK 300
Query: 386 VPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGGEILLDGVAIHKLQLKWLRSQMGLVSQE 445
+PSGKTVALVGGSGSGKSTV+SLLQRFYDP G+IL+D V+I+ +Q+KWLRSQMG+VSQE
Sbjct: 301 IPSGKTVALVGGSGSGKSTVISLLQRFYDPNEGDILIDSVSINNMQVKWLRSQMGMVSQE 360
Query: 446 PALFATSIKENILFGREDATYEEIVDAAKASNAHNFISMLPQGYDTQV 493
P+LFATSIKENILFG+EDA+++E+V+AAKASNAHNFIS P GY TQV
Sbjct: 361 PSLFATSIKENILFGKEDASFDEVVEAAKASNAHNFISQFPHGYQTQV 408
Score = 229 bits (583), Expect = 4e-60
Identities = 140/482 (29%), Positives = 250/482 (51%), Gaps = 7/482 (1%)
Query: 13 DKKKKNGSFKSIFMHADVLDWFFMAFGFFGAIGDGMMVPFVLFITSKIMNSVGSASGTSS 72
DKK SFK + M + +W G A G + P + +S +M SV T+
Sbjct: 566 DKKPLVPSFKRL-MAMNRPEWKHALCGCLSASLGGAVQPIYAY-SSGLMISVFFL--TNH 621
Query: 73 SNFVHDVNKNAVVVLYMACASFFVCFLEGYCWTRTGERQAARMRVRYLKAVLRQEVSYFD 132
+ ++ +A +FF + Y ++ GE R+R + L +L EV++FD
Sbjct: 622 EQIKENTRIYVLLFFGLALFTFFTSISQQYSFSYMGEYLTKRIREQMLSKILTFEVNWFD 681
Query: 133 LHVTSTTDVITSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFALLWRLAIVGFPFM 192
S+ + + ++ D+ V++ ++ +++ + S + + + + WR IV
Sbjct: 682 EEENSSGAICSRLAKDANVVRSLVGERMSLLVQTISTVMVACTIGLVIAWRFTIVMISVQ 741
Query: 193 VLLVIPGYMYKRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFVGESKTLAAFSNALE 252
++++ Y+ + + +++K +++ +A +A+S+IRT+ +F + + + E
Sbjct: 742 PVIIVCYYIQRVLLKNMSKKAIIAQDESSKLAAEAVSNIRTITTFSSQERIMKLLERVQE 801
Query: 253 GSVKLGLKQGLAKGLAIGSN-GVVYAIWSLIFYYGSIMVMYHGAKGGTVFVVGVTLAIGG 311
G + +Q G+ +G+ ++ +L F+YG ++ F + + G
Sbjct: 802 GPRRESARQSWLAGIMLGTTQSLITCTSALNFWYGGKLIADGKMVSKAFFELFLIFKTTG 861
Query: 312 LAFGTCFSNVRYFAEASVAGERIMEVIKRVPTIDSENMEGEIIEKVLGEVEFNNVEFVYP 371
A + A+ S + + + V+ R TI+ EN +G I+EK+ G++ F NV+F YP
Sbjct: 862 RAIAEAGTMTTDLAKGSNSVDSVFTVLDRRTTIEPENPDGYILEKIKGQITFLNVDFAYP 921
Query: 372 SRPESVILNDFCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGGEILLDGVAIHKLQ 431
+RP VI N+F +++ GK+ A+VG S SGKSTV+ L++RFYDP+ G + +DG I
Sbjct: 922 TRPNMVIFNNFSIEIHEGKSTAIVGPSRSGKSTVIGLIERFYDPLQGIVKIDGRDIRSYH 981
Query: 432 LKWLRSQMGLVSQEPALFATSIKENILFGREDATYE--EIVDAAKASNAHNFISMLPQGY 489
L+ LR M LVSQEP LFA +I+ENI++GR + EI++A K +NAH FI+ L GY
Sbjct: 982 LRSLRQHMSLVSQEPTLFAGTIRENIMYGRASNKIDESEIIEAGKTANAHEFITSLSDGY 1041
Query: 490 DT 491
DT
Sbjct: 1042 DT 1043
>At3g28860 P-glycoprotein, putative
Length = 1252
Score = 418 bits (1074), Expect = e-117
Identities = 219/492 (44%), Positives = 318/492 (64%), Gaps = 5/492 (1%)
Query: 5 DQKNVSINDKKKKNGS--FKSIFMHADVLDWFFMAFGFFGAIGDGMMVPFVLFITSKIMN 62
D K V +KKK S F +F AD D+ M G GAI G +P + +++N
Sbjct: 8 DAKTVPAEAEKKKEQSLPFFKLFSFADKFDYLLMFVGSLGAIVHGSSMPVFFLLFGQMVN 67
Query: 63 SVGSASGTSSSNFVHDVNKNAVVVLYMACASFFVCFLEGYCWTRTGERQAARMRVRYLKA 122
G + VH+V++ ++ +Y+ F + E CW +GERQ A +R +YL+A
Sbjct: 68 GFGK-NQMDLHQMVHEVSRYSLYFVYLGLVVCFSSYAEIACWMYSGERQVAALRKKYLEA 126
Query: 123 VLRQEVSYFDLHVTSTTDVITSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFALLW 182
VL+Q+V +FD T D++ SVS+D+L++QD +S+KV NF+ S FL+ +V F W
Sbjct: 127 VLKQDVGFFDTDAR-TGDIVFSVSTDTLLVQDAISEKVGNFIHYLSTFLAGLVVGFVSAW 185
Query: 183 RLAIVGFPFMVLLVIPGYMYKRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFVGESK 242
+LA++ + + G +Y + K RE Y AG IAEQAI+ +RTVYS+VGESK
Sbjct: 186 KLALLSVAVIPGIAFAGGLYAYTLTGITSKSRESYANAGVIAEQAIAQVRTVYSYVGESK 245
Query: 243 TLAAFSNALEGSVKLGLKQGLAKGLAIGSN-GVVYAIWSLIFYYGSIMVMYHGAKGGTVF 301
L A+S+A++ ++KLG K G+AKGL +G G+ W+L+F+Y + + GG F
Sbjct: 246 ALNAYSDAIQYTLKLGYKAGMAKGLGLGCTYGIACMSWALVFWYAGVFIRNGQTDGGKAF 305
Query: 302 VVGVTLAIGGLAFGTCFSNVRYFAEASVAGERIMEVIKRVPTIDSENMEGEIIEKVLGEV 361
+ +GG++ G FSN+ F++ AG ++ME+I + PTI + ++G+ +++V G +
Sbjct: 306 TAIFSAIVGGMSLGQSFSNLGAFSKGKAAGYKLMEIINQRPTIIQDPLDGKCLDQVHGNI 365
Query: 362 EFNNVEFVYPSRPESVILNDFCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGGEIL 421
EF +V F YPSRP+ +I +F + PSGKTVA+VGGSGSGKSTVVSL++RFYDP G+IL
Sbjct: 366 EFKDVTFSYPSRPDVMIFRNFNIFFPSGKTVAVVGGSGSGKSTVVSLIERFYDPNSGQIL 425
Query: 422 LDGVAIHKLQLKWLRSQMGLVSQEPALFATSIKENILFGREDATYEEIVDAAKASNAHNF 481
LDGV I LQLK+LR Q+GLV+QEPALFAT+I ENIL+G+ DAT E+ AA A+NAH+F
Sbjct: 426 LDGVEIKTLQLKFLREQIGLVNQEPALFATTILENILYGKPDATMVEVEAAASAANAHSF 485
Query: 482 ISMLPQGYDTQV 493
I++LP+GYDTQV
Sbjct: 486 ITLLPKGYDTQV 497
Score = 248 bits (633), Expect = 6e-66
Identities = 147/465 (31%), Positives = 251/465 (53%), Gaps = 8/465 (1%)
Query: 32 DWFFMAFGFFGAIGDGMMVPFVLFITSKIMNSVGSASGTSSSNFVHDVNKNAVVVLYMAC 91
+W + G G+I G + P + S ++ S + V +Y+
Sbjct: 683 EWPYSIMGAVGSILSGFIGPTFAIVMSNMIEVFYYTDYDSMERKTKEY-----VFIYIGA 737
Query: 92 ASFFV--CFLEGYCWTRTGERQAARMRVRYLKAVLRQEVSYFDLHVTSTTDVITSVSSDS 149
+ V ++ Y ++ GE R+R L A+LR EV +FD +++ + +++D+
Sbjct: 738 GLYAVGAYLIQHYFFSIMGENLTTRVRRMMLSAILRNEVGWFDEDEHNSSLIAARLATDA 797
Query: 150 LVIQDVLSDKVPNFLVNASRFLSSNIVAFALLWRLAIVGFPFMVLLVIPGYMYKRISMRL 209
++ +++++ L N + L+S IVAF + WR++++ LLV+ + +
Sbjct: 798 ADVKSAIAERISVILQNMTSLLTSFIVAFIVEWRVSLLILGTFPLLVLANFAQQLSLKGF 857
Query: 210 ARKIREEYNQAGTIAEQAISSIRTVYSFVGESKTLAAFSNALEGSVKLGLKQGLAKGLAI 269
A + + + IA + +S+IRTV +F +SK L+ F + L K L + G
Sbjct: 858 AGDTAKAHAKTSMIAGEGVSNIRTVAAFNAQSKILSLFCHELRVPQKRSLYRSQTSGFLF 917
Query: 270 G-SNGVVYAIWSLIFYYGSIMVMYHGAKGGTVFVVGVTLAIGGLAFGTCFSNVRYFAEAS 328
G S +Y +LI +YG+ +V + V V V L I + S
Sbjct: 918 GLSQLALYGSEALILWYGAHLVSKGVSTFSKVIKVFVVLVITANSVAETVSLAPEIIRGG 977
Query: 329 VAGERIMEVIKRVPTIDSENMEGEIIEKVLGEVEFNNVEFVYPSRPESVILNDFCLKVPS 388
A + V+ R ID ++ + + +E + G++EF +V+F YPSRP+ ++ DF L++ +
Sbjct: 978 EAVGSVFSVLDRQTRIDPDDADADPVETIRGDIEFRHVDFAYPSRPDVMVFRDFNLRIRA 1037
Query: 389 GKTVALVGGSGSGKSTVVSLLQRFYDPIGGEILLDGVAIHKLQLKWLRSQMGLVSQEPAL 448
G + ALVG SGSGKS+V+++++RFYDP+ G++++DG I +L LK LR ++GLV QEPAL
Sbjct: 1038 GHSQALVGASGSGKSSVIAMIERFYDPLAGKVMIDGKDIRRLNLKSLRLKIGLVQQEPAL 1097
Query: 449 FATSIKENILFGREDATYEEIVDAAKASNAHNFISMLPQGYDTQV 493
FA +I +NI +G++ AT E++DAA+A+NAH FIS LP+GY T V
Sbjct: 1098 FAATIFDNIAYGKDGATESEVIDAARAANAHGFISGLPEGYKTPV 1142
>At2g36910 putative ABC transporter
Length = 1286
Score = 402 bits (1033), Expect = e-112
Identities = 212/475 (44%), Positives = 307/475 (64%), Gaps = 3/475 (0%)
Query: 20 SFKSIFMHADVLDWFFMAFGFFGAIGDGMMVPFVLFITSKIMNSVGSASGTSSSNFVHDV 79
+FK +F AD LD+ M G GA G +P L + ++NS GS S + + +V
Sbjct: 28 AFKELFRFADGLDYVLMGIGSVGAFVHGCSLPLFLRFFADLVNSFGSNSN-NVEKMMEEV 86
Query: 80 NKNAVVVLYMACASFFVCFLEGYCWTRTGERQAARMRVRYLKAVLRQEVSYFDLHVTSTT 139
K A+ L + A + + E CW +GERQ +MR++YL+A L Q++ +FD V T+
Sbjct: 87 LKYALYFLVVGAAIWASSWAEISCWMWSGERQTTKMRIKYLEAALNQDIQFFDTEVR-TS 145
Query: 140 DVITSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFALLWRLAIVGFPFMVLLVIPG 199
DV+ ++++D++++QD +S+K+ NF+ + F+S IV F +W+LA+V + L+ + G
Sbjct: 146 DVVFAINTDAVMVQDAISEKLGNFIHYMATFVSGFIVGFTAVWQLALVTLAVVPLIAVIG 205
Query: 200 YMYKRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFVGESKTLAAFSNALEGSVKLGL 259
++ +L+ K +E +QAG I EQ + IR V +FVGES+ A+S+AL+ + KLG
Sbjct: 206 GIHTTTLSKLSNKSQESLSQAGNIVEQTVVQIRVVMAFVGESRASQAYSSALKIAQKLGY 265
Query: 260 KQGLAKGLAIGSNG-VVYAIWSLIFYYGSIMVMYHGAKGGTVFVVGVTLAIGGLAFGTCF 318
K GLAKG+ +G+ VV+ ++L+ +YG +V +H GG + IGGLA G
Sbjct: 266 KTGLAKGMGLGATYFVVFCCYALLLWYGGYLVRHHLTNGGLAIATMFAVMIGGLALGQSA 325
Query: 319 SNVRYFAEASVAGERIMEVIKRVPTIDSENMEGEIIEKVLGEVEFNNVEFVYPSRPESVI 378
++ FA+A VA +I +I PTI+ + G ++ V G VE NV+F YPSRP+ I
Sbjct: 326 PSMAAFAKAKVAAAKIFRIIDHKPTIERNSESGVELDSVTGLVELKNVDFSYPSRPDVKI 385
Query: 379 LNDFCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGGEILLDGVAIHKLQLKWLRSQ 438
LN+FCL VP+GKT+ALVG SGSGKSTVVSL++RFYDP G++LLDG + L+L+WLR Q
Sbjct: 386 LNNFCLSVPAGKTIALVGSSGSGKSTVVSLIERFYDPNSGQVLLDGQDLKTLKLRWLRQQ 445
Query: 439 MGLVSQEPALFATSIKENILFGREDATYEEIVDAAKASNAHNFISMLPQGYDTQV 493
+GLVSQEPALFATSIKENIL GR DA EI +AA+ +NAH+FI LP G+DTQV
Sbjct: 446 IGLVSQEPALFATSIKENILLGRPDADQVEIEEAARVANAHSFIIKLPDGFDTQV 500
Score = 222 bits (566), Expect = 3e-58
Identities = 137/489 (28%), Positives = 250/489 (51%), Gaps = 7/489 (1%)
Query: 7 KNVSINDKKKKNGSFKSIFMHADVLDWFFMAFGFFGAIGDGMMVPFVLFITSKIMNSVGS 66
+N + K + N ++ M++ +W + G G++ G + F ++ S +++ +
Sbjct: 673 RNEKLAFKDQANSFWRLAKMNSP--EWKYALLGSVGSVICGSLSAFFAYVLSAVLSVYYN 730
Query: 67 ASGTSSSNFVHDVNKNAVVVLYMACASFFVCFLEGYCWTRTGERQAARMRVRYLKAVLRQ 126
+ ++K +++ ++ A+ L+ W GE R+R + L AVL+
Sbjct: 731 PD---HEYMIKQIDKYCYLLIGLSSAALVFNTLQHSFWDIVGENLTKRVREKMLSAVLKN 787
Query: 127 EVSYFDLHVTSTTDVITSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFALLWRLAI 186
E+++FD + + ++ D+ ++ + D++ + N + L + F L WRLA+
Sbjct: 788 EMAWFDQEENESARIAARLALDANNVRSAIGDRISVIVQNTALMLVACTAGFVLQWRLAL 847
Query: 187 VGFPFMVLLVIPGYMYKRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFVGESKTLAA 246
V ++V + K + + + + +A +AI+++RTV +F E+K +
Sbjct: 848 VLVAVFPVVVAATVLQKMFMTGFSGDLEAAHAKGTQLAGEAIANVRTVAAFNSEAKIVRL 907
Query: 247 FSNALEGSVKLGLKQGLAKGLAIG-SNGVVYAIWSLIFYYGSIMVMYHGAKGGTVFVVGV 305
++ LE +K +G G G + +YA ++L +Y S +V + + V +
Sbjct: 908 YTANLEPPLKRCFWKGQIAGSGYGVAQFCLYASYALGLWYASWLVKHGISDFSKTIRVFM 967
Query: 306 TLAIGGLAFGTCFSNVRYFAEASVAGERIMEVIKRVPTIDSENMEGEII-EKVLGEVEFN 364
L + + F + A + E++ R I+ ++ + + +++ GEVE
Sbjct: 968 VLMVSANGAAETLTLAPDFIKGGQAMRSVFELLDRKTEIEPDDPDTTPVPDRLRGEVELK 1027
Query: 365 NVEFVYPSRPESVILNDFCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGGEILLDG 424
+++F YPSRP+ I D L+ +GKT+ALVG SG GKS+V+SL+QRFY+P G +++DG
Sbjct: 1028 HIDFSYPSRPDIQIFRDLSLRARAGKTLALVGPSGCGKSSVISLIQRFYEPSSGRVMIDG 1087
Query: 425 VAIHKLQLKWLRSQMGLVSQEPALFATSIKENILFGREDATYEEIVDAAKASNAHNFISM 484
I K LK +R + +V QEP LF T+I ENI +G E AT EI+ AA ++AH FIS
Sbjct: 1088 KDIRKYNLKAIRKHIAIVPQEPCLFGTTIYENIAYGHECATEAEIIQAATLASAHKFISA 1147
Query: 485 LPQGYDTQV 493
LP+GY T V
Sbjct: 1148 LPEGYKTYV 1156
>At4g25960 P-glycoprotein-2 (pgp2)
Length = 1233
Score = 374 bits (960), Expect = e-104
Identities = 201/475 (42%), Positives = 297/475 (62%), Gaps = 3/475 (0%)
Query: 20 SFKSIFMHADVLDWFFMAFGFFGAIGDGMMVPFVLFITSKIMNSVGSASGTSSSNFVHDV 79
S +F AD D M G GA G VP K++N +G A H V
Sbjct: 21 SLLKLFSFADFYDCVLMTLGSVGACIHGASVPIFFIFFGKLINIIGLAY-LFPKQASHRV 79
Query: 80 NKNAVVVLYMACASFFVCFLEGYCWTRTGERQAARMRVRYLKAVLRQEVSYFDLHVTSTT 139
K ++ +Y++ A F +LE CW TGERQAA+MR YL+++L Q++S FD ST
Sbjct: 80 AKYSLDFVYLSVAILFSSWLEVACWMHTGERQAAKMRRAYLRSMLSQDISLFDTEA-STG 138
Query: 140 DVITSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFALLWRLAIVGFPFMVLLVIPG 199
+VI++++SD LV+QD LS+KV NFL SRF++ + F +W++++V + L+ + G
Sbjct: 139 EVISAITSDILVVQDALSEKVGNFLHYISRFIAGFAIGFTSVWQISLVTLSIVPLIALAG 198
Query: 200 YMYKRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFVGESKTLAAFSNALEGSVKLGL 259
+Y +++ L ++R+ Y +AG IAE+ I ++RTV +F GE + + + ALE + K G
Sbjct: 199 GIYAFVAIGLIARVRKSYIKAGEIAEEVIGNVRTVQAFTGEERAVRLYREALENTYKYGR 258
Query: 260 KQGLAKGLAIGS-NGVVYAIWSLIFYYGSIMVMYHGAKGGTVFVVGVTLAIGGLAFGTCF 318
K GL KGL +GS + V++ W+L+ ++ S++V A GG F + + I GL+ G
Sbjct: 259 KAGLTKGLGLGSMHCVLFLSWALLVWFTSVVVHKDIADGGKSFTTMLNVVIAGLSLGQAA 318
Query: 319 SNVRYFAEASVAGERIMEVIKRVPTIDSENMEGEIIEKVLGEVEFNNVEFVYPSRPESVI 378
++ F A A I ++I+R + G + KV G ++F + F YPSRP+ VI
Sbjct: 319 PDISAFVRAKAAAYPIFKMIERNTVTKTSAKSGRKLGKVDGHIQFKDATFSYPSRPDVVI 378
Query: 379 LNDFCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGGEILLDGVAIHKLQLKWLRSQ 438
+ L +P+GK VALVGGSGSGKSTV+SL++RFY+PI G +LLDG I +L +KWLR Q
Sbjct: 379 FDRLNLAIPAGKIVALVGGSGSGKSTVISLIERFYEPISGAVLLDGNNISELDIKWLRGQ 438
Query: 439 MGLVSQEPALFATSIKENILFGREDATYEEIVDAAKASNAHNFISMLPQGYDTQV 493
+GLV+QEPALFAT+I+ENIL+G++DAT EEI AAK S A +FI+ LP+G++TQV
Sbjct: 439 IGLVNQEPALFATTIRENILYGKDDATAEEITRAAKLSEAISFINNLPEGFETQV 493
Score = 249 bits (637), Expect = 2e-66
Identities = 159/466 (34%), Positives = 256/466 (54%), Gaps = 13/466 (2%)
Query: 32 DWFFMAFGFFGAIGDGMMVP-FVLFITSKIMNSVGSASGTSSSNFVHDVNKNAVVVLYMA 90
DW + G A G +P F L ++ +++ T ++ K A++ +
Sbjct: 666 DWMYGVCGTICAFIAGSQMPLFALGVSQALVSYYSGWDETQK-----EIKKIAILFCCAS 720
Query: 91 CASFFVCFLEGYCWTRTGERQAARMRVRYLKAVLRQEVSYFDLHVTSTTDVITS-VSSDS 149
+ V +E C+ GER R+R +A+L+ E+ +FD V +T+ ++ S + SD+
Sbjct: 721 VITLIVYTIEHICFGTMGERLTLRVRENMFRAILKNEIGWFD-EVDNTSSMLASRLESDA 779
Query: 150 LVIQDVLSDKVPNFLVNASRFLSSNIVAFALLWRLAIVGFPFMVLLVIPGYMYKRISMR- 208
+++ ++ D+ L N ++S I+AF L WRL +V L VI G++ +++ M+
Sbjct: 780 TLLKTIVVDRSTILLQNLGLVVTSFIIAFILNWRLTLVVLATYPL-VISGHISEKLFMQG 838
Query: 209 LARKIREEYNQAGTIAEQAISSIRTVYSFVGESKTLAAFSNALEGSVKLGLKQGLAKGLA 268
+ + Y +A +A +++S+IRTV +F E K L +S L K ++G GL
Sbjct: 839 YGGDLNKAYLKANMLAGESVSNIRTVAAFCAEEKILELYSRELLEPSKSSFRRGQIAGLF 898
Query: 269 IG-SNGVVYAIWSLIFYYGSIMVMYHGAKGGTVFVVGVTLAIGGLAFGTCFSNVRYFAEA 327
G S +++ + L +YGS ++ A +V + L + LA G + +
Sbjct: 899 YGVSQFFIFSSYGLALWYGSTLMDKGLAGFKSVMKTFMVLIVTALAMGETLALAPDLLKG 958
Query: 328 SVAGERIMEVIKRVPTIDSENMEGEIIEKVLGEVEFNNVEFVYPSRPESVILNDFCLKVP 387
+ + E++ R I E E + V G +E V F YPSRP+ VI DF L V
Sbjct: 959 NQMVASVFEILDRKTQIVGETSEE--LNNVEGTIELKGVHFSYPSRPDVVIFRDFDLIVR 1016
Query: 388 SGKTVALVGGSGSGKSTVVSLLQRFYDPIGGEILLDGVAIHKLQLKWLRSQMGLVSQEPA 447
+GK++ALVG SGSGKS+V+SL+ RFYDP G+++++G I KL LK LR +GLV QEPA
Sbjct: 1017 AGKSMALVGQSGSGKSSVISLILRFYDPTAGKVMIEGKDIKKLDLKALRKHIGLVQQEPA 1076
Query: 448 LFATSIKENILFGREDATYEEIVDAAKASNAHNFISMLPQGYDTQV 493
LFAT+I ENIL+G E A+ E+V++A +NAH+FI+ LP+GY T+V
Sbjct: 1077 LFATTIYENILYGNEGASQSEVVESAMLANAHSFITSLPEGYSTKV 1122
>At1g10680 putative P-glycoprotein-2 emb|CAA71277
Length = 1227
Score = 369 bits (946), Expect = e-102
Identities = 197/482 (40%), Positives = 303/482 (61%), Gaps = 7/482 (1%)
Query: 13 DKKKKNGSFKSIFMHADVLDWFFMAFGFFGAIGDGMMVPFVLFITSKIMNSVGSASGTSS 72
+KK+ + SF +F AD D MA G GA G VP K++N +G A
Sbjct: 18 EKKRPSVSFLKLFSFADFYDCVLMALGSIGACIHGASVPVFFIFFGKLINIIGLAY-LFP 76
Query: 73 SNFVHDVNKNAVVVLYMACASFFVCFLEGYCWTRTGERQAARMRVRYLKAVLRQEVSYFD 132
H V K ++ +Y++ F +LE CW TGERQAA++R YL+++L Q++S FD
Sbjct: 77 QEASHKVAKYSLDFVYLSVVILFSSWLEVACWMHTGERQAAKIRKAYLRSMLSQDISLFD 136
Query: 133 LHVTSTTDVITSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFALLWRLAIVGFPFM 192
+ ST +VI++++S+ LV+QD +S+KV NF+ SRF++ + FA +W++++V +
Sbjct: 137 TEI-STGEVISAITSEILVVQDAISEKVGNFMHFISRFIAGFAIGFASVWQISLVTLSIV 195
Query: 193 VLLVIPGYMYKRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFVGESKTLAAFSNALE 252
+ + G +Y +S L ++R+ Y +A IAE+ I ++RTV +F GE K ++++ AL
Sbjct: 196 PFIALAGGIYAFVSSGLIVRVRKSYVKANEIAEEVIGNVRTVQAFTGEEKAVSSYQGALR 255
Query: 253 GSVKLGLKQGLAKGLAIGS-NGVVYAIWSLIFYYGSIMVMYHGAKGGTVFVVGVTLAIGG 311
+ G K GLAKGL +GS + V++ W+L+ ++ SI+V A GG F + + I G
Sbjct: 256 NTYNYGRKAGLAKGLGLGSLHFVLFLSWALLIWFTSIVVHKGIANGGESFTTMLNVVIAG 315
Query: 312 LAFGTCFSNVRYFAEASVAGERIMEVIKRVPTIDSENMEGEIIEKVLGEVEFNNVEFVYP 371
L+ G ++ F AS A I ++I+R ++E+ G + V G++ F +V F YP
Sbjct: 316 LSLGQAAPDISTFMRASAAAYPIFQMIER----NTEDKTGRKLGNVNGDILFKDVTFTYP 371
Query: 372 SRPESVILNDFCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGGEILLDGVAIHKLQ 431
SRP+ VI + +P+GK VALVGGSGSGKST++SL++RFY+P G ++LDG I L
Sbjct: 372 SRPDVVIFDKLNFVIPAGKVVALVGGSGSGKSTMISLIERFYEPTDGAVMLDGNDIRYLD 431
Query: 432 LKWLRSQMGLVSQEPALFATSIKENILFGREDATYEEIVDAAKASNAHNFISMLPQGYDT 491
LKWLR +GLV+QEP LFAT+I+ENI++G++DAT EEI +AAK S A +FI+ LP+G++T
Sbjct: 432 LKWLRGHIGLVNQEPVLFATTIRENIMYGKDDATSEEITNAAKLSEAISFINNLPEGFET 491
Query: 492 QV 493
QV
Sbjct: 492 QV 493
Score = 245 bits (626), Expect = 4e-65
Identities = 156/465 (33%), Positives = 258/465 (54%), Gaps = 11/465 (2%)
Query: 32 DWFFMAFGFFGAIGDGMMVP-FVLFITSKIMNSVGSASGTSSSNFVHDVNKNAVVVLYMA 90
DW + G G+ G +P F L I +++ T + +V + +++ +
Sbjct: 658 DWKYGLCGTLGSFIAGSQMPLFALGIAQALVSYYMDWETTQN-----EVKRISILFCCGS 712
Query: 91 CASFFVCFLEGYCWTRTGERQAARMRVRYLKAVLRQEVSYFDLHVTSTTDVITSVSSDSL 150
+ V +E + GER R+R + A+LR E+ +FD +++ + + + SD+
Sbjct: 713 VITVIVHTIEHTTFGIMGERLTLRVRQKMFSAILRNEIGWFDKVDNTSSMLASRLESDAT 772
Query: 151 VIQDVLSDKVPNFLVNASRFLSSNIVAFALLWRLAIVGFPFMVLLVIPGYMYKRISMR-L 209
+++ ++ D+ L N +++ I++F L WRL +V L +I G++ ++I M+
Sbjct: 773 LLRTIVVDRSTILLENLGLVVTAFIISFILNWRLTLVVLATYPL-IISGHISEKIFMQGY 831
Query: 210 ARKIREEYNQAGTIAEQAISSIRTVYSFVGESKTLAAFSNALEGSVKLGLKQGLAKGLAI 269
+ + Y +A +A ++IS+IRTV +F E K L +S L + ++G G+
Sbjct: 832 GGNLSKAYLKANMLAGESISNIRTVVAFCAEEKVLDLYSKELLEPSERSFRRGQMAGILY 891
Query: 270 G-SNGVVYAIWSLIFYYGSIMVMYHGAKGGTVFVVGVTLAIGGLAFGTCFSNVRYFAEAS 328
G S +++ + L +YGSI++ + +V + L + L G + + +
Sbjct: 892 GVSQFFIFSSYGLALWYGSILMEKGLSSFESVMKTFMVLIVTALVMGEVLALAPDLLKGN 951
Query: 329 VAGERIMEVIKRVPTIDSENMEGEIIEKVLGEVEFNNVEFVYPSRPESVILNDFCLKVPS 388
+ E++ R + + GE + V G +E V F YPSRP+ I +DF L VPS
Sbjct: 952 QMVVSVFELLDRRTQVVGDT--GEELSNVEGTIELKGVHFSYPSRPDVTIFSDFNLLVPS 1009
Query: 389 GKTVALVGGSGSGKSTVVSLLQRFYDPIGGEILLDGVAIHKLQLKWLRSQMGLVSQEPAL 448
GK++ALVG SGSGKS+V+SL+ RFYDP G I++DG I KL+LK LR +GLV QEPAL
Sbjct: 1010 GKSMALVGQSGSGKSSVLSLVLRFYDPTAGIIMIDGQDIKKLKLKSLRRHIGLVQQEPAL 1069
Query: 449 FATSIKENILFGREDATYEEIVDAAKASNAHNFISMLPQGYDTQV 493
FAT+I ENIL+G+E A+ E+++AAK +NAH+FIS LP+GY T+V
Sbjct: 1070 FATTIYENILYGKEGASESEVMEAAKLANAHSFISSLPEGYSTKV 1114
>At5g46540 multidrug resistance p-glycoprotein
Length = 1248
Score = 367 bits (943), Expect = e-102
Identities = 201/488 (41%), Positives = 300/488 (61%), Gaps = 12/488 (2%)
Query: 14 KKKKNG-------SFKSIFMHADVLDWFFMAFGFFGAIGDGMMVPFVLFITSKIMNSVGS 66
KKKKNG +F +F AD D M G A+ +G+ PF+ + +++N G
Sbjct: 6 KKKKNGGGGNQRIAFYKLFTFADRYDIVLMVIGTLSAMANGLTQPFMSILMGQLINVFGF 65
Query: 67 ASGTSSSNFVHDVNKNAVVVLYMACASFFVCFLEGYCWTRTGERQAARMRVRYLKAVLRQ 126
+ + +V+K AV LY+A + V FL+ CW TGERQ+ R+R YLK +LRQ
Sbjct: 66 SD---HDHVFKEVSKVAVKFLYLAAYAGVVSFLQVSCWMVTGERQSTRIRRLYLKTILRQ 122
Query: 127 EVSYFDLHVTSTTDVITSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFALLWRLAI 186
++ +FD T+T +VI +S D+++IQD + +KV F S F+ VAF + +L +
Sbjct: 123 DIGFFDTE-TNTGEVIGRMSGDTILIQDSMGEKVGKFTQLVSSFVGGFTVAFIVGMKLTL 181
Query: 187 VGFPFMVLLVIPGYMYKRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFVGESKTLAA 246
P + L+V G I + A++++ Y +AG + +QA+ SIRTV +F GE +++
Sbjct: 182 ALLPCVPLIVGTGGAMTYIMSKKAQRVQLAYTEAGNVVQQAVGSIRTVVAFTGEKQSMGK 241
Query: 247 FSNALEGSVKLGLKQGLAKGLAIG-SNGVVYAIWSLIFYYGSIMVMYHGAKGGTVFVVGV 305
+ LE + K +KQGL GL IG VVY + +YG+ ++ G GG V V
Sbjct: 242 YEKKLEIAYKSMVKQGLYSGLGIGIMMVVVYCTYGFAIWYGARQIIEKGYTGGQVMNVIT 301
Query: 306 TLAIGGLAFGTCFSNVRYFAEASVAGERIMEVIKRVPTIDSENMEGEIIEKVLGEVEFNN 365
++ GG+A G ++ FA + A ++ E IKR P ID+ +M GE++E++ G++E +
Sbjct: 302 SILTGGMALGQTLPSLNSFAAGTAAAYKMFETIKRKPKIDAYDMSGEVLEEIKGDIELRD 361
Query: 366 VEFVYPSRPESVILNDFCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGGEILLDGV 425
V F YP+RP+ I F L VP+G TVALVG SGSGKSTV+SL++RFYDP GE+L+DG+
Sbjct: 362 VYFRYPARPDVQIFVGFSLTVPNGMTVALVGQSGSGKSTVISLIERFYDPESGEVLIDGI 421
Query: 426 AIHKLQLKWLRSQMGLVSQEPALFATSIKENILFGREDATYEEIVDAAKASNAHNFISML 485
+ K Q+KW+RS++GLVSQEP LFAT+I+ENI++G++DA+ +EI A K +NA NFI L
Sbjct: 422 DLKKFQVKWIRSKIGLVSQEPILFATTIRENIVYGKKDASDQEIRTALKLANASNFIDKL 481
Query: 486 PQGYDTQV 493
PQG +T V
Sbjct: 482 PQGLETMV 489
Score = 265 bits (678), Expect = 3e-71
Identities = 154/413 (37%), Positives = 227/413 (54%), Gaps = 2/413 (0%)
Query: 83 AVVVLYMACASFFVCFLEGYCWTRTGERQAARMRVRYLKAVLRQEVSYFDLHVTSTTDVI 142
A++ + + V L+ Y + G + R+R VL Q++S+FD S+ +
Sbjct: 726 ALIFVALGLTDLIVIPLQNYLFAIAGAKLIKRIRSLSFDRVLHQDISWFDDTKNSSGVIG 785
Query: 143 TSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFALLWRLAIVGFPFMVLLVIPGYMY 202
+S+D+ ++ ++ D + + N + + + I+AF W LA++ ++ GY
Sbjct: 786 ARLSTDASTVKSIVGDVLGLIMQNMATIIGAFIIAFTANWLLALMALLVAPVMFFQGYYQ 845
Query: 203 KRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFVGESKTLAAFSNALEGSVKLGLKQG 262
+ K R +Y +A +A A+SSIRTV SF E K + + + + G K G
Sbjct: 846 IKFITGFGAKARGKYEEASQVASDAVSSIRTVASFCAEDKVMDLYQEKCDEPKQQGFKLG 905
Query: 263 LAKGLAIGSNGV-VYAIWSLIFYYGSIMVMYHGAKGGTVFVVGVTLAIGGLAFGTCFSNV 321
L GL G + + +Y I S+ F GS ++ A G F V L + + +
Sbjct: 906 LVSGLCYGGSYLALYVIESVCFLGGSWLIQNRRATFGEFFQVFFALTLTAVGVTQTSTMA 965
Query: 322 RYFAEASVAGERIMEVIKRVPTIDSENMEGEIIEKVLGEVEFNNVEFVYPSRPESVILND 381
+A + I +++ P IDS + +G I+ V G++E +V F YP RP+ I +D
Sbjct: 966 PDINKAKDSAASIFDILDSKPKIDSSSEKGTILPIVHGDIELQHVSFRYPMRPDIQIFSD 1025
Query: 382 FCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGGEILLDGVAIHKLQLKWLRSQMGL 441
CL + SG+TVALVG SGSGKSTV+SLL+RFYDP G+ILLD V I L+L WLR QMGL
Sbjct: 1026 LCLTISSGQTVALVGESGSGKSTVISLLERFYDPDSGKILLDQVEIQSLKLSWLREQMGL 1085
Query: 442 VSQEPALFATSIKENILFGR-EDATYEEIVDAAKASNAHNFISMLPQGYDTQV 493
VSQEP LF +I NI +G+ AT EEI+ AAKA+N HNFIS LPQGY+T V
Sbjct: 1086 VSQEPVLFNETIGSNIAYGKIGGATEEEIITAAKAANVHNFISSLPQGYETSV 1138
>At1g27940 hypothetical protein
Length = 1245
Score = 363 bits (931), Expect = e-100
Identities = 190/491 (38%), Positives = 300/491 (60%), Gaps = 10/491 (2%)
Query: 5 DQKNVSINDKKKKNGSFKSIFMHADVLDWFFMAFGFFGAIGDGMMVPFVLFITSKIMNSV 64
++KN+ KK++ S +F AD LD+F M G GA G +P K+++S+
Sbjct: 21 EKKNI-----KKESVSLMGLFSAADKLDYFLMLLGGLGACIHGATLPLFFVFFGKMLDSL 75
Query: 65 GSASGTSSSNFVHDVNKNAVVVLYMACASFFVCFLEGYCWTRTGERQAARMRVRYLKAVL 124
G+ S T V++NA+ ++Y+ +F ++ CW +TGERQ AR+R+ YLK++L
Sbjct: 76 GNLS-TDPKAISSRVSQNALYLVYLGLVNFVSAWIGVSCWMQTGERQTARLRINYLKSIL 134
Query: 125 RQEVSYFDLHVTSTTDVITSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFALLWRL 184
+++++FD + ++I +SSD++++QD + DK + L S+F++ ++ F +W+L
Sbjct: 135 AKDITFFDTEARDS-NLIFHISSDAILVQDAIGDKTDHVLRYLSQFIAGFVIGFLSVWQL 193
Query: 185 AIVGFPFMVLLVIPGYMYKRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFVGESKTL 244
++ + L+ I G Y + ++ K Y AG +AE+ +S +RTVY+FVGE K +
Sbjct: 194 TLLTLGVVPLIAIAGGGYAIVMSTISEKSETAYADAGKVAEEVMSQVRTVYAFVGEEKAV 253
Query: 245 AAFSNALEGSVKLGLKQGLAKGLAIG-SNGVVYAIWSLIFYYGSIMVMYHGAKGGTVFVV 303
++SN+L+ ++KLG + GLAKGL +G + +++ W+L+ +Y S++V + G F
Sbjct: 254 KSYSNSLKKALKLGKRSGLAKGLGVGLTYSLLFCAWALLLWYASLLVRHGKTNGAKAFTT 313
Query: 304 GVTLAIGGLAFGTCFSNVRYFAEASVAGERIMEVIKRVPTIDSENM-EGEIIEKVLGEVE 362
+ + G A G ++ A+ VA I +I + S+ + EG ++ V G +E
Sbjct: 314 ILNVIFSGFALGQAAPSLSAIAKGRVAAANIFRMIGNNNSESSQRLDEGTTLQNVAGRIE 373
Query: 363 FNNVEFVYPSRPESVILNDFCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGGEILL 422
F V F YPSRP V N + SGKT A VG SGSGKST++S++QRFY+P GEILL
Sbjct: 374 FQKVSFAYPSRPNMVFEN-LSFTIRSGKTFAFVGPSGSGKSTIISMVQRFYEPNSGEILL 432
Query: 423 DGVAIHKLQLKWLRSQMGLVSQEPALFATSIKENILFGREDATYEEIVDAAKASNAHNFI 482
DG I L+LKW R Q+GLVSQEPALFAT+I NIL G+E+A ++I++AAKA+NA +FI
Sbjct: 433 DGNDIKSLKLKWFREQLGLVSQEPALFATTIASNILLGKENANMDQIIEAAKAANADSFI 492
Query: 483 SMLPQGYDTQV 493
LP GY+TQV
Sbjct: 493 KSLPNGYNTQV 503
Score = 236 bits (601), Expect = 3e-62
Identities = 145/490 (29%), Positives = 250/490 (50%), Gaps = 6/490 (1%)
Query: 5 DQKNVSINDKKKKNGSFKSIF--MHADVLDWFFMAFGFFGAIGDGMMVPFVLFITSKIMN 62
DQ+ +D KK S I+ + + +W + G GA+ G P + ++
Sbjct: 648 DQEKTKNDDSKKDFSSSSMIWELIKLNSPEWPYALLGSIGAVLAGAQTPLFSMGIAYVLT 707
Query: 63 SVGSASGTSSSNFVHDVNKNAVVVLYMACASFFVCFLEGYCWTRTGERQAARMRVRYLKA 122
+ S + DV K A++ + + L+ Y +T GER +R+R+ A
Sbjct: 708 AFYSPF---PNVIKRDVEKVAIIFAGAGIVTAPIYLLQHYFYTLMGERLTSRVRLSLFSA 764
Query: 123 VLRQEVSYFDLHVTSTTDVITSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFALLW 182
+L E+ +FDL +T + + +++D+ +++ L+D++ + N S +++ +AF W
Sbjct: 765 ILSNEIGWFDLDENNTGSLTSILAADATLVRSALADRLSTIVQNLSLTVTALALAFFYSW 824
Query: 183 RLAIVGFPFMVLLVIPGYMYKRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFVGESK 242
R+A V LL+ + Y++A ++A +AI++IRTV ++ E +
Sbjct: 825 RVAAVVTACFPLLIAASLTEQLFLKGFGGDYTRAYSRATSVAREAIANIRTVAAYGAEKQ 884
Query: 243 TLAAFSNALEGSVKLGLKQGLAKGLAIG-SNGVVYAIWSLIFYYGSIMVMYHGAKGGTVF 301
F+ L K +G G G S + + ++L +Y S+++ + G
Sbjct: 885 ISEQFTCELSKPTKNAFVRGHISGFGYGLSQFLAFCSYALGLWYVSVLINHKETNFGDSI 944
Query: 302 VVGVTLAIGGLAFGTCFSNVRYFAEASVAGERIMEVIKRVPTIDSENMEGEIIEKVLGEV 361
+ L + + + + + A + V+ R I + ++ +V G++
Sbjct: 945 KSFMVLIVTAFSVSETLALTPDIVKGTQALGSVFRVLHRETKISPDQPNSRMVSQVKGDI 1004
Query: 362 EFNNVEFVYPSRPESVILNDFCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGGEIL 421
EF NV FVYP+RPE I + L+V +GK++A+VG SGSGKSTV+ L+ RFYDP G +
Sbjct: 1005 EFRNVSFVYPTRPEIDIFKNLNLRVSAGKSLAVVGPSGSGKSTVIGLIMRFYDPSNGNLC 1064
Query: 422 LDGVAIHKLQLKWLRSQMGLVSQEPALFATSIKENILFGREDATYEEIVDAAKASNAHNF 481
+DG I L L+ LR ++ LV QEPALF+T+I ENI +G E+A+ EI++AAKA+NAH F
Sbjct: 1065 IDGQDIKTLNLRSLRKKLALVQQEPALFSTTIYENIKYGNENASEAEIMEAAKAANAHEF 1124
Query: 482 ISMLPQGYDT 491
I + +GY T
Sbjct: 1125 IIKMEEGYKT 1134
>At4g18050 multidrug resistance protein/P-glycoprotein - like
Length = 1323
Score = 361 bits (927), Expect = e-100
Identities = 202/490 (41%), Positives = 295/490 (59%), Gaps = 5/490 (1%)
Query: 5 DQKNVSINDKKKKNGSFKSIFMHADVLDWFFMAFGFFGAIGDGMMVPFVLFITSKIMNSV 64
++K+ ND + SF +F AD D M G A G+G+ PF+ I +++N+
Sbjct: 2 EEKSSKKNDGGNQKVSFFKLFSFADKTDVVLMTVGTIAAAGNGLTQPFMTLIFGQLINAF 61
Query: 65 GSASGTSSSNFVHDVNKNAVVVLYMACASFFVCFLEGYCWTRTGERQAARMRVRYLKAVL 124
G+ T + V +V K AV +Y+A S V FL+ CW TGERQ+A +R YLK +L
Sbjct: 62 GT---TDPDHMVREVWKVAVKFIYLAVYSCVVAFLQVSCWMVTGERQSATIRGLYLKTIL 118
Query: 125 RQEVSYFDLHVTSTTDVITSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFALLWRL 184
RQ++ YFD T+T +VI +S D+++IQD + +KV F FL +AF L
Sbjct: 119 RQDIGYFDTE-TNTGEVIGRMSGDTILIQDAMGEKVGKFTQLLCTFLGGFAIAFYKGPLL 177
Query: 185 AIVGFPFMVLLVIPGYMYKRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFVGESKTL 244
A V + L+VI G I ++A + + Y +AG + EQ + +IRTV +F GE +
Sbjct: 178 AGVLCSCIPLIVIAGAAMSLIMSKMAGRGQVAYAEAGNVVEQTVGAIRTVVAFTGEKQAT 237
Query: 245 AAFSNALEGSVKLGLKQGLAKGLAIGSN-GVVYAIWSLIFYYGSIMVMYHGAKGGTVFVV 303
+ + LE + K ++QGL G +G+ V++ + L +YG+ ++M G GG V V
Sbjct: 238 EKYESKLEIAYKTVVQQGLISGFGLGTMLAVIFCSYGLAVWYGAKLIMEKGYNGGQVINV 297
Query: 304 GVTLAIGGLAFGTCFSNVRYFAEASVAGERIMEVIKRVPTIDSENMEGEIIEKVLGEVEF 363
+ GG++ G ++ FA A ++ E IKR P ID+ +M G ++E + G++E
Sbjct: 298 IFAVLTGGMSLGQTSPSLNAFAAGRAAAFKMFETIKRSPKIDAYDMSGSVLEDIRGDIEL 357
Query: 364 NNVEFVYPSRPESVILNDFCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGGEILLD 423
+V F YP+RP+ I F L VP+GKTVALVG SGSGKSTV+SL++RFYDP G++L+D
Sbjct: 358 KDVYFRYPARPDVQIFAGFSLFVPNGKTVALVGQSGSGKSTVISLIERFYDPESGQVLID 417
Query: 424 GVAIHKLQLKWLRSQMGLVSQEPALFATSIKENILFGREDATYEEIVDAAKASNAHNFIS 483
+ + KLQLKW+RS++GLVSQEP LFAT+IKENI +G+EDAT +EI A + +NA FI
Sbjct: 418 NIDLKKLQLKWIRSKIGLVSQEPVLFATTIKENIAYGKEDATDQEIRTAIELANAAKFID 477
Query: 484 MLPQGYDTQV 493
LPQG DT V
Sbjct: 478 KLPQGLDTMV 487
Score = 254 bits (649), Expect = 8e-68
Identities = 155/440 (35%), Positives = 230/440 (52%), Gaps = 29/440 (6%)
Query: 83 AVVVLYMACASFFVCFLEGYCWTRTGERQAARMRVRYLKAVLRQEVSYFD---------- 132
A++ + + +F + + Y + G + R+R V+ QE+S+FD
Sbjct: 729 ALIYIALGLTNFVMIPVPNYFFGIAGGKLIKRIRSMCFDKVVHQEISWFDDTANSRYYNF 788
Query: 133 --------LHVTSTTDVITSV---------SSDSLVIQDVLSDKVPNFLVNASRFLSSNI 175
L+V + + S+D+ ++ ++ D + + N + + I
Sbjct: 789 IYIINRRILYVLILIFICVLLPPVRLERECSTDASTVRSLVGDALALIVQNIATVTTGLI 848
Query: 176 VAFALLWRLAIVGFPFMVLLVIPGYMYKRISMRLARKIREEYNQAGTIAEQAISSIRTVY 235
+AF W LA++ +VI GY + + + Y +A +A A+SSIRTV
Sbjct: 849 IAFTANWILALIVLALSPFIVIQGYAQTKFLTGFSADAKAMYEEASQVANDAVSSIRTVA 908
Query: 236 SFVGESKTLAAFSNALEGSVKLGLKQGLAKGLAIG-SNGVVYAIWSLIFYYGSIMVMYHG 294
SF E K + + +G K G++ GL G G S +Y I + F G+ ++
Sbjct: 909 SFCAEEKVMDLYQQKCDGPKKNGVRLGLLSGAGFGFSFFFLYCINCVCFVSGAGLIQIGK 968
Query: 295 AKGGTVFVVGVTLAIGGLAFGTCFSNVRYFAEASVAGERIMEVIKRVPTIDSENMEGEII 354
A G VF V L I + + +A + I +++ P IDS + EG +
Sbjct: 969 ATFGEVFKVFFALTIMAIGVSQTSAMAPDSNKAKDSAASIFDILDSTPKIDSSSDEGTTL 1028
Query: 355 EKVLGEVEFNNVEFVYPSRPESVILNDFCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYD 414
+ V G++EF +V F YP RP+ I D CL +PSGKTVALVG SGSGKSTV+S+++RFY+
Sbjct: 1029 QNVNGDIEFRHVSFRYPMRPDVQIFRDLCLTIPSGKTVALVGESGSGKSTVISMIERFYN 1088
Query: 415 PIGGEILLDGVAIHKLQLKWLRSQMGLVSQEPALFATSIKENILFGRE-DATYEEIVDAA 473
P G+IL+D V I +L WLR QMGLVSQEP LF +I+ NI +G+ AT EEI+ AA
Sbjct: 1089 PDSGKILIDQVEIQTFKLSWLRQQMGLVSQEPILFNETIRSNIAYGKTGGATEEEIIAAA 1148
Query: 474 KASNAHNFISMLPQGYDTQV 493
KA+NAHNFIS LPQGYDT V
Sbjct: 1149 KAANAHNFISSLPQGYDTSV 1168
>At1g02520 P-glycoprotein, putative
Length = 1278
Score = 356 bits (914), Expect = 1e-98
Identities = 191/492 (38%), Positives = 294/492 (58%), Gaps = 4/492 (0%)
Query: 3 GGDQKNVSINDKKKKNGSFKSIFMHADVLDWFFMAFGFFGAIGDGMMVPFVLFITSKIMN 62
G + K +++K F +F AD D M G GAIG+GM +PF+ + +++
Sbjct: 25 GEETKKEEKSEEKANTVPFYKLFAFADSSDVLLMICGSIGAIGNGMSLPFMTLLFGDLID 84
Query: 63 SVGSASGTSSSNFVHDVNKNAVVVLYMACASFFVCFLEGYCWTRTGERQAARMRVRYLKA 122
S G ++ + V V+K + +Y+ + FL+ CW TGERQAAR+R YLK
Sbjct: 85 SFGK--NQNNKDIVDVVSKVCLKFVYLGLGTLGAAFLQVACWMITGERQAARIRSTYLKT 142
Query: 123 VLRQEVSYFDLHVTSTTDVITSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFALLW 182
+LRQ++ +FD+ T+T +V+ +S D+++IQD + +KV F+ S F+ ++AF W
Sbjct: 143 ILRQDIGFFDVE-TNTGEVVGRMSGDTVLIQDAMGEKVGKFIQLVSTFVGGFVLAFIKGW 201
Query: 183 RLAIVGFPFMVLLVIPGYMYKRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFVGESK 242
L +V + LL + G I R + + + Y +A T+ EQ I SIRTV SF GE +
Sbjct: 202 LLTLVMLTSIPLLAMAGAAMALIVTRASSRGQAAYAKAATVVEQTIGSIRTVASFTGEKQ 261
Query: 243 TLAAFSNALEGSVKLGLKQGLAKGLAIGSNGVVY-AIWSLIFYYGSIMVMYHGAKGGTVF 301
+ ++ + + K ++QG + GL +G V+ + ++L ++G M++ G GG V
Sbjct: 262 AINSYKKFITSAYKSSIQQGFSTGLGLGVMFFVFFSSYALAIWFGGKMILEKGYTGGAVI 321
Query: 302 VVGVTLAIGGLAFGTCFSNVRYFAEASVAGERIMEVIKRVPTIDSENMEGEIIEKVLGEV 361
V + + G ++ G V FA A ++ E IKR P ID+ ++ G+++E + G++
Sbjct: 322 NVIIIVVAGSMSLGQTSPCVTAFAAGQAAAYKMFETIKRKPLIDAYDVNGKVLEDIRGDI 381
Query: 362 EFNNVEFVYPSRPESVILNDFCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGGEIL 421
E +V F YP+RP+ I + F L +PSG T ALVG SGSGKSTV+SL++RFYDP G +L
Sbjct: 382 ELKDVHFSYPARPDEEIFDGFSLFIPSGATAALVGESGSGKSTVISLIERFYDPKSGAVL 441
Query: 422 LDGVAIHKLQLKWLRSQMGLVSQEPALFATSIKENILFGREDATYEEIVDAAKASNAHNF 481
+DGV + + QLKW+RS++GLVSQEP LF++SI ENI +G+E+AT EEI A + +NA F
Sbjct: 442 IDGVNLKEFQLKWIRSKIGLVSQEPVLFSSSIMENIAYGKENATVEEIKAATELANAAKF 501
Query: 482 ISMLPQGYDTQV 493
I LPQG DT V
Sbjct: 502 IDKLPQGLDTMV 513
Score = 273 bits (697), Expect = 2e-73
Identities = 154/415 (37%), Positives = 231/415 (55%), Gaps = 4/415 (0%)
Query: 83 AVVVLYMACASFFVCFLEGYCWTRTGERQAARMRVRYLKAVLRQEVSYFDLHVTSTTDVI 142
A++ + + S V + Y + G + R+R + + EV++FD S+ +
Sbjct: 753 AIIFVALGVTSLIVSPTQMYLFAVAGGKLIRRIRSMCFEKAVHMEVAWFDEPQNSSGTMG 812
Query: 143 TSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFALLWRLAIVGFPFMVLLVIPGYMY 202
+S+D+ +I+ ++ D + + N + S I+AF W LA++ + L+ I G++
Sbjct: 813 ARLSADATLIRALVGDALSLAVQNVASAASGLIIAFTASWELALIILVMLPLIGINGFVQ 872
Query: 203 KRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFVGESKTLAAFSNALEGSVKLGLKQG 262
+ + + +Y +A +A A+ SIRTV SF E K + + EG +K G+KQG
Sbjct: 873 VKFMKGFSADAKSKYEEASQVANDAVGSIRTVASFCAEEKVMQMYKKQCEGPIKDGIKQG 932
Query: 263 LAKGLAIG-SNGVVYAIWSLIFYYGSIMVMYHGAKGGTVFVVGVTLAIGGLAFGTCFSNV 321
GL G S +++ +++ FY G+ +V VF V L + + +
Sbjct: 933 FISGLGFGFSFFILFCVYATSFYAGARLVEDGKTTFNNVFQVFFALTMAAIGISQSSTFA 992
Query: 322 RYFAEASVAGERIMEVIKRVPTIDSENMEGEIIEKVLGEVEFNNVEFVYPSRPESVILND 381
++A VA I +I R IDS + G ++E V G++E ++ F YP+RP+ I D
Sbjct: 993 PDSSKAKVAAASIFAIIDRKSKIDSSDETGTVLENVKGDIELRHLSFTYPARPDIQIFRD 1052
Query: 382 FCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGGEILLDGVAIHKLQLKWLRSQMGL 441
CL + +GKTVALVG SGSGKSTV+SLLQRFYDP G I LDGV + KLQLKWLR QMGL
Sbjct: 1053 LCLTIRAGKTVALVGESGSGKSTVISLLQRFYDPDSGHITLDGVELKKLQLKWLRQQMGL 1112
Query: 442 VSQEPALFATSIKENILFGR---EDATYEEIVDAAKASNAHNFISMLPQGYDTQV 493
V QEP LF +I+ NI +G+ E AT EI+ AA+ +NAH FIS + QGYDT V
Sbjct: 1113 VGQEPVLFNDTIRANIAYGKGSEEAATESEIIAAAELANAHKFISSIQQGYDTVV 1167
>At3g62150 P-glycoprotein-like proetin
Length = 1292
Score = 353 bits (906), Expect = 1e-97
Identities = 185/489 (37%), Positives = 291/489 (58%), Gaps = 4/489 (0%)
Query: 6 QKNVSINDKKKKNGSFKSIFMHADVLDWFFMAFGFFGAIGDGMMVPFVLFITSKIMNSVG 65
+KN D+K K F +F AD D M G GA+G+G+ P + + +++ G
Sbjct: 50 EKNKQEEDEKTKTVPFHKLFAFADSFDIILMILGTIGAVGNGLGFPIMTILFGDVIDVFG 109
Query: 66 SASGTSSSNFVHDVNKNAVVVLYMACASFFVCFLEGYCWTRTGERQAARMRVRYLKAVLR 125
+SS+ + K A+ +Y+ + L+ W +GERQA R+R YL+ +LR
Sbjct: 110 Q--NQNSSDVSDKIAKVALKFVYLGLGTLVAALLQVSGWMISGERQAGRIRSLYLQTILR 167
Query: 126 QEVSYFDLHVTSTTDVITSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFALLWRLA 185
Q++++FD+ T+T +V+ +S D+++IQD + +KV + S F+ ++AF W L
Sbjct: 168 QDIAFFDVE-TNTGEVVGRMSGDTVLIQDAMGEKVGKAIQLVSTFIGGFVIAFTEGWLLT 226
Query: 186 IVGFPFMVLLVIPGYMYKRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFVGESKTLA 245
+V + LLV+ G + ++A + + Y +A + EQ + SIRTV SF GE + ++
Sbjct: 227 LVMVSSIPLLVMSGAALAIVISKMASRGQTSYAKAAVVVEQTVGSIRTVASFTGEKQAIS 286
Query: 246 AFSNALEGSVKLGLKQGLAKGLAIGS-NGVVYAIWSLIFYYGSIMVMYHGAKGGTVFVVG 304
++ L + + G+ +G + GL +G+ N V++ ++L +YG M++ G GG V ++
Sbjct: 287 NYNKHLVSAYRAGVFEGASTGLGLGTLNIVIFCTYALAVWYGGKMILEKGYTGGQVLIII 346
Query: 305 VTLAIGGLAFGTCFSNVRYFAEASVAGERIMEVIKRVPTIDSENMEGEIIEKVLGEVEFN 364
+ G ++ G + FA A ++ E IKR P ID+ + G++++ + G++E N
Sbjct: 347 FAVLTGSMSLGQASPCLSAFAAGQAAAYKMFEAIKRKPEIDASDTTGKVLDDIRGDIELN 406
Query: 365 NVEFVYPSRPESVILNDFCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGGEILLDG 424
NV F YP+RPE I F L + SG TVALVG SGSGKSTVVSL++RFYDP GE+ +DG
Sbjct: 407 NVNFSYPARPEEQIFRGFSLSISSGSTVALVGQSGSGKSTVVSLIERFYDPQSGEVRIDG 466
Query: 425 VAIHKLQLKWLRSQMGLVSQEPALFATSIKENILFGREDATYEEIVDAAKASNAHNFISM 484
+ + + QLKW+RS++GLVSQEP LF +SIKENI +G+E+AT EEI A + +NA FI
Sbjct: 467 INLKEFQLKWIRSKIGLVSQEPVLFTSSIKENIAYGKENATVEEIRKATELANASKFIDK 526
Query: 485 LPQGYDTQV 493
LPQG DT V
Sbjct: 527 LPQGLDTMV 535
Score = 273 bits (699), Expect = 1e-73
Identities = 170/487 (34%), Positives = 263/487 (53%), Gaps = 12/487 (2%)
Query: 9 VSINDKKKKNGSFKSIFMHADVLDWFFMAFGFFGAIGDGMMVPFVLFITSKIMNSVGSAS 68
VS K+KK F+ ++ + + G A+ +G+++P I +++SV A
Sbjct: 705 VSTPIKEKKVSFFRVAALNKPEIPMLIL--GSIAAVLNGVILP----IFGILISSVIKAF 758
Query: 69 GTSSSNFVHDVNKNAVVVLYMACASFFVCFLEGYCWTRTGERQAARMRVRYLKAVLRQEV 128
D A++ + + AS V + ++ G + R+R + V+R EV
Sbjct: 759 FKPPEQLKSDTRFWAIIFMLLGVASMVVFPAQTIFFSIAGCKLVQRIRSMCFEKVVRMEV 818
Query: 129 SYFDLHVTSTTDVITSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFALLWRLAIVG 188
+FD S+ + +S+D+ ++ ++ D + + N + + ++AF W+LA +
Sbjct: 819 GWFDETENSSGAIGARLSADAATVRGLVGDALAQTVQNLASVTAGLVIAFVASWQLAFIV 878
Query: 189 FPFMVLLVIPGYMYKRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFVGESKTLAAFS 248
+ L+ + GY+Y + + + +E A +A A+ SIRTV SF E K + +
Sbjct: 879 LAMLPLIGLNGYIYMKFMVGFSADAKE----ASQVANDAVGSIRTVASFCAEEKVMKMYK 934
Query: 249 NALEGSVKLGLKQGLAKGLAIG-SNGVVYAIWSLIFYYGSIMVMYHGAKGGTVFVVGVTL 307
EG ++ G++QG+ G+ G S V+++ ++ FY G+ +V +VF V L
Sbjct: 935 KKCEGPMRTGIRQGIVSGIGFGVSFFVLFSSYAASFYAGARLVDDGKTTFDSVFRVFFAL 994
Query: 308 AIGGLAFGTCFSNVRYFAEASVAGERIMEVIKRVPTIDSENMEGEIIEKVLGEVEFNNVE 367
+ +A S ++AS A I VI R ID + G +++ V G++E ++
Sbjct: 995 TMAAVAISQSSSLSPDSSKASNAAASIFAVIDRESKIDPSDESGRVLDNVKGDIELRHIS 1054
Query: 368 FVYPSRPESVILNDFCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGGEILLDGVAI 427
F YPSRP+ I D CL + +GKT+ALVG SGSGKSTV++LLQRFYDP G+I LDGV I
Sbjct: 1055 FKYPSRPDVQIFQDLCLSIRAGKTIALVGESGSGKSTVIALLQRFYDPDSGQITLDGVEI 1114
Query: 428 HKLQLKWLRSQMGLVSQEPALFATSIKENILFGR-EDATYEEIVDAAKASNAHNFISMLP 486
LQLKWLR Q GLVSQEP LF +I+ NI +G+ DAT EIV AA+ SNAH FIS L
Sbjct: 1115 KTLQLKWLRQQTGLVSQEPVLFNETIRANIAYGKGGDATETEIVSAAELSNAHGFISGLQ 1174
Query: 487 QGYDTQV 493
QGYDT V
Sbjct: 1175 QGYDTMV 1181
>At1g28010 hypothetical protein
Length = 1247
Score = 352 bits (902), Expect = 4e-97
Identities = 185/481 (38%), Positives = 292/481 (60%), Gaps = 5/481 (1%)
Query: 15 KKKNGSFKSIFMHADVLDWFFMAFGFFGAIGDGMMVPFVLFITSKIMNSVGSASGTSSSN 74
KK++ S +F AD +D+F M G G G +P +++S+G S T +
Sbjct: 27 KKESVSLMGLFSAADNVDYFLMFLGGLGTCIHGGTLPLFFVFFGGMLDSLGKLS-TDPNA 85
Query: 75 FVHDVNKNAVVVLYMACASFFVCFLEGYCWTRTGERQAARMRVRYLKAVLRQEVSYFDLH 134
V++NA+ ++Y+ + ++ CW +TGERQ AR+R+ YLK++L +++++FD
Sbjct: 86 ISSRVSQNALYLVYLGLVNLVSAWIGVACWMQTGERQTARLRINYLKSILAKDITFFDTE 145
Query: 135 VTSTTDVITSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFALLWRLAIVGFPFMVL 194
+ + I +SSD++++QD + DK + L +F++ ++ F +W+L ++ + L
Sbjct: 146 ARDS-NFIFHISSDAILVQDAIGDKTGHVLRYLCQFIAGFVIGFLSVWQLTLLTLGVVPL 204
Query: 195 LVIPGYMYKRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFVGESKTLAAFSNALEGS 254
+ I G Y + ++ K Y AG +AE+ +S +RTVY+FVGE K + ++SN+L+ +
Sbjct: 205 IAIAGGGYAIVMSTISEKSEAAYADAGKVAEEVMSQVRTVYAFVGEEKAVKSYSNSLKKA 264
Query: 255 VKLGLKQGLAKGLAIG-SNGVVYAIWSLIFYYGSIMVMYHGAKGGTVFVVGVTLAIGGLA 313
+KL + GLAKGL +G + +++ W+L+F+Y S++V + G F + + G A
Sbjct: 265 LKLSKRSGLAKGLGVGLTYSLLFCAWALLFWYASLLVRHGKTNGAKAFTTILNVIYSGFA 324
Query: 314 FGTCFSNVRYFAEASVAGERIMEVIKRVPTIDSENME-GEIIEKVLGEVEFNNVEFVYPS 372
G ++ ++ VA I ++I SE +E G ++ V+G++EF V F YPS
Sbjct: 325 LGQAVPSLSAISKGRVAAANIFKMIGNNNLESSERLENGTTLQNVVGKIEFCGVSFAYPS 384
Query: 373 RPESVILNDFCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGGEILLDGVAIHKLQL 432
RP V N + SGKT A VG SGSGKST++S++QRFY+P GEILLDG I L+L
Sbjct: 385 RPNMVFEN-LSFTIHSGKTFAFVGPSGSGKSTIISMVQRFYEPRSGEILLDGNDIKNLKL 443
Query: 433 KWLRSQMGLVSQEPALFATSIKENILFGREDATYEEIVDAAKASNAHNFISMLPQGYDTQ 492
KWLR QMGLVSQEPALFAT+I NIL G+E A ++I++AAKA+NA +FI LP GY+TQ
Sbjct: 444 KWLREQMGLVSQEPALFATTIASNILLGKEKANMDQIIEAAKAANADSFIKSLPNGYNTQ 503
Query: 493 V 493
V
Sbjct: 504 V 504
Score = 232 bits (592), Expect = 3e-61
Identities = 138/463 (29%), Positives = 239/463 (50%), Gaps = 4/463 (0%)
Query: 32 DWFFMAFGFFGAIGDGMMVPFVLFITSKIMNSVGSASGTSSSNFVHDVNKNAVVVLYMAC 91
+W + G GA+ G + ++ + S S +V+K A++ +
Sbjct: 679 EWLYALLGSIGAVLAGSQPALFSMGLAYVLTTFYSPF---PSLIKREVDKVAIIFVGAGI 735
Query: 92 ASFFVCFLEGYCWTRTGERQAARMRVRYLKAVLRQEVSYFDLHVTSTTDVITSVSSDSLV 151
+ + L+ Y +T GER +R+R+ A+L E+ +FDL +T + + +++D+ +
Sbjct: 736 VTAPIYILQHYFYTLMGERLTSRVRLSLFSAILSNEIGWFDLDENNTGSLTSILAADATL 795
Query: 152 IQDVLSDKVPNFLVNASRFLSSNIVAFALLWRLAIVGFPFMVLLVIPGYMYKRISMRLAR 211
++ ++D++ + N S +++ +AF WR+A V LL+ +
Sbjct: 796 VRSAIADRLSTIVQNLSLTITALALAFFYSWRVAAVVTACFPLLIAASLTEQLFLKGFGG 855
Query: 212 KIREEYNQAGTIAEQAISSIRTVYSFVGESKTLAAFSNALEGSVKLGLKQGLAKGLAIG- 270
Y++A ++A +AIS+IRTV +F E + F+ L K L +G G G
Sbjct: 856 DYTRAYSRATSLAREAISNIRTVAAFSAEKQISEQFTCELSKPTKSALLRGHISGFGYGL 915
Query: 271 SNGVVYAIWSLIFYYGSIMVMYHGAKGGTVFVVGVTLAIGGLAFGTCFSNVRYFAEASVA 330
S + + ++L +Y S+++ + + L + + + + + A
Sbjct: 916 SQCLAFCSYALGLWYISVLIKRNETNFEDSIKSFMVLLVTAYSVAETLALTPDIVKGTQA 975
Query: 331 GERIMEVIKRVPTIDSENMEGEIIEKVLGEVEFNNVEFVYPSRPESVILNDFCLKVPSGK 390
+ V+ R I + ++ + G++EF NV F YP+RPE I + L+V +GK
Sbjct: 976 LGSVFRVLHRETEIPPDQPNSRLVTHIKGDIEFRNVSFAYPTRPEIAIFKNLNLRVSAGK 1035
Query: 391 TVALVGGSGSGKSTVVSLLQRFYDPIGGEILLDGVAIHKLQLKWLRSQMGLVSQEPALFA 450
++A+VG SGSGKSTV+ L+ RFYDP G + +DG I + L+ LR ++ LV QEPALF+
Sbjct: 1036 SLAVVGPSGSGKSTVIGLIMRFYDPSNGNLCIDGHDIKSVNLRSLRKKLALVQQEPALFS 1095
Query: 451 TSIKENILFGREDATYEEIVDAAKASNAHNFISMLPQGYDTQV 493
TSI ENI +G E+A+ EI++AAKA+NAH FIS + +GY T V
Sbjct: 1096 TSIHENIKYGNENASEAEIIEAAKAANAHEFISRMEEGYMTHV 1138
>At1g02530 hypothetical protein
Length = 1273
Score = 346 bits (888), Expect = 2e-95
Identities = 187/497 (37%), Positives = 295/497 (58%), Gaps = 8/497 (1%)
Query: 2 GGGD----QKNVSINDKKKKNGSFKSIFMHADVLDWFFMAFGFFGAIGDGMMVPFVLFIT 57
G GD + + S D+K K +F AD D F M G GAIG+G+ +P + +
Sbjct: 7 GEGDSVSHEHSTSKTDEKAKTVPLYKLFAFADSFDVFLMICGSLGAIGNGVCLPLMTLLF 66
Query: 58 SKIMNSVGSASGTSSSNFVHDVNKNAVVVLYMACASFFVCFLEGYCWTRTGERQAARMRV 117
+++S G ++ + V V+K + +Y+ FL+ CW TGERQAA++R
Sbjct: 67 GDLIDSFGK--NQNNKDIVDVVSKVCLKFVYLGLGRLGAAFLQVACWMITGERQAAKIRS 124
Query: 118 RYLKAVLRQEVSYFDLHVTSTTDVITSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVA 177
YLK +LRQ++ +FD+ T+T +V+ +S D++ IQD + +KV F+ S F+ +A
Sbjct: 125 NYLKTILRQDIGFFDVE-TNTGEVVGRMSGDTVHIQDAMGEKVGKFIQLVSTFVGGFALA 183
Query: 178 FALLWRLAIVGFPFMVLLVIPGYMYKRISMRLARKIREEYNQAGTIAEQAISSIRTVYSF 237
FA W L +V + L + G + R + + + Y +A T+ EQ I SIRTV SF
Sbjct: 184 FAKGWLLTLVMLTSIPFLAMAGAAMALLVTRASSRGQAAYAKAATVVEQTIGSIRTVASF 243
Query: 238 VGESKTLAAFSNALEGSVKLGLKQGLAKGLAIGSN-GVVYAIWSLIFYYGSIMVMYHGAK 296
GE + + ++ + + K ++QG + GL +G V ++ ++L ++G M++ G
Sbjct: 244 TGEKQAINSYKKYITSAYKSSIQQGFSTGLGLGVMIYVFFSSYALAIWFGGKMILEKGYT 303
Query: 297 GGTVFVVGVTLAIGGLAFGTCFSNVRYFAEASVAGERIMEVIKRVPTIDSENMEGEIIEK 356
GG+V V + + G ++ G V FA A ++ E IKR P ID+ ++ G+++
Sbjct: 304 GGSVINVIIIVVAGSMSLGQTSPCVTAFAAGQAAAYKMFETIKRKPLIDAYDVNGKVLGD 363
Query: 357 VLGEVEFNNVEFVYPSRPESVILNDFCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPI 416
+ G++E +V F YP+RP+ I + F L +PSG T ALVG SGSGKSTV++L++RFYDP
Sbjct: 364 IRGDIELKDVHFSYPARPDEEIFDGFSLFIPSGATAALVGESGSGKSTVINLIERFYDPK 423
Query: 417 GGEILLDGVAIHKLQLKWLRSQMGLVSQEPALFATSIKENILFGREDATYEEIVDAAKAS 476
GE+L+DG+ + + QLKW+RS++GLV QEP LF++SI ENI +G+E+AT +EI A + +
Sbjct: 424 AGEVLIDGINLKEFQLKWIRSKIGLVCQEPVLFSSSIMENIAYGKENATLQEIKVATELA 483
Query: 477 NAHNFISMLPQGYDTQV 493
NA FI+ LPQG DT+V
Sbjct: 484 NAAKFINNLPQGLDTKV 500
Score = 269 bits (687), Expect = 3e-72
Identities = 154/415 (37%), Positives = 232/415 (55%), Gaps = 4/415 (0%)
Query: 83 AVVVLYMACASFFVCFLEGYCWTRTGERQAARMRVRYLKAVLRQEVSYFDLHVTSTTDVI 142
A++ + + S V + Y + G + R++ + + EVS+FD S+ +
Sbjct: 748 AIIFVALGVTSLIVSPSQMYLFAVAGGKLIRRIQSMCFEKAVHMEVSWFDEPENSSGTMG 807
Query: 143 TSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFALLWRLAIVGFPFMVLLVIPGYMY 202
+S+D+ +I+ ++ D + + NA+ S I+AF W LA++ + L+ I G++
Sbjct: 808 ARLSTDAALIRALVGDALSLAVQNAASAASGLIIAFTASWELALIILVMLPLIGINGFLQ 867
Query: 203 KRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFVGESKTLAAFSNALEGSVKLGLKQG 262
+ + + +Y +A +A A+ SIRTV SF E K + ++ EG +K G+KQG
Sbjct: 868 VKFMKGFSADAKSKYEEASQVANDAVGSIRTVASFCAEEKVMQMYNKQCEGPIKDGVKQG 927
Query: 263 LAKGLAIG-SNGVVYAIWSLIFYYGSIMVMYHGAKGGTVFVVGVTLAIGGLAFGTCFSNV 321
GL G S +++ +++ FY + +V VF V L + + +
Sbjct: 928 FISGLGFGFSFFILFCVYATSFYAAARLVEDGKTTFIDVFQVFFALTMAAIGISQSSTFA 987
Query: 322 RYFAEASVAGERIMEVIKRVPTIDSENMEGEIIEKVLGEVEFNNVEFVYPSRPESVILND 381
++A VA I +I R IDS + G ++E V G++E ++ F YP+RP I D
Sbjct: 988 PDSSKAKVAAASIFAIIDRKSKIDSSDETGTVLENVKGDIELRHLSFTYPARPGIQIFRD 1047
Query: 382 FCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGGEILLDGVAIHKLQLKWLRSQMGL 441
CL + +GKTVALVG SGSGKSTV+SLLQRFYDP G+I LDGV + KLQLKWLR QMGL
Sbjct: 1048 LCLTIRAGKTVALVGESGSGKSTVISLLQRFYDPDSGQITLDGVELKKLQLKWLRQQMGL 1107
Query: 442 VSQEPALFATSIKENILFGR---EDATYEEIVDAAKASNAHNFISMLPQGYDTQV 493
V QEP LF +I+ NI +G+ E AT EI+ AA+ +NAH FIS + QGYDT V
Sbjct: 1108 VGQEPVLFNDTIRANIAYGKGSEEAATESEIIAAAELANAHKFISSIQQGYDTVV 1162
>At2g47000 putative ABC transporter
Length = 1286
Score = 344 bits (882), Expect = 8e-95
Identities = 184/481 (38%), Positives = 292/481 (60%), Gaps = 6/481 (1%)
Query: 14 KKKKNGSFKSIFMHADVLDWFFMAFGFFGAIGDGMMVPFVLFITSKIMNSVGSASGTSSS 73
+K K F +F AD D+ M G G+IG+G+ P + + +++ A G + +
Sbjct: 41 EKTKTVPFYKLFAFADSFDFLLMILGTLGSIGNGLGFPLMTLLFGDLID----AFGENQT 96
Query: 74 NFVHDVNKNAVVVLYMACASFFVCFLEGYCWTRTGERQAARMRVRYLKAVLRQEVSYFDL 133
N V+K A+ +++ +F FL+ W +GERQAAR+R YLK +LRQ++++FD+
Sbjct: 97 NTTDKVSKVALKFVWLGIGTFAAAFLQLSGWMISGERQAARIRSLYLKTILRQDIAFFDI 156
Query: 134 HVTSTTDVITSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFALLWRLAIVGFPFMV 193
T+T +V+ +S D+++IQD + +KV + + F+ ++AF W L +V +
Sbjct: 157 D-TNTGEVVGRMSGDTVLIQDAMGEKVGKAIQLLATFVGGFVIAFVRGWLLTLVMLSSIP 215
Query: 194 LLVIPGYMYKRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFVGESKTLAAFSNALEG 253
LLV+ G + + + A + + Y +A T+ EQ I SIRTV SF GE + ++ ++ L
Sbjct: 216 LLVMAGALLAIVIAKTASRGQTAYAKAATVVEQTIGSIRTVASFTGEKQAISNYNKHLVT 275
Query: 254 SVKLGLKQGLAKGLAIGSNG-VVYAIWSLIFYYGSIMVMYHGAKGGTVFVVGVTLAIGGL 312
+ K G+ +G + GL +G+ VV+ ++L +YG +++ G GG V + + + G +
Sbjct: 276 AYKAGVIEGGSTGLGLGTLFLVVFCSYALAVWYGGKLILDKGYTGGQVLNIIIAVLTGSM 335
Query: 313 AFGTCFSNVRYFAEASVAGERIMEVIKRVPTIDSENMEGEIIEKVLGEVEFNNVEFVYPS 372
+ G + FA A ++ E I+R P IDS + G++++ + G++E +V F YP+
Sbjct: 336 SLGQTSPCLSAFAAGQAAAYKMFETIERRPNIDSYSTNGKVLDDIKGDIELKDVYFTYPA 395
Query: 373 RPESVILNDFCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGGEILLDGVAIHKLQL 432
RP+ I F L + SG TVALVG SGSGKSTVVSL++RFYDP G++L+DG+ + + QL
Sbjct: 396 RPDEQIFRGFSLFISSGTTVALVGQSGSGKSTVVSLIERFYDPQAGDVLIDGINLKEFQL 455
Query: 433 KWLRSQMGLVSQEPALFATSIKENILFGREDATYEEIVDAAKASNAHNFISMLPQGYDTQ 492
KW+RS++GLVSQEP LF SIK+NI +G+EDAT EEI AA+ +NA F+ LPQG DT
Sbjct: 456 KWIRSKIGLVSQEPVLFTASIKDNIAYGKEDATTEEIKAAAELANASKFVDKLPQGLDTM 515
Query: 493 V 493
V
Sbjct: 516 V 516
Score = 280 bits (715), Expect = 2e-75
Identities = 165/460 (35%), Positives = 252/460 (53%), Gaps = 6/460 (1%)
Query: 36 MAFGFFGAIGDGMMVPFVLFITSKIMNSVGSASGTSSSNFVHDVNKNAVVVLYMACASFF 95
+ G A +G+++P I +++SV A D + A++ + + AS
Sbjct: 720 LILGSISAAANGVILP----IFGILISSVIKAFFQPPKKLKEDTSFWAIIFMVLGFASII 775
Query: 96 VCFLEGYCWTRTGERQAARMRVRYLKAVLRQEVSYFDLHVTSTTDVITSVSSDSLVIQDV 155
+ + + G + R+R + V+ EV +FD S+ + +S+D+ I+ +
Sbjct: 776 AYPAQTFFFAIAGCKLVQRIRSMCFEKVVHMEVGWFDEPENSSGTIGARLSADAATIRGL 835
Query: 156 LSDKVPNFLVNASRFLSSNIVAFALLWRLAIVGFPFMVLLVIPGYMYKRISMRLARKIRE 215
+ D + + N S L+ I+AF W+LA V + L+ + G++Y + + ++
Sbjct: 836 VGDSLAQTVQNLSSILAGLIIAFLACWQLAFVVLAMLPLIALNGFLYMKFMKGFSADAKK 895
Query: 216 EYNQAGTIAEQAISSIRTVYSFVGESKTLAAFSNALEGSVKLGLKQGLAKGLAIG-SNGV 274
Y +A +A A+ SIRTV SF E K + +S EG +K G++QG+ G+ G S V
Sbjct: 896 MYGEASQVANDAVGSIRTVASFCAEDKVMNMYSKKCEGPMKNGIRQGIVSGIGFGFSFFV 955
Query: 275 VYAIWSLIFYYGSIMVMYHGAKGGTVFVVGVTLAIGGLAFGTCFSNVRYFAEASVAGERI 334
+++ ++ FY G+ +V +VF V L + +A S ++A VA I
Sbjct: 956 LFSSYAASFYVGARLVDDGKTTFDSVFRVFFALTMAAMAISQSSSLSPDSSKADVAAASI 1015
Query: 335 MEVIKRVPTIDSENMEGEIIEKVLGEVEFNNVEFVYPSRPESVILNDFCLKVPSGKTVAL 394
++ R ID G +++ V G++E +V F YP+RP+ I D CL + +GKTVAL
Sbjct: 1016 FAIMDRESKIDPSVESGRVLDNVKGDIELRHVSFKYPARPDVQIFQDLCLSIRAGKTVAL 1075
Query: 395 VGGSGSGKSTVVSLLQRFYDPIGGEILLDGVAIHKLQLKWLRSQMGLVSQEPALFATSIK 454
VG SGSGKSTV++LLQRFYDP GEI LDGV I L+LKWLR Q GLVSQEP LF +I+
Sbjct: 1076 VGESGSGKSTVIALLQRFYDPDSGEITLDGVEIKSLRLKWLRQQTGLVSQEPILFNETIR 1135
Query: 455 ENILFGR-EDATYEEIVDAAKASNAHNFISMLPQGYDTQV 493
NI +G+ DA+ EIV +A+ SNAH FIS L QGYDT V
Sbjct: 1136 ANIAYGKGGDASESEIVSSAELSNAHGFISGLQQGYDTMV 1175
>At4g01820 P-glycoprotein-like protein pgp3
Length = 1229
Score = 341 bits (874), Expect = 6e-94
Identities = 184/482 (38%), Positives = 288/482 (59%), Gaps = 4/482 (0%)
Query: 13 DKKKKNGSFKSIFMHADVLDWFFMAFGFFGAIGDGMMVPFVLFITSKIMNSVGSASGTSS 72
++K K F +F +D D M G GAIG+G+ P + + +++S+G S+
Sbjct: 2 EEKTKTVPFYKLFSFSDSTDVLLMIVGSIGAIGNGVGFPLMTLLFGDLIDSIGQ--NQSN 59
Query: 73 SNFVHDVNKNAVVVLYMACASFFVCFLEGYCWTRTGERQAARMRVRYLKAVLRQEVSYFD 132
+ V V+K + +Y+ + FL+ CW TGERQAAR+R YLK +LRQ++ +FD
Sbjct: 60 KDIVEIVSKVCLKFVYLGLGTLGAAFLQVACWMITGERQAARIRSLYLKTILRQDIGFFD 119
Query: 133 LHVTSTTDVITSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFALLWRLAIVGFPFM 192
+ TST +V+ +S D+++I + + +KV F+ + F+ ++AF W L +V +
Sbjct: 120 VE-TSTGEVVGRMSGDTVLILEAMGEKVGKFIQLIATFVGGFVLAFVKGWLLTLVMLVSI 178
Query: 193 VLLVIPGYMYKRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFVGESKTLAAFSNALE 252
LL I G I R + + + Y +A T+ EQ + SIRTV SF GE + + ++ +
Sbjct: 179 PLLAIAGAAMPIIVTRASSREQAAYAKASTVVEQTLGSIRTVASFTGEKQAMKSYREFIN 238
Query: 253 GSVKLGLKQGLAKGLAIGSNGVVY-AIWSLIFYYGSIMVMYHGAKGGTVFVVGVTLAIGG 311
+ + +KQG + GL +G V+ ++L ++G M++ G GG V V VT+
Sbjct: 239 LAYRASVKQGFSMGLGLGVVFFVFFCSYALAIWFGGEMILKKGYTGGEVVNVMVTVVASS 298
Query: 312 LAFGTCFSNVRYFAEASVAGERIMEVIKRVPTIDSENMEGEIIEKVLGEVEFNNVEFVYP 371
++ G + FA A ++ E I+R P+ID+ ++ G+++E + GE+E +V F YP
Sbjct: 299 MSLGQTTPCLTAFAAGKAAAYKMFETIERKPSIDAFDLNGKVLEDIRGEIELRDVCFSYP 358
Query: 372 SRPESVILNDFCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGGEILLDGVAIHKLQ 431
+RP + F L +PSG T ALVG SGSGKS+V+SL++RFYDP G +L+DGV + + Q
Sbjct: 359 ARPMEEVFGGFSLLIPSGATAALVGESGSGKSSVISLIERFYDPSSGSVLIDGVNLKEFQ 418
Query: 432 LKWLRSQMGLVSQEPALFATSIKENILFGREDATYEEIVDAAKASNAHNFISMLPQGYDT 491
LKW+R ++GLVSQEP LF++SI ENI +G+E+AT EEI AAK +NA NFI LP+G +T
Sbjct: 419 LKWIRGKIGLVSQEPVLFSSSIMENIGYGKENATVEEIQAAAKLANAANFIDKLPRGLET 478
Query: 492 QV 493
V
Sbjct: 479 LV 480
Score = 272 bits (696), Expect = 3e-73
Identities = 166/497 (33%), Positives = 266/497 (53%), Gaps = 16/497 (3%)
Query: 4 GDQKNVSINDKKKKNGSFKSIFMHADVLDWFFMAFGFFGAIGDGMMVPFVLFITSKIMNS 63
G Q+N I+ ++ +N S I + GA+ +G + P + +K++ +
Sbjct: 631 GRQENTEISREQSRNVSITRIAALNKPETTILILGTLLGAV-NGTIFPIFGILFAKVIEA 689
Query: 64 VGSASGTSSSNFVHDVNKNA----VVVLYMACASFFVCFLEGYCWTRTGERQAARMRVRY 119
HD+ +++ ++ + + AS V + Y + G R R+RV
Sbjct: 690 FFKPP--------HDMKRDSRFWSMIFVLLGVASLIVYPMHTYLFAVAGGRLIQRIRVMC 741
Query: 120 LKAVLRQEVSYFDLHVTSTTDVITSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFA 179
+ V+ EV +FD S+ + + +S+D+ +I+ ++ D + + NA+ +S I+AF
Sbjct: 742 FEKVVHMEVGWFDDPENSSGTIGSRLSADAALIKTLVGDSLSLSVKNAAAAVSGLIIAFT 801
Query: 180 LLWRLAIVGFPFMVLLVIPGYMYKRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFVG 239
W+LA++ + L+ I GY+ + + +Y +A +A A+ SIRTV SF
Sbjct: 802 ASWKLAVIILVMIPLIGINGYLQIKFIKGFTADAKAKYEEASQVANDAVGSIRTVASFCA 861
Query: 240 ESKTLAAFSNALEGSVKLGLKQGLAKGLAIG-SNGVVYAIWSLIFYYGSIMVMYHGAKGG 298
E K + + E ++K G+KQGL G+ G S V+Y++++ FY G+ +V
Sbjct: 862 EEKVMEMYKKRCEDTIKSGIKQGLISGVGFGISFFVLYSVYASCFYVGARLVKAGRTNFN 921
Query: 299 TVFVVGVTLAIGGLAFGTCFSNVRYFAEASVAGERIMEVIKRVPTIDSENMEGEIIEKVL 358
VF V + L + + S ++A A I +I IDS + G ++E V
Sbjct: 922 DVFQVFLALTMTAIGISQASSFAPDSSKAKGAAASIFGIIDGKSMIDSRDESGLVLENVK 981
Query: 359 GEVEFNNVEFVYPSRPESVILNDFCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGG 418
G++E ++ F Y +RP+ I D C + +G+TVALVG SGSGKSTV+SLLQRFYDP G
Sbjct: 982 GDIELCHISFTYQTRPDVQIFRDLCFAIRAGQTVALVGESGSGKSTVISLLQRFYDPDSG 1041
Query: 419 EILLDGVAIHKLQLKWLRSQMGLVSQEPALFATSIKENILFGR--EDATYEEIVDAAKAS 476
I LD V + KLQLKW+R QMGLV QEP LF +I+ NI +G+ ++A+ EI+ AA+ +
Sbjct: 1042 HITLDRVELKKLQLKWVRQQMGLVGQEPVLFNDTIRSNIAYGKGGDEASEAEIIAAAELA 1101
Query: 477 NAHNFISMLPQGYDTQV 493
NAH FIS + QGYDT V
Sbjct: 1102 NAHGFISSIQQGYDTVV 1118
>At4g01830 putative P-glycoprotein-like protein
Length = 1230
Score = 333 bits (853), Expect = 2e-91
Identities = 179/478 (37%), Positives = 283/478 (58%), Gaps = 4/478 (0%)
Query: 17 KNGSFKSIFMHADVLDWFFMAFGFFGAIGDGMMVPFVLFITSKIMNSVGSASGTSSSNFV 76
K F +F +D D M G GAI +G+ P + + ++++++G ++ V
Sbjct: 11 KTVPFYKLFFFSDSTDVLLMIVGSIGAIANGVCSPLMTLLFGELIDAMGP--NQNNEEIV 68
Query: 77 HDVNKNAVVVLYMACASFFVCFLEGYCWTRTGERQAARMRVRYLKAVLRQEVSYFDLHVT 136
V+K + ++Y+ + FL+ CW TGERQAAR+R YLK +LRQ++ +FD+ +T
Sbjct: 69 ERVSKVCLSLVYLGLGALGAAFLQVACWMITGERQAARIRSLYLKTILRQDIGFFDVEMT 128
Query: 137 STTDVITSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFALLWRLAIVGFPFMVLLV 196
T +V+ +S D+++I D + +KV F+ S F+ ++AF W L +V + LL
Sbjct: 129 -TGEVVGRMSGDTVLILDAMGEKVGKFIQLISTFVGGFVIAFLRGWLLTLVMLTSIPLLA 187
Query: 197 IPGYMYKRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFVGESKTLAAFSNALEGSVK 256
+ G I R + + + Y +A + EQ + SIRTV SF GE + ++++ + + K
Sbjct: 188 MSGAAIAIIVTRASSQEQAAYAKASNVVEQTLGSIRTVASFTGEKQAMSSYKELINLAYK 247
Query: 257 LGLKQGLAKGLAIGSNGVVY-AIWSLIFYYGSIMVMYHGAKGGTVFVVGVTLAIGGLAFG 315
+KQG GL +G +V+ + ++L ++G M++ G GG V V VT+ +A G
Sbjct: 248 SNVKQGFVTGLGLGVMFLVFFSTYALGTWFGGEMILRKGYTGGAVINVMVTVVSSSIALG 307
Query: 316 TCFSNVRYFAEASVAGERIMEVIKRVPTIDSENMEGEIIEKVLGEVEFNNVEFVYPSRPE 375
+ F A ++ E I+R P ID+ ++ G+++E + GE+E +V F YP+RP+
Sbjct: 308 QASPCLTAFTAGKAAAYKMFETIEREPLIDTFDLNGKVLEDIRGEIELRDVCFSYPARPK 367
Query: 376 SVILNDFCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGGEILLDGVAIHKLQLKWL 435
+ F L +PSG T ALVG SGSGKSTV+SL++RFYDP G++L+DGV + + QLKW+
Sbjct: 368 EEVFGGFSLLIPSGTTTALVGESGSGKSTVISLIERFYDPNSGQVLIDGVDLKEFQLKWI 427
Query: 436 RSQMGLVSQEPALFATSIKENILFGREDATYEEIVDAAKASNAHNFISMLPQGYDTQV 493
R ++GLVSQEP LF++SI ENI +G+E AT EEI A+K +NA FI LP G +T V
Sbjct: 428 RGKIGLVSQEPVLFSSSIMENIGYGKEGATVEEIQAASKLANAAKFIDKLPLGLETLV 485
Score = 265 bits (678), Expect = 3e-71
Identities = 148/414 (35%), Positives = 233/414 (55%), Gaps = 3/414 (0%)
Query: 83 AVVVLYMACASFFVCFLEGYCWTRTGERQAARMRVRYLKAVLRQEVSYFDLHVTSTTDVI 142
+++ + + A+ V Y + G R R+R + V+ EV +FD S+ +
Sbjct: 706 SMIFVLLGVAAVIVYPTTNYLFAIAGGRLIRRIRSMCFEKVVHMEVGWFDEPGNSSGAMG 765
Query: 143 TSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFALLWRLAIVGFPFMVLLVIPGYMY 202
+S+D+ +I+ ++ D + + N + ++ I+AF W +AI+ + + I GY+
Sbjct: 766 ARLSADAALIRTLVGDSLCLSVKNVASLVTGLIIAFTASWEVAIIILVIIPFIGINGYIQ 825
Query: 203 KRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFVGESKTLAAFSNALEGSVKLGLKQG 262
+ + + +Y +A +A A+ SIRTV SF E K + + E ++K G+KQG
Sbjct: 826 IKFMKGFSADAKAKYEEASQVANDAVGSIRTVASFCAEEKVMEMYKKRCEDTIKSGIKQG 885
Query: 263 LAKGLAIG-SNGVVYAIWSLIFYYGSIMVMYHGAKGGTVFVVGVTLAIGGLAFGTCFSNV 321
L G+ G S V+Y++++ FY G+ +V VF V + L + + S
Sbjct: 886 LISGVGFGISFFVLYSVYASCFYVGARLVKAGRTNFNDVFQVFLALTLTAVGISQASSFA 945
Query: 322 RYFAEASVAGERIMEVIKRVPTIDSENMEGEIIEKVLGEVEFNNVEFVYPSRPESVILND 381
++ A I +I R+ IDS + G ++E V G++E ++ F Y +RP+ + D
Sbjct: 946 PDSSKGKGAAVSIFRIIDRISKIDSRDESGMVLENVKGDIELCHISFTYQTRPDVQVFRD 1005
Query: 382 FCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGGEILLDGVAIHKLQLKWLRSQMGL 441
CL + +G+TVALVG SGSGKSTV+SLLQRFYDP G I LDGV + KL+LKWLR QMGL
Sbjct: 1006 LCLSIRAGQTVALVGESGSGKSTVISLLQRFYDPDSGHITLDGVELKKLRLKWLRQQMGL 1065
Query: 442 VSQEPALFATSIKENILFGR--EDATYEEIVDAAKASNAHNFISMLPQGYDTQV 493
V QEP LF +I+ NI +G+ E+AT EI+ A++ +NAH FIS + +GYDT V
Sbjct: 1066 VGQEPVLFNDTIRANIAYGKGGEEATEAEIIAASELANAHRFISSIQKGYDTVV 1119
>At2g39480 putative ABC transporter
Length = 1407
Score = 327 bits (837), Expect = 1e-89
Identities = 184/479 (38%), Positives = 284/479 (58%), Gaps = 12/479 (2%)
Query: 21 FKSIFMHADVLDWFFMAFGFFGAIGDGMMVPFVLFITSKIMNSVGSASGTSSSNFVHDVN 80
F +F AD DW M FG A G + L +KI+ + A T S + + D
Sbjct: 71 FSQLFACADRFDWVLMVFGSVAAAAHGTALIVYLHYFAKIVQVL--AFPTDSDHLISDDQ 128
Query: 81 KN-----AVVVLYMACASFFVCFLEGYCWTRTGERQAARMRVRYLKAVLRQEVSYFDLHV 135
N ++ ++Y+A F ++E CW TGERQ A +R +Y++ +L Q++S+FD +
Sbjct: 129 FNRLLELSLTIVYIAGGVFISGWIEVSCWILTGERQTAVIRSKYVQVLLNQDMSFFDTY- 187
Query: 136 TSTTDVITSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAFALLWRLAIVGFPFMVLL 195
+ D+++ V SD L+IQ LS+KV N++ N + F+S I+ F W +A++ +
Sbjct: 188 GNNGDIVSQVLSDVLLIQSALSEKVGNYIHNMATFISGLIIGFVNCWEIALITLATGPFI 247
Query: 196 VIPGYMYKRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFVGESKTLAAFSNALEGSV 255
V G + RLA I++ Y +A +IAEQA+S +RT+Y+F E+ +++ +L+ ++
Sbjct: 248 VAAGGISNIFLHRLAENIQDAYAEAASIAEQAVSYVRTLYAFTNETLAKYSYATSLQATL 307
Query: 256 KLGLKQGLAKGLAIG-SNGVVYAIWSLIFYYGSIMVMYHGAKGGTVFVVGVTLAIGGLAF 314
+ G+ L +GL +G + G+ ++ + G V++H A GG + + + GL
Sbjct: 308 RYGILISLVQGLGLGFTYGLAICSCAMQLWIGRFFVIHHRANGGEIITALFAVILSGLGL 367
Query: 315 GTCFSNVRYFAEASVAGERIMEVIKRVPTIDSENMEGEIIEKVLGEVEFNNVEFVYPSRP 374
+N F + +A R+ E+I R + N EG I+ V G +EF NV F Y SRP
Sbjct: 368 NQAATNFYSFDQGRIAAYRLFEMISR--SSSGTNQEGIILSAVQGNIEFRNVYFSYLSRP 425
Query: 375 ESVILNDFCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIGGEILLDGVAIHKLQLKW 434
E IL+ F L VP+ K VALVG +GSGKS+++ L++RFYDP GE+LLDG I L+L+W
Sbjct: 426 EIPILSGFYLTVPAKKAVALVGRNGSGKSSIIPLMERFYDPTLGEVLLDGENIKNLKLEW 485
Query: 435 LRSQMGLVSQEPALFATSIKENILFGREDATYEEIVDAAKASNAHNFISMLPQGYDTQV 493
LRSQ+GLV+QEPAL + SI+ENI +GR DAT ++I +AAK ++AH FIS L +GY+TQV
Sbjct: 486 LRSQIGLVTQEPALLSLSIRENIAYGR-DATLDQIEEAAKKAHAHTFISSLEKGYETQV 543
Score = 263 bits (671), Expect = 2e-70
Identities = 149/498 (29%), Positives = 270/498 (53%), Gaps = 9/498 (1%)
Query: 4 GDQKNVSINDKKKKNGSFK---SIFMHADVL--DWFFMAFGFFGAIGDGMMVPFVLFITS 58
G + S + K K+G K S + A + +W + G GA G P + ++ +
Sbjct: 798 GHSDDTSASVKVAKDGQHKEPPSFWRLAQLSFPEWLYAVLGSIGAAIFGSFNPLLAYVIA 857
Query: 59 KIMNSVGSASGTSSSNFVHDVNKNAVVVLYMACASFFVCFLEGYCWTRTGERQAARMRVR 118
++ + ++ G S+ +V+K +++ M + FL+ + + GE+ R+R
Sbjct: 858 LVVTTYYTSKG---SHLREEVDKWCLIIACMGIVTVVANFLQHFYFGIMGEKMTERVRRM 914
Query: 119 YLKAVLRQEVSYFDLHVTSTTDVITSVSSDSLVIQDVLSDKVPNFLVNASRFLSSNIVAF 178
A+LR EV ++D S + +++D+ ++ S+++ F+ ++ + + ++
Sbjct: 915 MFSAMLRNEVGWYDEEENSPDTLSMRLANDATFVRAAFSNRLSIFIQDSFAVIVAILIGL 974
Query: 179 ALLWRLAIVGFPFMVLLVIPGYMYKRISMRLARKIREEYNQAGTIAEQAISSIRTVYSFV 238
L WRLA+V + +L + K ++ I+E + +A + E A+ +I TV +F
Sbjct: 975 LLGWRLALVALATLPVLTLSAIAQKLWLAGFSKGIQEMHRKASLVLEDAVRNIYTVVAFC 1034
Query: 239 GESKTLAAFSNALEGSVKLGLKQGLAKGLAIG-SNGVVYAIWSLIFYYGSIMVMYHGAKG 297
+K + + L+ ++ G+A G A G S +++A +L+ +Y ++ V K
Sbjct: 1035 AGNKVMELYRLQLQRILRQSFFHGMAIGFAFGFSQFLLFACNALLLWYTALSVDRRYMKL 1094
Query: 298 GTVFVVGVTLAIGGLAFGTCFSNVRYFAEASVAGERIMEVIKRVPTIDSENMEGEIIEKV 357
T + + A F Y + + + E+I RVPTI+ ++ V
Sbjct: 1095 STALTEYMVFSFATFALVEPFGLAPYILKRRRSLASVFEIIDRVPTIEPDDTSALSPPNV 1154
Query: 358 LGEVEFNNVEFVYPSRPESVILNDFCLKVPSGKTVALVGGSGSGKSTVVSLLQRFYDPIG 417
G +E N++F YP+RPE ++L++F LKV G+TVA+VG SGSGKST++SL++R+YDP+
Sbjct: 1155 YGSIELKNIDFCYPTRPEVLVLSNFSLKVNGGQTVAVVGVSGSGKSTIISLIERYYDPVA 1214
Query: 418 GEILLDGVAIHKLQLKWLRSQMGLVSQEPALFATSIKENILFGREDATYEEIVDAAKASN 477
G++LLDG + L+WLRS MGL+ QEP +F+T+I+ENI++ R +A+ E+ +AA+ +N
Sbjct: 1215 GQVLLDGRDLKSYNLRWLRSHMGLIQQEPIIFSTTIRENIIYARHNASEAEMKEAARIAN 1274
Query: 478 AHNFISMLPQGYDTQVSV 495
AH+FIS LP GYDT + +
Sbjct: 1275 AHHFISSLPHGYDTHIGM 1292
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.138 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,404,868
Number of Sequences: 26719
Number of extensions: 422459
Number of successful extensions: 1647
Number of sequences better than 10.0: 114
Number of HSP's better than 10.0 without gapping: 86
Number of HSP's successfully gapped in prelim test: 28
Number of HSP's that attempted gapping in prelim test: 1387
Number of HSP's gapped (non-prelim): 168
length of query: 509
length of database: 11,318,596
effective HSP length: 104
effective length of query: 405
effective length of database: 8,539,820
effective search space: 3458627100
effective search space used: 3458627100
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 62 (28.5 bits)
Medicago: description of AC141109.8


