

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141106.8 - phase: 0
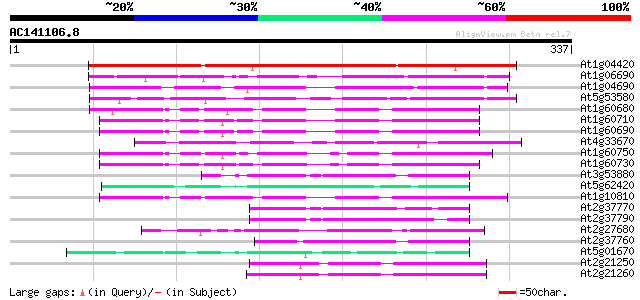
(337 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g04420 unknown protein 227 8e-60
At1g06690 unknown protein 84 1e-16
At1g04690 putative K+ channel, beta subunit 82 5e-16
At5g53580 aldo/keto reductase-like protein 78 8e-15
At1g60680 unknown protein 69 4e-12
At1g60710 unknown protein 64 9e-11
At1g60690 64 9e-11
At4g33670 putative protein 63 2e-10
At1g60750 63 2e-10
At1g60730 unknown protein 63 3e-10
At3g53880 putative alcohol dehydrogenase 59 3e-09
At5g62420 aldose reductase-like protein 59 5e-09
At1g10810 putative auxin-induced protein 57 1e-08
At2g37770 putative aldo/keto reductase 54 2e-07
At2g37790 putative alcohol dehydrogenase 52 6e-07
At2g27680 unknown protein 51 1e-06
At2g37760 putative alcohol dehydrogenase 49 3e-06
At5g01670 aldose reductase-like protein 48 9e-06
At2g21250 putative NADPH dependent mannose 6-phosphate reductase 43 3e-04
At2g21260 putative NADPH dependent mannose 6-phosphate reductase 42 5e-04
>At1g04420 unknown protein
Length = 412
Score = 227 bits (578), Expect = 8e-60
Identities = 122/263 (46%), Positives = 170/263 (64%), Gaps = 9/263 (3%)
Query: 48 NLNVSRLCLGTMTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYF 107
+LN+S + +GTMTFGEQNT +S ++L A GIN D+AE YP+P + +T G ++ Y
Sbjct: 65 DLNISEVTMGTMTFGEQNTEKESHEMLSYAIEEGINCIDTAEAYPIPMKKETQGKTDLYI 124
Query: 108 GHWIKHRNIPRDRLVIATKVAGPSGQMTWIRGGPKSL--DATNISQAIDNSLLRMQLDYI 165
W+K + RD++V+ATKV G S + +IR + L DA NI ++++ SL R+ DYI
Sbjct: 125 SSWLKSQQ--RDKIVLATKVCGYSERSAYIRDSGEILRVDAANIKESVEKSLKRLGTDYI 182
Query: 166 DLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGL 225
DL QIHWPDRYVP+FG+ Y+ + S+ EQL A + +GK+RYIG+SNET YG+
Sbjct: 183 DLLQIHWPDRYVPLFGDFYYETSKWRPSVPFAEQLRAFQDLIVEGKVRYIGVSNETSYGV 242
Query: 226 MKFIQVAEKSSSYPKIVSLQNSYSLLCR-TFDSAMAECCHQE--SISLLAYSPLAMGILS 282
+F+ A K PKIVS+QN YSLL R ++ + E CH + ++ LLAYSPL G LS
Sbjct: 243 TEFVNTA-KLEGLPKIVSIQNGYSLLVRCRYEVDLVEVCHPKNCNVGLLAYSPLGGGSLS 301
Query: 283 GKYF-SHGNGPADARLNLFKGLL 304
GKY + +ARLNLF G +
Sbjct: 302 GKYLATDQEATKNARLNLFPGYM 324
>At1g06690 unknown protein
Length = 377
Score = 83.6 bits (205), Expect = 1e-16
Identities = 78/267 (29%), Positives = 127/267 (47%), Gaps = 47/267 (17%)
Query: 48 NLNVSRLCLGTMTFGEQNTLSQSFQLLDEAYHA-----------GINFFDSAEMYPVPQR 96
+L V++L +G ++G+ N+ FQ D A GI+FFD+AE+Y
Sbjct: 55 DLKVTKLGIGVWSWGD-NSYWNDFQWDDRKLKAAKGAFDTSLDNGIDFFDTAEVYGSKFS 113
Query: 97 AQTWGMSEEYFGHWIKHRN--IPRDRLVIATKVAGPSGQMTWIRGGPKSLDATNISQAID 154
SE G +I+ R P + +ATK A + W R G +S + A+
Sbjct: 114 LGAIS-SETLLGRFIRERKERYPGAEVSVATKFAA----LPW-RFGRES-----VVTALK 162
Query: 155 NSLLRMQLDYIDLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRY 214
+SL R++L +DLYQ+HWP ++G Y LD L AV G ++
Sbjct: 163 DSLSRLELSSVDLYQLHWPG----LWGNEGY--------------LDGLGDAVEQGLVKA 204
Query: 215 IGLSNETPYGLMKFIQVAEKSSSYPKIVSLQNSYSLLCRTFD-SAMAECCHQESISLLAY 273
+G+SN + L + +K + S Q +YSL+ R + + + C + ++L+AY
Sbjct: 205 VGVSNYSEKRLRDAYERLKKRGI--PLASNQVNYSLIYRAPEQTGVKAACDELGVTLIAY 262
Query: 274 SPLAMGILSGKYFSHGNGPADARLNLF 300
SP+A G L+GKY + N P+ R ++
Sbjct: 263 SPIAQGALTGKY-TPENPPSGPRGRIY 288
>At1g04690 putative K+ channel, beta subunit
Length = 328
Score = 81.6 bits (200), Expect = 5e-16
Identities = 73/255 (28%), Positives = 113/255 (43%), Gaps = 37/255 (14%)
Query: 49 LNVSRLCLGT-MTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYF 107
L VS L G +TFG Q + ++ +L G+NFFD+AE+Y G +EE
Sbjct: 11 LKVSTLSFGAWVTFGNQLDVKEAKSILQCCRDHGVNFFDNAEVY-------ANGRAEEIM 63
Query: 108 GHWIKHRNIPRDRLVIATKVAGPSGQMTWIRGGP--KSLDATNISQAIDNSLLRMQLDYI 165
G I+ R +VI+TK+ W GP K L +I + SL R+ +DY+
Sbjct: 64 GQAIRELGWRRSDIVISTKI-------FWGGPGPNDKGLSRKHIVEGTKASLKRLDMDYV 116
Query: 166 DLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGL 225
D+ H PD P I+E + A++ ++ G Y G S + +
Sbjct: 117 DVLYCHRPDASTP-----------------IEETVRAMNYVIDKGWAFYWGTSEWSAQQI 159
Query: 226 MKFIQVAEKSSSYPKIVSLQNSYSLLCR-TFDSAMAECCHQESISLLAYSPLAMGILSGK 284
+ A++ IV Q Y++ R ++ I L +SPLA G+L+GK
Sbjct: 160 TEAWGAADRLDLVGPIVE-QPEYNMFARHKVETEFLPLYTNHGIGLTTWSPLASGVLTGK 218
Query: 285 YFSHGNGPADARLNL 299
Y + G P+D+R L
Sbjct: 219 Y-NKGAIPSDSRFAL 232
>At5g53580 aldo/keto reductase-like protein
Length = 365
Score = 77.8 bits (190), Expect = 8e-15
Identities = 80/270 (29%), Positives = 120/270 (43%), Gaps = 47/270 (17%)
Query: 49 LNVSRLCLGTMTFGEQ----------NTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQ 98
L+VS + GT +G Q + L Q+F+L A GIN FD+A+ Y +
Sbjct: 45 LSVSPMGFGTWAWGNQLLWGYQTSMDDQLQQAFEL---ALENGINLFDTADSYGT---GR 98
Query: 99 TWGMSEEYFGHWIKHRNI---PRDRLVIATKVAGPSGQMTWIRGGPKSLDATNISQAIDN 155
G SE G +IK ++ +V+ATK A P L + A
Sbjct: 99 LNGQSERLLGKFIKESQGLKGKQNEVVVATKFAAY----------PWRLTSGQFVNACRA 148
Query: 156 SLLRMQLDYIDLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYI 215
SL R+Q+D + + Q+HW Y P+Q+ D L + G +R +
Sbjct: 149 SLDRLQIDQLGIGQLHW--------STASYAPLQELVL------WDGLVQMYEKGLVRAV 194
Query: 216 GLSNETPYGLMKFIQVAEKSSSYPKIVSLQNSYSLLCRTFDSA-MAECCHQESISLLAYS 274
G+SN P L+K I K+ P + S Q +SLL + + C + I L++YS
Sbjct: 195 GVSNYGPQQLVK-IHDYLKTRGVP-LCSAQVQFSLLSMGKEQLEIKSICDELGIRLISYS 252
Query: 275 PLAMGILSGKYFSHGNGPADARLNLFKGLL 304
PL +G+L+GKY S P R LF+ +L
Sbjct: 253 PLGLGMLTGKY-SSSKLPTGPRSLLFRQIL 281
>At1g60680 unknown protein
Length = 346
Score = 68.9 bits (167), Expect = 4e-12
Identities = 65/240 (27%), Positives = 109/240 (45%), Gaps = 44/240 (18%)
Query: 49 LNVSRLCLGTMT----FGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSE 104
L VS LG M +G + + LL A ++G+ FFD+++MY P+ +E
Sbjct: 17 LEVSAQGLGCMALSARYGAPKPETDAIALLHHAINSGVTFFDTSDMYG-PET------NE 69
Query: 105 EYFGHWIKHRNIPRDRLVIATKVAG--PSGQMTWIRGGPKSLDATNISQAIDNSLLRMQL 162
G +K ++++ +ATK G+++ +RG P+ + A A + SL R+ +
Sbjct: 70 LLLGKALKDG--VKEKVELATKFGFFIVEGEISEVRGDPEYVRA-----ACEASLKRLDI 122
Query: 163 DYIDLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETP 222
IDLY H D VP I+ + L + V +GKI+YIGLS +
Sbjct: 123 ACIDLYYQHRIDTRVP-----------------IEITMRELKKLVEEGKIKYIGLSEASA 165
Query: 223 YGLMKFIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGILS 282
++ + I ++Q +SL R + + C + I ++AYSPL G L+
Sbjct: 166 -------STIRRAHAVHPITAVQIEWSLWSRDAEEDIIPICRELGIGIVAYSPLGRGFLA 218
>At1g60710 unknown protein
Length = 345
Score = 64.3 bits (155), Expect = 9e-11
Identities = 59/231 (25%), Positives = 104/231 (44%), Gaps = 42/231 (18%)
Query: 55 CLGTMTF-GEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYFGHWIKH 113
C+G F G +++ L+ A H+G+ D++++Y P+ +E G +K
Sbjct: 26 CMGLSAFYGAPKPENEAIALIHHAIHSGVTLLDTSDIYG-PET------NEVLLGKALKD 78
Query: 114 RNIPRDRLVIATK--VAGPSGQMTWIRGGPKSLDATNISQAIDNSLLRMQLDYIDLYQIH 171
R+++ +ATK ++ G+ +RG P+ + A A + SL R+ + IDLY H
Sbjct: 79 G--VREKVELATKFGISYAEGKRE-VRGDPEYVRA-----ACEASLKRLDIACIDLYYQH 130
Query: 172 WPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGLMKFIQV 231
D VP I+ + L + V +GKI+YIGLS +
Sbjct: 131 RVDTRVP-----------------IEITMGELKKLVEEGKIKYIGLSEASA-------ST 166
Query: 232 AEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGILS 282
++ + I ++Q +SL R + + C + I ++AYSPL G +
Sbjct: 167 IRRAHAVHPITAVQIEWSLWTRDVEEEIIPTCRELGIGIVAYSPLGRGFFA 217
>At1g60690
Length = 345
Score = 64.3 bits (155), Expect = 9e-11
Identities = 59/231 (25%), Positives = 104/231 (44%), Gaps = 42/231 (18%)
Query: 55 CLG-TMTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYFGHWIKH 113
C+G T +G +++ L+ A H+G+ F D+++MY P+ +E G +K
Sbjct: 26 CMGLTGHYGASKPETEAIALIHHAIHSGVTFLDTSDMYG-PET------NEILLGKALKD 78
Query: 114 RNIPRDRLVIATK--VAGPSGQMTWIRGGPKSLDATNISQAIDNSLLRMQLDYIDLYQIH 171
R+++ +ATK ++ G I+G P + A A + SL R+ + IDLY H
Sbjct: 79 G--VREKVELATKFGISYAEGNRE-IKGDPAYVRA-----ACEASLKRLDVTCIDLYYQH 130
Query: 172 WPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGLMKFIQV 231
D VP I+ + L + + +GKI+YIGLS +
Sbjct: 131 RIDTRVP-----------------IEITMGELKKLIEEGKIKYIGLSEASA-------ST 166
Query: 232 AEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGILS 282
++ + I ++Q +SL R + + C + I +++YSPL G +
Sbjct: 167 IRRAHTVHPITAVQLEWSLWTRDVEEEIVPTCRELGIGIVSYSPLGRGFFA 217
>At4g33670 putative protein
Length = 319
Score = 63.2 bits (152), Expect = 2e-10
Identities = 56/236 (23%), Positives = 101/236 (42%), Gaps = 40/236 (16%)
Query: 76 EAYHAGINFFDSAEMYPVPQRAQTWGMSEEYFGHWIKHRNIPRDRLVIATKVAGPSGQMT 135
EA+ GINFFD++ Y +SE+ G +K +PR ++ATK
Sbjct: 44 EAFRLGINFFDTSPYYGGT-------LSEKMLGKGLKALQVPRSDYIVATKCGRYKEGFD 96
Query: 136 WIRGGPKSLDATNISQAIDNSLLRMQLDYIDLYQIHWPDRYVPMFGETEYDPVQQYSSIS 195
+ A + ++ID SL R+QLDY+D+ H + E+ + Q
Sbjct: 97 F--------SAERVRKSIDESLERLQLDYVDILHCH----------DIEFGSLDQI---- 134
Query: 196 IDEQLDALSRAVNDGKIRYIGLSNETPYGLMKFIQVAEKSSSYPKIVSL----QNSYSLL 251
+ E + AL + +GK R+IG++ P + ++ + I+S N +LL
Sbjct: 135 VSETIPALQKLKQEGKTRFIGITG-LPLDIFTYVLDRVPPGTVDVILSYCHYGVNDSTLL 193
Query: 252 CRTFDSAMAECCHQESISLLAYSPLAMGILSGKYFSHGNGPADARLNLFKGLLTHC 307
+ + + +++ SPLAMG+L+ + + + + K + HC
Sbjct: 194 ------DLLPYLKSKGVGVISASPLAMGLLTEQGPPEWHPASPELKSASKAAVAHC 243
>At1g60750
Length = 330
Score = 63.2 bits (152), Expect = 2e-10
Identities = 66/240 (27%), Positives = 106/240 (43%), Gaps = 43/240 (17%)
Query: 55 CLGTMTF-GEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYFGHWIKH 113
C+G F G + + LL A +AG+ F D++++Y P+ +E G +K
Sbjct: 27 CMGLSDFYGAPTPETNAVALLRHAINAGVTFLDTSDIYG-PET------NELLLGKALKD 79
Query: 114 RNIPRDRLVIATK---VAGPSGQMTWIRGGPKSLDATNISQAIDNSLLRMQLDYIDLYQI 170
RD++ +ATK A G+ + RG P+ + A + SL R+ + IDLY
Sbjct: 80 GL--RDKVELATKFGITASEDGKFGF-RGDPEY-----VRIACEASLKRLGVTCIDLYYQ 131
Query: 171 HWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGLMKFIQ 230
H D +P+ I+I E L + V +GKI+YIGLS +
Sbjct: 132 HRIDTTLPI-------------EITIGE----LKKLVEEGKIKYIGLSEASA-------S 167
Query: 231 VAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGILSGKYFSHGN 290
++ + I ++Q +SL R + + C + I ++AYSPL G L F N
Sbjct: 168 TIRRAHAVHPITAVQIEWSLWSRDVEEDIIPTCRELGIGIVAYSPLGRGFLGLPRFQQEN 227
>At1g60730 unknown protein
Length = 345
Score = 62.8 bits (151), Expect = 3e-10
Identities = 61/231 (26%), Positives = 106/231 (45%), Gaps = 42/231 (18%)
Query: 55 CLGTMTF-GEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYFGHWIKH 113
C+G F G +++ L+ A H+G+ F D++++Y P+ +E +K
Sbjct: 26 CMGLSAFYGTPKPETEAIALIHHAIHSGVTFLDTSDIYG-PET------NELLLSKALKD 78
Query: 114 RNIPRDRLVIATK--VAGPSGQMTWIRGGPKSLDATNISQAIDNSLLRMQLDYIDLYQIH 171
R+++ +ATK + G++ + +G P + A A + SL+R+ + IDLY H
Sbjct: 79 G--VREKVELATKYGIRYAEGKVEF-KGDPAYVRA-----ACEASLMRVDVACIDLYYQH 130
Query: 172 WPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGLMKFIQV 231
D VP+ I+I E L + V +GKI+YIGLS +
Sbjct: 131 RIDTRVPI-------------EITIGE----LKKLVEEGKIKYIGLSEASA-------ST 166
Query: 232 AEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGILS 282
++ + I +LQ +SL R + + C + I ++AYSPL G +
Sbjct: 167 IRRAHAVHPITALQIEWSLWSRDVEEDIIPTCRELGIGIVAYSPLGRGFFA 217
>At3g53880 putative alcohol dehydrogenase
Length = 315
Score = 59.3 bits (142), Expect = 3e-09
Identities = 44/161 (27%), Positives = 71/161 (43%), Gaps = 22/161 (13%)
Query: 116 IPRDRLVIATKVAGPSGQMTWIRGGPKSLDATNISQAIDNSLLRMQLDYIDLYQIHWPDR 175
+ R++L I +K+ W+ LD ++ A++ +L +QLDY+DLY +HWP R
Sbjct: 71 VKREKLFITSKI--------WLT----DLDPPDVQDALNRTLQDLQLDYVDLYLMHWPVR 118
Query: 176 YVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGLMKFIQVAEKS 235
G ++ P + I I A+ V+ GK R IG+SN + L ++ A
Sbjct: 119 LKK--GTVDFKP-ENIMPIDIPSTWKAMEALVDSGKARAIGVSNFSTKKLSDLVEAA--- 172
Query: 236 SSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPL 276
++ N + E C + I L YSPL
Sbjct: 173 ----RVPPAVNQVECHPSWQQHKLHEFCKSKGIHLSGYSPL 209
>At5g62420 aldose reductase-like protein
Length = 316
Score = 58.5 bits (140), Expect = 5e-09
Identities = 52/221 (23%), Positives = 88/221 (39%), Gaps = 27/221 (12%)
Query: 56 LGTMTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYFGHWIKHRN 115
LG T+ Q + + +A G FD+A++Y + T G I +
Sbjct: 17 LGMGTYCPQKDRESTISAVHQAIKIGYRHFDTAKIYGSEEALGT------ALGQAISYGT 70
Query: 116 IPRDRLVIATKVAGPSGQMTWIRGGPKSLDATNISQAIDNSLLRMQLDYIDLYQIHWPDR 175
+ RD L + +K+ W S D + A+ +L M LDY+D Y +HWP +
Sbjct: 71 VQRDDLFVTSKL--------W------SSDHHDPISALIQTLKTMGLDYLDNYLVHWPIK 116
Query: 176 YVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGLMKFIQVAEKS 235
P E + + I+E + R + G R IG+SN + K + + +
Sbjct: 117 LKPGVSEPIPKEDEFEKDLGIEETWQGMERCLEMGLCRSIGVSN---FSSKKIFDLLDFA 173
Query: 236 SSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPL 276
S P + N + + + C + +I + YSPL
Sbjct: 174 SVSPSV----NQVEMHPLWRQRKLRKVCEENNIHVSGYSPL 210
>At1g10810 putative auxin-induced protein
Length = 344
Score = 57.0 bits (136), Expect = 1e-08
Identities = 63/248 (25%), Positives = 102/248 (40%), Gaps = 41/248 (16%)
Query: 55 CLGTMTF-GEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVPQRAQTWGMSEEYFGHWIKH 113
C+G F G + L+ A ++GI D++++Y P+ +E G +K
Sbjct: 26 CMGLSIFDGTTKVETDLIALIHHAINSGITLLDTSDIYG-PET------NELLLGQALKD 78
Query: 114 RNIPRDRLVIATKVAGP-SGQMTWIRGGPKSLDATNISQAIDNSLLRMQLDYIDLYQIHW 172
R+++ +ATK Q RG P + A A + SL R+ + IDLY H
Sbjct: 79 GM--REKVELATKFGLLLKDQKLGYRGDPAYVRA-----ACEASLRRLGVSCIDLYYQHR 131
Query: 173 PDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAVNDGKIRYIGLSNETPYGLMKFIQVA 232
D VP I+ + L + V +GKI+YIGLS
Sbjct: 132 IDTTVP-----------------IEVTIGELKKLVEEGKIKYIGLSEACA-------STI 167
Query: 233 EKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESISLLAYSPLAMGIL-SGKYFSHGNG 291
++ + + ++Q +SL R + + C + I ++AYSPL +G +G F
Sbjct: 168 RRAHAVHPLTAVQLEWSLWSRDVEEDIIPTCRELGIGIVAYSPLGLGFFAAGPKFIESMD 227
Query: 292 PADARLNL 299
D R L
Sbjct: 228 NGDYRKGL 235
>At2g37770 putative aldo/keto reductase
Length = 315
Score = 53.5 bits (127), Expect = 2e-07
Identities = 38/132 (28%), Positives = 62/132 (46%), Gaps = 10/132 (7%)
Query: 145 DATNISQAIDNSLLRMQLDYIDLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALS 204
D ++ +A++ +L +QL+Y+DLY IHWP R G P + + I A+
Sbjct: 88 DPQDVPEALNRTLKDLQLEYVDLYLIHWPARIKK--GSVGIKP-ENLLPVDIPSTWKAME 144
Query: 205 RAVNDGKIRYIGLSNETPYGLMKFIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCH 264
+ GK R IG+SN + L +++A P V+ + +T + E C
Sbjct: 145 ALYDSGKARAIGVSNFSTKKLADLLELAR----VPPAVNQVECHPSWRQT---KLQEFCK 197
Query: 265 QESISLLAYSPL 276
+ + L AYSPL
Sbjct: 198 SKGVHLSAYSPL 209
>At2g37790 putative alcohol dehydrogenase
Length = 350
Score = 51.6 bits (122), Expect = 6e-07
Identities = 37/132 (28%), Positives = 58/132 (43%), Gaps = 10/132 (7%)
Query: 145 DATNISQAIDNSLLRMQLDYIDLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALS 204
D + +A++ +L +QLDY+DLY IHWP G T + P + I A+
Sbjct: 88 DPQEVPEALNRTLQDLQLDYVDLYLIHWPVSLKK--GSTGFKP-ENILPTDIPSTWKAME 144
Query: 205 RAVNDGKIRYIGLSNETPYGLMKFIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCH 264
+ GK R IG+SN + L + VA + ++ + + R F C
Sbjct: 145 SLFDSGKARAIGVSNFSSKKLADLLVVARVPPAVNQVECHPSWQQNVLRDF-------CK 197
Query: 265 QESISLLAYSPL 276
+ + L YSPL
Sbjct: 198 SKGVHLSGYSPL 209
>At2g27680 unknown protein
Length = 384
Score = 50.8 bits (120), Expect = 1e-06
Identities = 52/208 (25%), Positives = 91/208 (43%), Gaps = 40/208 (19%)
Query: 80 AGINFFDSAEMYPVPQRAQTWGMSEEYFGHWIKH--RNIPRDRLVIATKVAGPSGQMTWI 137
AG++ FD A+ Y G +E+ +G +I R P + L K+ G + W+
Sbjct: 90 AGLSTFDMADHY---------GPAEDLYGIFINRVRRERPPEYL---EKIKGLT---KWV 134
Query: 138 RGGPKSLDATNISQAIDNSLLRMQLDYIDLYQIHWPDRYVPMFGETEYDPVQQYSSISID 197
P + ++ + Q ID S RM + +D+ Q HW D Y++
Sbjct: 135 PP-PIKMTSSYVRQNIDISRKRMDVAALDMLQFHWWD----------------YANDGYL 177
Query: 198 EQLDALSRAVNDGKIRYIGLSNETPYGLMKFIQVAEKSSSYPKIVSLQNSYSLLCRTFDS 257
+ L L+ +GKI+ + L+N L K ++ + P +VS Q +S++
Sbjct: 178 DALKHLTDLKEEGKIKTVALTNFDTERLQKILE-----NGIP-VVSNQVQHSIVDMRPQQ 231
Query: 258 AMAECCHQESISLLAYSPLAMGILSGKY 285
MA+ C + L+ Y + G+LS K+
Sbjct: 232 RMAQLCELTGVKLITYGTVMGGLLSEKF 259
>At2g37760 putative alcohol dehydrogenase
Length = 311
Score = 49.3 bits (116), Expect = 3e-06
Identities = 33/129 (25%), Positives = 56/129 (42%), Gaps = 10/129 (7%)
Query: 148 NISQAIDNSLLRMQLDYIDLYQIHWPDRYVPMFGETEYDPVQQYSSISIDEQLDALSRAV 207
++ +A++ +L +Q+DY+DLY IHWP + E+ + + I A+
Sbjct: 87 DVPKALEKTLQDLQIDYVDLYLIHWP---ASLKKESLMPTPEMLTKPDITSTWKAMEALY 143
Query: 208 NDGKIRYIGLSNETPYGLMKFIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQES 267
+ GK R IG+SN + L + VA ++ N + E C +
Sbjct: 144 DSGKARAIGVSNFSSKKLTDLLNVA-------RVTPAVNQVECHPVWQQQGLHELCKSKG 196
Query: 268 ISLLAYSPL 276
+ L YSPL
Sbjct: 197 VHLSGYSPL 205
>At5g01670 aldose reductase-like protein
Length = 322
Score = 47.8 bits (112), Expect = 9e-06
Identities = 60/248 (24%), Positives = 95/248 (38%), Gaps = 44/248 (17%)
Query: 35 SGGGGVPVFNLAPNLNVSRLCLGTMTFGEQNTLSQSFQLLDEAYHAGINFFDSAEMYPVP 94
S G + F L + + LGT G Q + ++ G D+A Y
Sbjct: 8 SEGQNMESFRLLSGHKIPAVGLGTWRSGSQ----AAHAVVTAIVEGGYRHIDTAWEYG-D 62
Query: 95 QRAQTWGMSEEYFGHWIKHRNIPRDRLVIATKVAGPSGQMTWIRGGPKSLDATNISQAID 154
QR G+ H + R L + +K+ W L + A+
Sbjct: 63 QREVGQGIKRAM------HAGLERRDLFVTSKL--------WCT----ELSPERVRPALQ 104
Query: 155 NSLLRMQLDYIDLYQIHWPDRY------VPMFGETEYDPVQQYSSISIDEQLDALSRAVN 208
N+L +QL+Y+DLY IHWP R P G+ V + + +++ LS+
Sbjct: 105 NTLKELQLEYLDLYLIHWPIRLREGASKPPKAGD-----VLDFDMEGVWREMENLSK--- 156
Query: 209 DGKIRYIGLSNETPYGLMKFIQVAEKSSSYPKIVSLQNSYSLLCRTFDSAMAECCHQESI 268
D +R IG+ N T L K + AE P + ++ + + E C + I
Sbjct: 157 DSLVRNIGVCNFTVTKLNKLLGFAE---LIPAVCQMEMHPGWR----NDRILEFCKKNEI 209
Query: 269 SLLAYSPL 276
+ AYSPL
Sbjct: 210 HVTAYSPL 217
>At2g21250 putative NADPH dependent mannose 6-phosphate reductase
Length = 309
Score = 42.7 bits (99), Expect = 3e-04
Identities = 34/153 (22%), Positives = 68/153 (44%), Gaps = 23/153 (15%)
Query: 145 DATNISQAIDNSLLRMQLDYIDLYQIHWP-----------DRYVPMFGETEYDPVQQYSS 193
D ++ +A +SL ++QLDY+DL+ +H+P D + G + D ++
Sbjct: 81 DHGHVIEACKDSLKKLQLDYLDLFLVHFPVATKHTGVGTTDSALGDDGVLDID-----TT 135
Query: 194 ISIDEQLDALSRAVNDGKIRYIGLSNETPYGLMKFIQVAEKSSSYPKIVSLQNSYSLLCR 253
IS++ + + V+ G +R IG+SN + + +Y KI N
Sbjct: 136 ISLETTWHDMEKLVSMGLVRSIGISNYDVF-------LTRDCLAYSKIKPAVNQIETHPY 188
Query: 254 TFDSAMAECCHQESISLLAYSPLAMGILSGKYF 286
++ + C + I + A++PL + ++F
Sbjct: 189 FQRDSLVKFCQKHGICVTAHTPLGGATANAEWF 221
>At2g21260 putative NADPH dependent mannose 6-phosphate reductase
Length = 309
Score = 42.0 bits (97), Expect = 5e-04
Identities = 35/155 (22%), Positives = 68/155 (43%), Gaps = 23/155 (14%)
Query: 143 SLDATNISQAIDNSLLRMQLDYIDLYQIHWP-----------DRYVPMFGETEYDPVQQY 191
S D ++ +A +SL ++QLDY+DL+ +H P D + G + D
Sbjct: 79 SSDHGHVIEACKDSLKKLQLDYLDLFLVHIPIATKHTGIGTTDSALGDDGVLDID----- 133
Query: 192 SSISIDEQLDALSRAVNDGKIRYIGLSNETPYGLMKFIQVAEKSSSYPKIVSLQNSYSLL 251
++IS++ + + V+ G +R IG+SN + + +Y KI N
Sbjct: 134 TTISLETTWHDMEKLVSMGLVRSIGISNYDVF-------LTRDCLAYSKIKPAVNQIETH 186
Query: 252 CRTFDSAMAECCHQESISLLAYSPLAMGILSGKYF 286
++ + C + I + A++PL + ++F
Sbjct: 187 PYFQRDSLVKFCQKHGICVTAHTPLGGATANAEWF 221
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.138 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,471,843
Number of Sequences: 26719
Number of extensions: 309330
Number of successful extensions: 748
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 24
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 687
Number of HSP's gapped (non-prelim): 33
length of query: 337
length of database: 11,318,596
effective HSP length: 100
effective length of query: 237
effective length of database: 8,646,696
effective search space: 2049266952
effective search space used: 2049266952
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 60 (27.7 bits)
Medicago: description of AC141106.8


