

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140104.6 - phase: 0
(216 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
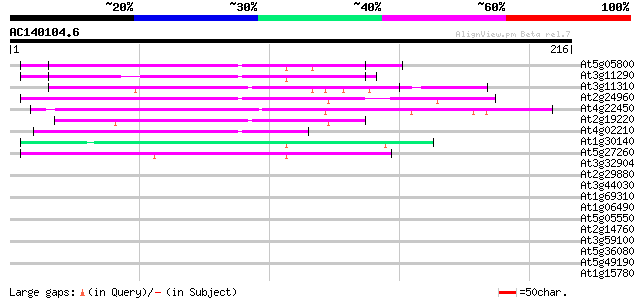
Score E
Sequences producing significant alignments: (bits) Value
At5g05800 putative protein 68 3e-12
At3g11290 unknown protein 60 9e-10
At3g11310 hypothetical protein 57 6e-09
At2g24960 hypothetical protein 54 5e-08
At4g22450 hypothetical protein 46 2e-05
At2g19220 hypothetical protein 45 3e-05
At4g02210 unknown protein 44 6e-05
At1g30140 hypothetical protein 42 2e-04
At5g27260 putative protein 42 2e-04
At3g32904 putative protein 40 0.001
At2g29880 hypothetical protein 37 0.006
At3g44030 putative protein 37 0.010
At1g69310 putative WRKY transcription factor 34 0.051
At1g06490 glucan synthase, putative 34 0.067
At5g05550 unknown protein 32 0.20
At2g14760 hypothetical protein 32 0.20
At3g59100 putative protein 32 0.25
At5g36080 putative protein 31 0.43
At5g49190 sucrose synthase 29 1.7
At1g15780 unknown protein 29 1.7
>At5g05800 putative protein
Length = 449
Score = 68.2 bits (165), Expect = 3e-12
Identities = 41/148 (27%), Positives = 67/148 (44%), Gaps = 2/148 (1%)
Query: 5 ESLPNSSENLGKAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKN 64
+S+ +SS K W + K+ LD+ + E K + F + W+ I N+
Sbjct: 156 QSMCSSSNPQTKGYWSPSTHKLFLDLLVQETLKGNRPDTHFNKEGWKTILGTINENTGLG 215
Query: 65 YTQKQLKNRMDTLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIR-NVKYAKFR 123
YT+ QLKN D R W IW QL+G + + W+ + A W IR N + +FR
Sbjct: 216 YTRPQLKNHWDCTRKAWKIWCQLVG-ASSMKWDPESRSFGATEEEWRIYIRENPRAGQFR 274
Query: 124 FQGLEFCDELEFIFGEIVATSQCACAPA 151
+ + D+L IF ++ + P+
Sbjct: 275 HKEVPHADQLAIIFNGVIEPGETYTPPS 302
Score = 55.1 bits (131), Expect = 3e-08
Identities = 31/123 (25%), Positives = 56/123 (45%), Gaps = 2/123 (1%)
Query: 16 KAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKNYTQKQLKNRMD 75
KA W+ ++ +D+C+ + K G F + W I + ++ Y + QLKN D
Sbjct: 4 KAVWEPEYHRVFVDLCVEQTMLGNKPGTHFSKEGWRNILISFQEQTGAMYDRMQLKNHWD 63
Query: 76 TLRGEWTIWKQLLGKETGLGWNHHIGNIDA-DSAWWDAKIRNVKYAKFRFQGLEFCDELE 134
T+ +W IW++L+ + + + WN A D W + N ++R +LE
Sbjct: 64 TMSRQWKIWRRLV-ETSFMNWNPESNRFRATDDDWANYLQENPDAGQYRLSVPHDLKKLE 122
Query: 135 FIF 137
+F
Sbjct: 123 ILF 125
>At3g11290 unknown protein
Length = 460
Score = 60.1 bits (144), Expect = 9e-10
Identities = 38/138 (27%), Positives = 62/138 (44%), Gaps = 2/138 (1%)
Query: 5 ESLPNSSENLGKAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKN 64
+S N S K W S ++ +D+ E K + + + W+ I + N+ K+
Sbjct: 154 QSASNFSSPQSKGYWSPSSHELFVDLLFQEALKGNRPDSHYPKETWKMILETINQNTGKS 213
Query: 65 YTQKQLKNRMDTLRGEWTIWKQLLGKETGLGWNHHIGNIDA-DSAWWDAKIRNVKYAKFR 123
+T+ QLKN D R W IW Q++G + W+ A D W + N + A FR
Sbjct: 214 FTRPQLKNHWDCTRKSWKIWCQVIGAPV-MKWDATSRTFGATDEDWKNYLKENHRAAPFR 272
Query: 124 FQGLEFCDELEFIFGEIV 141
+ L D+L IF ++
Sbjct: 273 RKQLPHADKLATIFKGLI 290
Score = 45.1 bits (105), Expect = 3e-05
Identities = 30/123 (24%), Positives = 57/123 (45%), Gaps = 9/123 (7%)
Query: 16 KAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKNYTQKQLKNRMD 75
KA W+ ++ +D+C+ + + G + I + +R +T+ QLKN D
Sbjct: 4 KAAWEPEYHRVFVDLCVEQKMLGNQPGT-------QHILKPFLQRTGARFTRNQLKNHWD 56
Query: 76 TLRGEWTIWKQLLGKETGLGWNHHIGNIDA-DSAWWDAKIRNVKYAKFRFQGLEFCDELE 134
T+ +W IW +L+ + + + W+ A D W + N + ++R F ++LE
Sbjct: 57 TMIKQWKIWCRLV-QCSDMQWDPQTNTFGANDQDWANYLHVNPEAGQYRLNPPSFLEKLE 115
Query: 135 FIF 137
IF
Sbjct: 116 LIF 118
>At3g11310 hypothetical protein
Length = 539
Score = 57.4 bits (137), Expect = 6e-09
Identities = 40/141 (28%), Positives = 64/141 (45%), Gaps = 7/141 (4%)
Query: 16 KAKWDTFSTKMLLDICMVEIHKCGKLGIAFRN-----KKWEEIRDEYNKRANKNYTQKQL 70
KA W + S ++ +D+ E K + A RN + W + + +N++ YT+KQL
Sbjct: 173 KAYWSSSSHEIFVDLLFTESLKENRPKPARRNGYYAKETWNMMVESFNQKTGLRYTRKQL 232
Query: 71 KNRMDTLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIRNVKYA-KFRFQGLEF 129
KN + R W W Q +G L W+ + A S W+ + K A +FR + +
Sbjct: 233 KNHWNITRDAWRRWCQAVGSPL-LKWDANTKTFGATSEDWENYSKENKRAEQFRLKHIPH 291
Query: 130 CDELEFIFGEIVATSQCACAP 150
D+L IF V + A P
Sbjct: 292 ADKLAIIFKGHVEPGKTALRP 312
Score = 42.0 bits (97), Expect = 2e-04
Identities = 41/177 (23%), Positives = 75/177 (42%), Gaps = 21/177 (11%)
Query: 16 KAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKNYTQKQLKNRMD 75
K W+ K+ +D+C+ + K+ + FR I + + + +T+ QLKN D
Sbjct: 5 KVMWEPELHKVFVDLCVEQ-----KM-LGFRLPGLNRIWESFVQNTGARFTRDQLKNHWD 58
Query: 76 TLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIR-NVKYAKFRFQGL--EFCDE 132
T+ W W +L+ + + + W+ A + W R N K ++RF+ F +
Sbjct: 59 TMLRLWRAWCRLV-ECSEMKWDPQTKKFGASTEVWTNYFRVNPKAKQYRFRSSPPPFLKD 117
Query: 133 LEFIF-----GEIVATSQCACAPAMGVPLESTGKNTTADVAREIIESDDELNIDELS 184
L+ IF G+ TS C +P N T D + + D+ D+++
Sbjct: 118 LKMIFEGTDLGDEEGTS---CGKRKRIP---DADNDTGDEDNDTGDDDNYTGDDDIT 168
>At2g24960 hypothetical protein
Length = 797
Score = 54.3 bits (129), Expect = 5e-08
Identities = 45/185 (24%), Positives = 82/185 (44%), Gaps = 12/185 (6%)
Query: 5 ESLPNSSENLGKAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKN 64
ES+ S + K +W + ++I + +I + K G AF + W ++ +N R +
Sbjct: 158 ESVVLSGKESSKTEWTLEMDQYFVEIMVDQIGRGNKTGNAFSKQAWIDMLVLFNARFSGQ 217
Query: 65 YTQKQLKNRMDTLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIRNVKYAK-FR 123
Y ++ L++R + L + + +L KE G W+ I AD A WD+ I++ A+ +R
Sbjct: 218 YGKRVLRHRYNKLLKYYKDMEAIL-KEDGFSWDETRLMISADDAVWDSYIKDHPLARTYR 276
Query: 124 FQGLEFCDELEFIFGEIVATSQCACAPAMGVPLESTGKNT-TADVAREIIESDDELNIDE 182
+ L ++L+ IF AC G G T++ ++ D I
Sbjct: 277 MKSLPSYNDLDTIF---------ACQAEQGTDHRDDGSAAQTSETKASQEQNSDRTRIFW 327
Query: 183 LSPME 187
PM+
Sbjct: 328 TPPMD 332
Score = 39.7 bits (91), Expect = 0.001
Identities = 26/125 (20%), Positives = 53/125 (41%), Gaps = 2/125 (1%)
Query: 19 WDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKNYTQKQLKNRMDTLR 78
W + +D+ + +H+ + G F + W E+ +N + Y + LK+R L
Sbjct: 15 WTPTMERFFIDLMLEHLHRGNRTGHTFNKQAWNEMLTVFNSKFGSQYDKDVLKSRYTNLW 74
Query: 79 GEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIRNVKYAK-FRFQGLEFCDELEFIF 137
++ K LL G W+ + D + W ++ A+ ++ + + +L I+
Sbjct: 75 KQYNDVKCLL-DHGGFVWDQTHQTVIGDDSLWSLYLKAHPEARVYKTKPVLNFSDLCLIY 133
Query: 138 GEIVA 142
G VA
Sbjct: 134 GYTVA 138
Score = 39.3 bits (90), Expect = 0.002
Identities = 22/89 (24%), Positives = 42/89 (46%), Gaps = 1/89 (1%)
Query: 27 LLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKNYTQKQLKNRMDTLRGEWTIWKQ 86
L+D+ + +++ ++G F W E+ +N + + + LKNR LR + K
Sbjct: 335 LIDLLVEQVNNGNRVGQTFITSAWNEMVTAFNAKFGSQHNKDVLKNRYKHLRRLYNDIKF 394
Query: 87 LLGKETGLGWNHHIGNIDADSAWWDAKIR 115
LL ++ G W+ + AD W+ I+
Sbjct: 395 LL-EQNGFSWDARRDMVIADDDIWNTYIQ 422
Score = 38.9 bits (89), Expect = 0.002
Identities = 24/112 (21%), Positives = 47/112 (41%), Gaps = 1/112 (0%)
Query: 4 KESLPNSSENLGKAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANK 63
K P S+ +W L+D+ + ++ + K+G F + W ++ + +N +
Sbjct: 519 KNDYPCSNIGPPCIEWTRVMDHCLIDLMLEQVSRGNKIGETFTEQAWADMAESFNAKFGL 578
Query: 64 NYTQKQLKNRMDTLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIR 115
L+NR L E +L + G W+ I A+ +W+A I+
Sbjct: 579 QTDMFMLENRYILLMKERDDINNILNLD-GFTWDVEKQTIVAEDEYWEAYIK 629
>At4g22450 hypothetical protein
Length = 457
Score = 45.8 bits (107), Expect = 2e-05
Identities = 48/224 (21%), Positives = 100/224 (44%), Gaps = 27/224 (12%)
Query: 9 NSSENLGKAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKNYTQK 68
N ++NL W+ T+ L++ + E K + + I D + + + +T +
Sbjct: 5 NGTDNL---VWNDEQTRFYLELRIEEKLKGNIRNQNVNDVGRQTIIDRFYEVYGERHTWR 61
Query: 69 QLKNRMDTLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIRNVKYA-KFRFQGL 127
+ + T + ++ +K+L TGLG+ + G+I WW+ + + ++ A KF+ + +
Sbjct: 62 KFGVKFTTCKKQYEGFKKLTHNRTGLGY-YPNGSIRMSDDWWNERCKELRGARKFKDKPV 120
Query: 128 EFCDELEFIFGEIVATSQCACAPAMG-VPLESTGKNTTADVAREIIESDDE-------LN 179
D +E +FG + + AP G L+S ++ D + I DD+ LN
Sbjct: 121 ANVDLMEKVFGTVHISGAEGWAPKQGEEKLDSMHQDDDGDSTQIPITQDDDGESRQIPLN 180
Query: 180 IDE--------------LSPMENTQSKNKRKVSPRMRKQQRVRQ 209
+D+ P +++SK+ +K S ++ +Q V +
Sbjct: 181 LDDDEATHILPTQSNVGNGPSYSSKSKSSKKRSRSVQDEQGVAE 224
>At2g19220 hypothetical protein
Length = 439
Score = 45.1 bits (105), Expect = 3e-05
Identities = 32/124 (25%), Positives = 52/124 (41%), Gaps = 5/124 (4%)
Query: 18 KWDTFSTKMLLDICMVEIHKCG---KLGIAFRNKKWEEIRDEYNKRANKNYTQKQLKNRM 74
KW S +++D C E K K F + W+ I ++ N+ YT KQL+N
Sbjct: 164 KWSPSSHAIVVDTCFQESLKGIRPIKRNHLFTKESWKMILEKINRITGLGYTHKQLENHF 223
Query: 75 DTLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIRNVKYAK-FRFQGLEFCDEL 133
R W W + + + W+ + A WD + K A+ F+ + + D+L
Sbjct: 224 TRTRTSWKHWCETIASPI-MKWDANTRKFGATEEDWDKYLMINKRARVFKRRHIPHADKL 282
Query: 134 EFIF 137
IF
Sbjct: 283 ATIF 286
Score = 36.2 bits (82), Expect = 0.014
Identities = 39/171 (22%), Positives = 73/171 (41%), Gaps = 25/171 (14%)
Query: 16 KAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKNYTQKQLKNRMD 75
KA W+ ++ +D+C VE G + + I + + + + +T QL N D
Sbjct: 4 KAAWEPEHDEVFVDLC-VEQKMLGN------QPEMQHILEAFQEMGVR-FTIDQLINHWD 55
Query: 76 TLRGEWTIWKQLLGKETGLGWNHHIGNIDA-DSAWWDAKIRNVKYAKFRFQGLEFCDELE 134
T+ +W IW +L+ + + W+ A D W + N + ++R F ++LE
Sbjct: 56 TMIKQWKIWCRLV-QCKDIKWDSLTNTFGATDQEWANYLEVNPEAGQYRCNPPLFLEKLE 114
Query: 135 FIFGEIVATSQCACAPAMGVPLESTGKNTTADVAREIIESDDELNIDELSP 185
IF G+ L+ G ++ + + ++I E DE N+ P
Sbjct: 115 IIFA--------------GMNLDGEGTSSGSKM-KQICEHRDEENVTGYVP 150
>At4g02210 unknown protein
Length = 439
Score = 43.9 bits (102), Expect = 6e-05
Identities = 24/106 (22%), Positives = 49/106 (45%), Gaps = 1/106 (0%)
Query: 10 SSENLGKAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKNYTQKQ 69
SS + W + +D+ + + + ++ FR + W E+ + +N + N+
Sbjct: 177 SSVTRCRTTWHPPMDRYFIDLMLDQARRGNQIEGVFRKQAWTEMVNLFNAKFESNFDVDV 236
Query: 70 LKNRMDTLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIR 115
LKNR +LR ++ K +L + G W++ + AD+ W I+
Sbjct: 237 LKNRYKSLRRQFNAIKSIL-RSDGFAWDNERQMVTADNNVWQDYIK 281
Score = 31.6 bits (70), Expect = 0.33
Identities = 27/133 (20%), Positives = 56/133 (41%), Gaps = 4/133 (3%)
Query: 9 NSSENLGKAKWDTFSTKMLLDICMVEIHKCGKL-GIAFRNKKWEEIRDEYNKRANKNYTQ 67
N +E L + W + +++ + ++ K + F + W+ + + + Y +
Sbjct: 5 NGNERL-RTVWTPEMDQYFIELMVEQVRKGNRFEDHLFSKRAWKFMSCSFTAKFKFLYGK 63
Query: 68 KQLKNRMDTLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIR-NVKYAKFRFQG 126
LKNR TLR + LL E G W+ + AD+ WD ++ + FR +
Sbjct: 64 DVLKNRHKTLRNLFKSVNNLL-IEDGFSWDDTRQMVVADNCVWDEYLKIHPDSRSFRIKS 122
Query: 127 LEFCDELEFIFGE 139
+ +L ++ +
Sbjct: 123 IPCYKDLCLVYSD 135
>At1g30140 hypothetical protein
Length = 221
Score = 42.4 bits (98), Expect = 2e-04
Identities = 37/167 (22%), Positives = 65/167 (38%), Gaps = 10/167 (5%)
Query: 5 ESLPNSSENLGKAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKN 64
E P E +W T +L+++ + + GI + ++ NKR N
Sbjct: 2 EDPPRGKEKGPYNQWTPDETDVLIEL--IRQNWRDSSGIIGKLTVESKLLPALNKRLGCN 59
Query: 65 YTQKQLKNRMDTLRGEWTIWKQLLGKETGLGWNHHIGNIDA-DSAWWDAKIRNVKYAKFR 123
K +R+ L+ + + L +G GW+ A D W D + + +
Sbjct: 60 KNHKNYMSRLKFLKNLYQSYLDLKRFSSGFGWDPETKKFTAPDEVWRDYLKAHPNHKHMQ 119
Query: 124 FQGLEFCDELEFIFGEIVAT-------SQCACAPAMGVPLESTGKNT 163
+ ++ ++L+ IFG++VAT S C V S GK T
Sbjct: 120 TESIDHFEDLQIIFGDVVATGSFAVGMSDSTCPRIYTVGERSQGKET 166
>At5g27260 putative protein
Length = 303
Score = 42.0 bits (97), Expect = 2e-04
Identities = 37/146 (25%), Positives = 60/146 (40%), Gaps = 3/146 (2%)
Query: 5 ESLPNSSENLGKAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIR--DEYNKRAN 62
+S P + W TK+L+ + + I+ + +K E + E NK
Sbjct: 3 DSQPKKRKKGDYNPWSPEETKLLVQLLVEGINNNWRDSNGTISKLTVETKFMPEINKEFC 62
Query: 63 KNYTQKQLKNRMDTLRGEWTIWKQLLGKETGLGWNHHIGNIDA-DSAWWDAKIRNVKYAK 121
++ +RM L+ ++ L +G GW+ A D W D + +
Sbjct: 63 RSKNYNHYLSRMKYLKIQYQSCLDLQRFSSGFGWDPLTKRFTASDEVWSDYLKAHPNNKQ 122
Query: 122 FRFQGLEFCDELEFIFGEIVATSQCA 147
R+ EF DEL+ IFGE VAT + A
Sbjct: 123 LRYDTFEFFDELQIIFGEGVATGKNA 148
>At3g32904 putative protein
Length = 330
Score = 40.0 bits (92), Expect = 0.001
Identities = 17/63 (26%), Positives = 34/63 (52%)
Query: 51 EEIRDEYNKRANKNYTQKQLKNRMDTLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWW 110
E I D++NK N N + +K ++D L+ ++ +K+L+ TG+ + I ++WW
Sbjct: 15 EFILDKFNKEFNLNINYQFVKEKLDQLKRKYKKYKELMKNSTGISVDTTTSVISTSNSWW 74
Query: 111 DAK 113
+
Sbjct: 75 QER 77
>At2g29880 hypothetical protein
Length = 308
Score = 37.4 bits (85), Expect = 0.006
Identities = 32/148 (21%), Positives = 63/148 (41%), Gaps = 8/148 (5%)
Query: 58 NKRANKNYTQKQLKNRMDTLRGEWTIWKQLLGKETGLGWNHHIGNIDA-DSAWWDAKIRN 116
NK+ N T +RM +++ E++++ L +G GW+ A D W + +
Sbjct: 63 NKKFKCNKTYTNYLSRMKSMKKEYSVYAALFWFSSGFGWDPITKQFTAPDDVWAAYLMGH 122
Query: 117 VKYAKFRFQGLEFCDELEFIFGEIVATSQCACAPAMGVPLESTGKNTTADVAREIIESDD 176
+ R E ++L+ IF +A A G+ +S N + +++ D
Sbjct: 123 PNHHHMRTSTFEDFEDLQLIFESAIAKGN----NAFGLGGDS---NAETFEEEDDLQAGD 175
Query: 177 ELNIDELSPMENTQSKNKRKVSPRMRKQ 204
+N E++ E ++ K K+ R R +
Sbjct: 176 NVNHMEINDDEVNETLPKEKLPTRKRSK 203
>At3g44030 putative protein
Length = 775
Score = 36.6 bits (83), Expect = 0.010
Identities = 21/71 (29%), Positives = 34/71 (47%), Gaps = 1/71 (1%)
Query: 51 EEIRDEYNKRANKNYTQKQLKNRMDTLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWW 110
E + + +N N N T KN++D + + WK L K TG+ + + I+A +AWW
Sbjct: 72 EYMVENFNIAFNMNITYGFFKNKLDEFKKSYKRWKALTHK-TGITVDPNTSFINASNAWW 130
Query: 111 DAKIRNVKYAK 121
+ K K
Sbjct: 131 TEQEFGCKLTK 141
>At1g69310 putative WRKY transcription factor
Length = 287
Score = 34.3 bits (77), Expect = 0.051
Identities = 30/130 (23%), Positives = 55/130 (42%), Gaps = 9/130 (6%)
Query: 77 LRGEWTIWKQL-LGKETGLGWNHHIGNIDADSAWWDAKIRNVKYAKFRFQGLEFCDELEF 135
L + + W++L L + ++ NI +D W N+ ++ L F +L
Sbjct: 9 LSNDDSAWRELTLTAQDSDFFDRDTSNILSDFGW------NLHHSSDHPHSLRFDSDLTQ 62
Query: 136 IFG--EIVATSQCACAPAMGVPLESTGKNTTADVAREIIESDDELNIDELSPMENTQSKN 193
G TS C+ + A+ V + ST N +A + +++ E +P T K
Sbjct: 63 TTGVKPTTVTSSCSSSAAVSVAVTSTNNNPSATSSSSEDPAENSTASAEKTPPPETPVKE 122
Query: 194 KRKVSPRMRK 203
K+K R+R+
Sbjct: 123 KKKAQKRIRQ 132
>At1g06490 glucan synthase, putative
Length = 1933
Score = 33.9 bits (76), Expect = 0.067
Identities = 17/57 (29%), Positives = 29/57 (50%), Gaps = 4/57 (7%)
Query: 80 EWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIRNVKYAKFRFQGLEFCDELEFI 136
+WT WK+ +G G+G I + +WWD + ++K+ R + LE L F+
Sbjct: 1701 DWTDWKRWMGNRGGIG----IVLDKSWESWWDIEQEHLKHTNLRGRVLEILLALRFL 1753
>At5g05550 unknown protein
Length = 246
Score = 32.3 bits (72), Expect = 0.20
Identities = 18/55 (32%), Positives = 27/55 (48%), Gaps = 3/55 (5%)
Query: 46 RNKKWEEIRDEYNKRANKNYTQK---QLKNRMDTLRGEWTIWKQLLGKETGLGWN 97
R W+++ D N R N +K Q KNR+DTL+ ++ K L T +N
Sbjct: 49 RQNDWKDVADAVNSRHGDNSRKKTDLQCKNRVDTLKKKYKTEKAKLSPSTWRFYN 103
>At2g14760 hypothetical protein
Length = 519
Score = 32.3 bits (72), Expect = 0.20
Identities = 18/71 (25%), Positives = 34/71 (47%)
Query: 14 LGKAKWDTFSTKMLLDICMVEIHKCGKLGIAFRNKKWEEIRDEYNKRANKNYTQKQLKNR 73
+ K W T+ LL++ +E G + K E + ++ KR ++KNR
Sbjct: 313 VAKIIWSDEMTRFLLELITLEKQAGNYRGKSLIEKGKENVLVKFKKRFPITLNWNKVKNR 372
Query: 74 MDTLRGEWTIW 84
+DTL+ ++ I+
Sbjct: 373 LDTLKKQYEIY 383
>At3g59100 putative protein
Length = 1808
Score = 32.0 bits (71), Expect = 0.25
Identities = 17/56 (30%), Positives = 28/56 (49%), Gaps = 4/56 (7%)
Query: 80 EWTIWKQLLGKETGLGWNHHIGNIDADSAWWDAKIRNVKYAKFRFQGLEFCDELEF 135
+WT WK+ LG G+G I + +WW+ + ++K+ R + LE L F
Sbjct: 1576 DWTDWKRWLGDRGGIG----IPVEKSWESWWNVEQEHLKHTSIRGRILEITLALRF 1627
>At5g36080 putative protein
Length = 153
Score = 31.2 bits (69), Expect = 0.43
Identities = 18/56 (32%), Positives = 27/56 (48%), Gaps = 1/56 (1%)
Query: 55 DEYNKRANKNYTQKQLKNRMDTLRGEWTIWKQLLGKETGLGWNHHIGNIDADSAWW 110
+++N+ N N T KN+ D L+ + WK L K TG+ + I A WW
Sbjct: 19 EKFNQVFNMNRTYVFFKNKHDELKKSYKGWKFLTHK-TGISVDPKTSMIFAYDVWW 73
>At5g49190 sucrose synthase
Length = 805
Score = 29.3 bits (64), Expect = 1.7
Identities = 13/38 (34%), Positives = 20/38 (52%)
Query: 50 WEEIRDEYNKRANKNYTQKQLKNRMDTLRGEWTIWKQL 87
W +I + KR + YT K+ R+ TL G + WK +
Sbjct: 734 WVKISEGGLKRIYERYTWKKYSERLLTLAGVYAFWKHV 771
>At1g15780 unknown protein
Length = 1335
Score = 29.3 bits (64), Expect = 1.7
Identities = 17/76 (22%), Positives = 38/76 (49%), Gaps = 1/76 (1%)
Query: 140 IVATSQCACAPAMGVPLESTGKNTTADVAREIIESDDELNIDELSPMENTQSKNKRKVSP 199
++ +Q + GVP +T +T + + ++ + + +LS + TQS ++ S
Sbjct: 264 VLQPNQLHSSQQPGVPTSATQPSTVNSAPLQGLHTNQQSS-PQLSSQQTTQSMLRQHQSS 322
Query: 200 RMRKQQRVRQRLGLHQ 215
+R+ + +Q G+HQ
Sbjct: 323 MLRQHPQSQQASGIHQ 338
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.132 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,923,681
Number of Sequences: 26719
Number of extensions: 204952
Number of successful extensions: 705
Number of sequences better than 10.0: 31
Number of HSP's better than 10.0 without gapping: 21
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 665
Number of HSP's gapped (non-prelim): 42
length of query: 216
length of database: 11,318,596
effective HSP length: 95
effective length of query: 121
effective length of database: 8,780,291
effective search space: 1062415211
effective search space used: 1062415211
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC140104.6


