

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141109.13 - phase: 0
(208 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
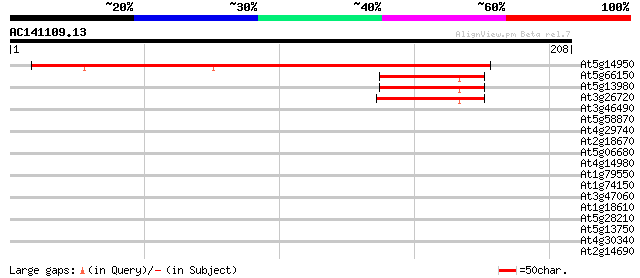
Score E
Sequences producing significant alignments: (bits) Value
At5g14950 alpha-mannosidase -like protein 222 1e-58
At5g66150 alpha-mannosidase 43 1e-04
At5g13980 alpha-mannosidase 42 3e-04
At3g26720 alpha-mannosidase, putative 41 4e-04
At3g46490 putative protein 33 0.11
At5g58870 cell division protein - like 30 0.69
At4g29740 cytokinin oxidase - like protein 30 0.69
At2g18670 putative RING zinc finger protein 28 3.4
At5g06680 gamma-tubulin interacting protein-like 28 4.5
At4g14980 unknown protein 27 5.9
At1g79550 phosphoglycerate kinase (EC 2.7.2.3) like protein 27 5.9
At1g74150 hypothetical protein 27 5.9
At3g47060 FtsH metalloprotease - like protein 27 7.7
At1g18610 unknown protein 27 7.7
At5g28210 mRNA capping enzyme - like 27 10.0
At5g13750 transporter-like protein 27 10.0
At4g30340 unknown protein 27 10.0
At2g14690 1,4-beta-xylan endohydrolase 27 10.0
>At5g14950 alpha-mannosidase -like protein
Length = 1173
Score = 222 bits (566), Expect = 1e-58
Identities = 110/173 (63%), Positives = 131/173 (75%), Gaps = 3/173 (1%)
Query: 9 GNWAQSILPSSNPKSKQP--RKSKRRTLVKDFIFSNFFIIGLIISLLFFLIVLLRFGVPK 66
G W QS+LP++ KSK RK ++RTLV +FIF+NFF+I L +SLLFFL+ L FGVP
Sbjct: 20 GGWGQSLLPTALSKSKLAINRKPRKRTLVVNFIFANFFVIALTVSLLFFLLTLFHFGVPG 79
Query: 67 PITTHFRT-RTSRFRKPKKLSLNGSSTIFGGFASVDLTTKGLYDKIEFLDVDGGAWKQGW 125
PI++ F T R++R KP+K A VD+TTK LYD+IEFLD DGG WKQGW
Sbjct: 80 PISSRFLTSRSNRIVKPRKNINRRPLNDSNSGAVVDITTKDLYDRIEFLDTDGGPWKQGW 139
Query: 126 SVSYRGDEWDNEKLKVFVVPHSHNDPGWKLTVEEYYDRQSRHILDTIVETLSK 178
V+Y+ DEW+ EKLK+FVVPHSHNDPGWKLTVEEYY RQSRHILDTIVETLSK
Sbjct: 140 RVTYKDDEWEKEKLKIFVVPHSHNDPGWKLTVEEYYQRQSRHILDTIVETLSK 192
>At5g66150 alpha-mannosidase
Length = 1047
Score = 43.1 bits (100), Expect = 1e-04
Identities = 20/47 (42%), Positives = 31/47 (65%), Gaps = 8/47 (17%)
Query: 138 KLKVFVVPHSHNDPGWKLTVEEYYDRQS--------RHILDTIVETL 176
KL V +VPHSH+D GW TV++YY + R++LD++V++L
Sbjct: 46 KLNVHLVPHSHDDVGWLKTVDQYYVGSNNRIQNACVRNVLDSVVDSL 92
>At5g13980 alpha-mannosidase
Length = 1024
Score = 41.6 bits (96), Expect = 3e-04
Identities = 21/47 (44%), Positives = 29/47 (61%), Gaps = 8/47 (17%)
Query: 138 KLKVFVVPHSHNDPGWKLTVEEYYDRQS--------RHILDTIVETL 176
KL V VVPHSH+D GW TV++YY + +++LD+IV L
Sbjct: 36 KLNVHVVPHSHDDVGWLKTVDQYYVGSNNSIQVACVQNVLDSIVPAL 82
>At3g26720 alpha-mannosidase, putative
Length = 1019
Score = 41.2 bits (95), Expect = 4e-04
Identities = 18/48 (37%), Positives = 31/48 (64%), Gaps = 8/48 (16%)
Query: 137 EKLKVFVVPHSHNDPGWKLTVEEYYDRQS--------RHILDTIVETL 176
EK+ V +VPHSH+D GW TV++YY + +++LD+++ +L
Sbjct: 36 EKINVHLVPHSHDDVGWLKTVDQYYVGSNNSIRGACVQNVLDSVIASL 83
>At3g46490 putative protein
Length = 306
Score = 33.1 bits (74), Expect = 0.11
Identities = 15/52 (28%), Positives = 27/52 (51%), Gaps = 4/52 (7%)
Query: 119 GAWKQGWSVSYRGDE----WDNEKLKVFVVPHSHNDPGWKLTVEEYYDRQSR 166
G +K+G+++ + G + WD + P+ PGW+ T+E+YY R
Sbjct: 102 GDYKEGFTIGFEGSKDGPHWDKPFHSPNIWPNPDVLPGWRETMEKYYQEALR 153
>At5g58870 cell division protein - like
Length = 806
Score = 30.4 bits (67), Expect = 0.69
Identities = 28/78 (35%), Positives = 38/78 (47%), Gaps = 8/78 (10%)
Query: 43 FFIIGLIISLLFFLIVLL----RFGV--PKPITTHFRTRTSRFRKPKKLSLNGSSTIFGG 96
FF GLI+ LF++ VL RF V + T RTR S K+S +G + F
Sbjct: 273 FFNSGLIV--LFYIAVLAGLLHRFPVNFSQSTTGQLRTRKSGGPGGGKVSGDGETITFAD 330
Query: 97 FASVDLTTKGLYDKIEFL 114
A VD + L + +EFL
Sbjct: 331 VAGVDEAKEELEEIVEFL 348
>At4g29740 cytokinin oxidase - like protein
Length = 524
Score = 30.4 bits (67), Expect = 0.69
Identities = 15/42 (35%), Positives = 25/42 (58%), Gaps = 7/42 (16%)
Query: 155 LTVEEYYDRQSRHILDTIVETLSKVKFCLFPFLGFGDFYVYV 196
L V +YYDR + I+D +++TLS+ LGF +++V
Sbjct: 322 LEVAKYYDRTTLPIIDQVIDTLSRT-------LGFAPGFMFV 356
>At2g18670 putative RING zinc finger protein
Length = 181
Score = 28.1 bits (61), Expect = 3.4
Identities = 26/91 (28%), Positives = 43/91 (46%), Gaps = 14/91 (15%)
Query: 10 NWAQSILPSSNPKSKQPRKSKRRTLVKDFIFSNFFIIGLIISLLFFLIVLLRFGV----- 64
N+ S P S S P K K R L + F++G+I+ +FFL ++L G+
Sbjct: 6 NYRISGEPPSTTPSHPPPKPKTRIL-------SLFLVGVIMFSIFFLFLVL-IGIASVLI 57
Query: 65 -PKPITTHFRTRTSRFRKPKKLSLNGSSTIF 94
P +++ R R R ++ S +G S+ F
Sbjct: 58 LPLLLSSLHRHHRRRRRNRRQESSDGLSSRF 88
>At5g06680 gamma-tubulin interacting protein-like
Length = 838
Score = 27.7 bits (60), Expect = 4.5
Identities = 17/52 (32%), Positives = 29/52 (55%), Gaps = 4/52 (7%)
Query: 137 EKLKVFVVPHSHNDPGWKLTVEEYYDRQSRHILDTI-VETLSKVKFCLFPFL 187
++L+V ++PH D GW + EY ++R LDT+ E++ +F FL
Sbjct: 561 DRLRVKMMPHGSGDRGWDVFSLEY---EARVPLDTVFTESVLSKYLRVFNFL 609
>At4g14980 unknown protein
Length = 470
Score = 27.3 bits (59), Expect = 5.9
Identities = 13/28 (46%), Positives = 16/28 (56%), Gaps = 3/28 (10%)
Query: 182 CLFPFL---GFGDFYVYVCCYCCLFYEA 206
C FPF+ GDF YVC C+F +A
Sbjct: 441 CKFPFIMWGAVGDFTGYVCSINCVFPKA 468
>At1g79550 phosphoglycerate kinase (EC 2.7.2.3) like protein
Length = 401
Score = 27.3 bits (59), Expect = 5.9
Identities = 17/38 (44%), Positives = 21/38 (54%), Gaps = 1/38 (2%)
Query: 84 KLSLNGSSTIFGGFASVDLTTK-GLYDKIEFLDVDGGA 120
+LS G +TI GG SV K GL DK+ + GGA
Sbjct: 345 ELSGKGVTTIIGGGDSVAAVEKVGLADKMSHISTGGGA 382
>At1g74150 hypothetical protein
Length = 552
Score = 27.3 bits (59), Expect = 5.9
Identities = 12/39 (30%), Positives = 22/39 (55%), Gaps = 4/39 (10%)
Query: 85 LSLNGSSTIFGGFASVDLTTKGLYDKIEFLDVDGGAWKQ 123
++L + +FGGF ++ LYD + LD++ G W +
Sbjct: 238 VALERNLFVFGGFTD----SQNLYDDLYVLDLETGVWSK 272
>At3g47060 FtsH metalloprotease - like protein
Length = 802
Score = 26.9 bits (58), Expect = 7.7
Identities = 22/78 (28%), Positives = 37/78 (47%), Gaps = 4/78 (5%)
Query: 41 SNFFIIGLIISLLFFLIV--LLRFGVPKPITTHFRTRTSRFRKPK--KLSLNGSSTIFGG 96
S F +I+L + ++ L+RF V ++ + RT + P K+S G + F
Sbjct: 267 SGGFFNSALIALFYIAVLAGLIRFPVSFSTSSTGQLRTRKAGGPDGGKVSGGGETITFAD 326
Query: 97 FASVDLTTKGLYDKIEFL 114
A VD + L + +EFL
Sbjct: 327 VAGVDEAKEELEEIVEFL 344
>At1g18610 unknown protein
Length = 574
Score = 26.9 bits (58), Expect = 7.7
Identities = 14/39 (35%), Positives = 20/39 (50%), Gaps = 4/39 (10%)
Query: 85 LSLNGSSTIFGGFASVDLTTKGLYDKIEFLDVDGGAWKQ 123
+SL + +FGGF + LYD + LDVD W +
Sbjct: 259 VSLGRNFFVFGGFTDA----QNLYDDLYVLDVDTCIWSK 293
>At5g28210 mRNA capping enzyme - like
Length = 625
Score = 26.6 bits (57), Expect = 10.0
Identities = 8/15 (53%), Positives = 13/15 (86%)
Query: 123 QGWSVSYRGDEWDNE 137
+G++V +RGD WDN+
Sbjct: 523 EGFAVEFRGDGWDND 537
>At5g13750 transporter-like protein
Length = 478
Score = 26.6 bits (57), Expect = 10.0
Identities = 15/49 (30%), Positives = 24/49 (48%), Gaps = 2/49 (4%)
Query: 25 QPRKSKRRTLVKDFIFSNF--FIIGLIISLLFFLIVLLRFGVPKPITTH 71
QP K +D IF F F+ L IS+ FL+ ++ +P+ + H
Sbjct: 190 QPAKQYPSLFSQDSIFGKFPFFLPCLAISVFAFLVTIVSSRIPETLHNH 238
>At4g30340 unknown protein
Length = 490
Score = 26.6 bits (57), Expect = 10.0
Identities = 12/30 (40%), Positives = 15/30 (50%)
Query: 7 RGGNWAQSILPSSNPKSKQPRKSKRRTLVK 36
RGG+W + L KQP KS T V+
Sbjct: 447 RGGDWKNAFLQMDGEPWKQPMKSDYSTFVE 476
>At2g14690 1,4-beta-xylan endohydrolase
Length = 552
Score = 26.6 bits (57), Expect = 10.0
Identities = 12/43 (27%), Positives = 20/43 (45%)
Query: 108 YDKIEFLDVDGGAWKQGWSVSYRGDEWDNEKLKVFVVPHSHND 150
+DK+ D D + G + EW E +++ + H HND
Sbjct: 454 FDKLTLADKDFKNTQAGDLIDKLLQEWKQEPVEIPIQHHEHND 496
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.143 0.458
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,017,732
Number of Sequences: 26719
Number of extensions: 217492
Number of successful extensions: 779
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 763
Number of HSP's gapped (non-prelim): 21
length of query: 208
length of database: 11,318,596
effective HSP length: 95
effective length of query: 113
effective length of database: 8,780,291
effective search space: 992172883
effective search space used: 992172883
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC141109.13


