

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140104.12 - phase: 0
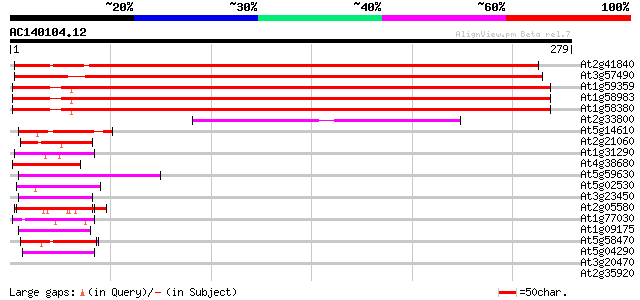
(279 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g41840 40S ribosomal protein S2 472 e-133
At3g57490 40S ribosomal protein S2 homolog 465 e-131
At1g59359 ribosomal protein S2, putative 459 e-130
At1g58983 ribosomal protein S2, putative 459 e-130
At1g58380 unknown protein 459 e-130
At2g33800 30S ribosomal protein S5 75 4e-14
At5g14610 DRH1 DEAD box protein - like 47 1e-05
At2g21060 glycine-rich protein (AtGRP2) 46 3e-05
At1g31290 hypothetical protein 44 7e-05
At4g38680 glycine-rich protein 2 (GRP2) 43 2e-04
At5g59630 glycine-rich protein - like 42 5e-04
At5g02530 unknown protein 42 5e-04
At3g23450 hypothetical protein 42 5e-04
At2g05580 unknown protein 42 5e-04
At1g77030 hypothetical protein 42 5e-04
At1g09175 putative GPI-anchored protein 42 5e-04
At5g58470 RNA/ssDNA-binding protein - like 41 6e-04
At5g04290 glycine-rich protein 41 6e-04
At3g20470 glycine-rich protein, putative 40 0.001
At2g35920 ATP-dependent RNA helicase A like protein 40 0.001
>At2g41840 40S ribosomal protein S2
Length = 285
Score = 472 bits (1214), Expect = e-133
Identities = 233/261 (89%), Positives = 243/261 (92%), Gaps = 3/261 (1%)
Query: 3 ERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVKEG 62
ERGG+RG FG GFGGRGG RG RGGRG GRGGRR GGR EEEKWVPVTKLGRLV G
Sbjct: 11 ERGGERGDFGRGFGGRGG-RGDRGGRGRGGRGGRR--GGRASEEEKWVPVTKLGRLVAAG 67
Query: 63 KIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGDN 122
I+ +EQIYLHSLP+KE+QIID L+GPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGD
Sbjct: 68 HIKQIEQIYLHSLPVKEYQIIDMLIGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGDG 127
Query: 123 NGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSVTV 182
NGHVGLGVKCSKEVATAIRGAIILAKLSV+PVRRGYWGNKIGKPHTVPCKVTGKCGSVTV
Sbjct: 128 NGHVGLGVKCSKEVATAIRGAIILAKLSVVPVRRGYWGNKIGKPHTVPCKVTGKCGSVTV 187
Query: 183 RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFLTP 242
RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCL KTYGFLTP
Sbjct: 188 RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLQKTYGFLTP 247
Query: 243 EFWKETRFSKSPFQEYTDLLA 263
EFWKETRFS+SP+QE+TD LA
Sbjct: 248 EFWKETRFSRSPYQEHTDFLA 268
>At3g57490 40S ribosomal protein S2 homolog
Length = 276
Score = 465 bits (1196), Expect = e-131
Identities = 229/263 (87%), Positives = 239/263 (90%), Gaps = 8/263 (3%)
Query: 3 ERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVKEG 62
ERGGDRG FG GFGGRGG RGG GRG R GR EEEKWVPVTKLGRLVKEG
Sbjct: 7 ERGGDRGDFGRGFGGRGGGRGGPRGRG--------RRAGRAPEEEKWVPVTKLGRLVKEG 58
Query: 63 KIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGDN 122
KI +EQIYLHSLP+KE+QIID LVGP+LKDEVMKIMPVQKQTRAGQRTRFKAF+VVGD+
Sbjct: 59 KITKIEQIYLHSLPVKEYQIIDLLVGPSLKDEVMKIMPVQKQTRAGQRTRFKAFIVVGDS 118
Query: 123 NGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSVTV 182
NGHVGLGVKCSKEVATAIRGAIILAKLSV+P+RRGYWGNKIGKPHTVPCKVTGKCGSVTV
Sbjct: 119 NGHVGLGVKCSKEVATAIRGAIILAKLSVVPIRRGYWGNKIGKPHTVPCKVTGKCGSVTV 178
Query: 183 RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFLTP 242
RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCL KTYGFLTP
Sbjct: 179 RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLQKTYGFLTP 238
Query: 243 EFWKETRFSKSPFQEYTDLLAKP 265
EFWKETRFSKSP+QE+TD L P
Sbjct: 239 EFWKETRFSKSPYQEHTDFLLIP 261
>At1g59359 ribosomal protein S2, putative
Length = 284
Score = 459 bits (1181), Expect = e-130
Identities = 231/269 (85%), Positives = 239/269 (87%), Gaps = 6/269 (2%)
Query: 2 AERGGDRGGFGSGFGGRGGDRGGRGGRG-DRGRGGRRRAGGRREEEEKWVPVTKLGRLVK 60
AERGGDRG FG GFGG G GGRG DRG GR R GGR EE KWVPVTKLGRLV
Sbjct: 10 AERGGDRGDFGRGFGGGRG-----GGRGRDRGPRGRGRRGGRASEETKWVPVTKLGRLVA 64
Query: 61 EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 120
+ KI LEQIYLHSLP+KE+QIID LVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG
Sbjct: 65 DNKITKLEQIYLHSLPVKEYQIIDHLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 124
Query: 121 DNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSV 180
D NGHVGLGVKCSKEVATAIRGAIILAKLSV+PVRRGYWGNKIGKPHTVPCKVTGKCGSV
Sbjct: 125 DGNGHVGLGVKCSKEVATAIRGAIILAKLSVVPVRRGYWGNKIGKPHTVPCKVTGKCGSV 184
Query: 181 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL 240
TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCL KTYGFL
Sbjct: 185 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLQKTYGFL 244
Query: 241 TPEFWKETRFSKSPFQEYTDLLAKPTAKA 269
TPEFWKETRFS+SP+QE+TD L+ A
Sbjct: 245 TPEFWKETRFSRSPYQEHTDFLSTKAVSA 273
>At1g58983 ribosomal protein S2, putative
Length = 284
Score = 459 bits (1181), Expect = e-130
Identities = 231/269 (85%), Positives = 239/269 (87%), Gaps = 6/269 (2%)
Query: 2 AERGGDRGGFGSGFGGRGGDRGGRGGRG-DRGRGGRRRAGGRREEEEKWVPVTKLGRLVK 60
AERGGDRG FG GFGG G GGRG DRG GR R GGR EE KWVPVTKLGRLV
Sbjct: 10 AERGGDRGDFGRGFGGGRG-----GGRGRDRGPRGRGRRGGRASEETKWVPVTKLGRLVA 64
Query: 61 EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 120
+ KI LEQIYLHSLP+KE+QIID LVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG
Sbjct: 65 DNKITKLEQIYLHSLPVKEYQIIDHLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 124
Query: 121 DNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSV 180
D NGHVGLGVKCSKEVATAIRGAIILAKLSV+PVRRGYWGNKIGKPHTVPCKVTGKCGSV
Sbjct: 125 DGNGHVGLGVKCSKEVATAIRGAIILAKLSVVPVRRGYWGNKIGKPHTVPCKVTGKCGSV 184
Query: 181 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL 240
TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCL KTYGFL
Sbjct: 185 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLQKTYGFL 244
Query: 241 TPEFWKETRFSKSPFQEYTDLLAKPTAKA 269
TPEFWKETRFS+SP+QE+TD L+ A
Sbjct: 245 TPEFWKETRFSRSPYQEHTDFLSTKAVSA 273
>At1g58380 unknown protein
Length = 284
Score = 459 bits (1181), Expect = e-130
Identities = 231/269 (85%), Positives = 239/269 (87%), Gaps = 6/269 (2%)
Query: 2 AERGGDRGGFGSGFGGRGGDRGGRGGRG-DRGRGGRRRAGGRREEEEKWVPVTKLGRLVK 60
AERGGDRG FG GFGG G GGRG DRG GR R GGR EE KWVPVTKLGRLV
Sbjct: 10 AERGGDRGDFGRGFGGGRG-----GGRGRDRGPRGRGRRGGRASEETKWVPVTKLGRLVA 64
Query: 61 EGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 120
+ KI LEQIYLHSLP+KE+QIID LVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG
Sbjct: 65 DNKITKLEQIYLHSLPVKEYQIIDHLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVG 124
Query: 121 DNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSV 180
D NGHVGLGVKCSKEVATAIRGAIILAKLSV+PVRRGYWGNKIGKPHTVPCKVTGKCGSV
Sbjct: 125 DGNGHVGLGVKCSKEVATAIRGAIILAKLSVVPVRRGYWGNKIGKPHTVPCKVTGKCGSV 184
Query: 181 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFL 240
TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCL KTYGFL
Sbjct: 185 TVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLQKTYGFL 244
Query: 241 TPEFWKETRFSKSPFQEYTDLLAKPTAKA 269
TPEFWKETRFS+SP+QE+TD L+ A
Sbjct: 245 TPEFWKETRFSRSPYQEHTDFLSTKAVSA 273
>At2g33800 30S ribosomal protein S5
Length = 303
Score = 75.1 bits (183), Expect = 4e-14
Identities = 41/133 (30%), Positives = 73/133 (54%), Gaps = 7/133 (5%)
Query: 92 KDEVMKIMPVQKQTRAGQRTRFKAFVVVGDNNGHVGLGVKCSKEVATAIRGAIILAKLSV 151
++ V+++ V K + G++ +F+A VVVGD G+VG+G +KEV A++ + I A+ ++
Sbjct: 148 EERVVQVRRVTKVVKGGKQLKFRAIVVVGDKQGNVGVGCAKAKEVVAAVQKSAIDARRNI 207
Query: 152 IPVRRGYWGNKIGKPHTVPCKVTGKCGSVTVRMVPAPRGSGIVAARVPKKVLQFAGIDDV 211
+ V + K T P + G G+ V + PA G+G++A + VL+ AG+++
Sbjct: 208 VQV-------PMTKYSTFPHRSEGDYGAAKVMLRPASPGTGVIAGGAVRIVLEMAGVENA 260
Query: 212 FTSSRGSTKTLGN 224
GS L N
Sbjct: 261 LGKQLGSNNALNN 273
>At5g14610 DRH1 DEAD box protein - like
Length = 713
Score = 47.0 bits (110), Expect = 1e-05
Identities = 26/48 (54%), Positives = 29/48 (60%), Gaps = 7/48 (14%)
Query: 5 GGDRGGFG-SGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVP 51
GG RGG+G SG+GGRG G G RGD G GGR +GGR W P
Sbjct: 628 GGGRGGYGDSGYGGRG--ESGYGSRGDSGYGGRGDSGGR----GSWAP 669
Score = 32.7 bits (73), Expect = 0.22
Identities = 22/46 (47%), Positives = 23/46 (49%), Gaps = 11/46 (23%)
Query: 1 MAERGGD------RGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAG 40
MA RGG R G S GG GGRGG GD G GGR +G
Sbjct: 606 MATRGGGGMNKFRRWGTPSSGGG-----GGRGGYGDSGYGGRGESG 646
>At2g21060 glycine-rich protein (AtGRP2)
Length = 201
Score = 45.8 bits (107), Expect = 3e-05
Identities = 25/39 (64%), Positives = 25/39 (64%), Gaps = 4/39 (10%)
Query: 6 GDRGGFGSGFGGRGGDRGG---RGGRGDRGRGGRRRAGG 41
G RGGFG G GGRGG RGG GG G RG GGR GG
Sbjct: 97 GGRGGFGGG-GGRGGGRGGGSYGGGYGGRGSGGRGGGGG 134
Score = 38.5 bits (88), Expect = 0.004
Identities = 22/34 (64%), Positives = 22/34 (64%), Gaps = 3/34 (8%)
Query: 4 RGGDRGGFGSGFGGRGGDR--GGRGGRGDRGRGG 35
RGG GG G G GGRGG GG GGRG GRGG
Sbjct: 99 RGGFGGGGGRG-GGRGGGSYGGGYGGRGSGGRGG 131
Score = 35.8 bits (81), Expect = 0.026
Identities = 18/28 (64%), Positives = 20/28 (71%), Gaps = 4/28 (14%)
Query: 4 RGGDRGG--FGSGFGGRGGDRGGRGGRG 29
RGG RGG +G G+GGRG GGRGG G
Sbjct: 108 RGGGRGGGSYGGGYGGRGS--GGRGGGG 133
Score = 34.3 bits (77), Expect = 0.076
Identities = 22/36 (61%), Positives = 23/36 (63%), Gaps = 3/36 (8%)
Query: 6 GDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
G+ GG GS GGRGG GG GGRG GRGG GG
Sbjct: 88 GNSGGGGSS-GGRGGFGGG-GGRGG-GRGGGSYGGG 120
Score = 32.3 bits (72), Expect = 0.29
Identities = 18/39 (46%), Positives = 18/39 (46%)
Query: 3 ERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
E G G G G GG GGRGG G G G R GG
Sbjct: 77 EVSGPDGAPVQGNSGGGGSSGGRGGFGGGGGRGGGRGGG 115
Score = 28.1 bits (61), Expect = 5.5
Identities = 14/26 (53%), Positives = 14/26 (53%)
Query: 10 GFGSGFGGRGGDRGGRGGRGDRGRGG 35
G G GG GG R G GG G G GG
Sbjct: 154 GGGGYSGGGGGGRYGSGGGGGGGGGG 179
Score = 27.3 bits (59), Expect = 9.3
Identities = 13/23 (56%), Positives = 13/23 (56%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGG 27
GG GG G G G GG GG GG
Sbjct: 156 GGYSGGGGGGRYGSGGGGGGGGG 178
>At1g31290 hypothetical protein
Length = 1194
Score = 44.3 bits (103), Expect = 7e-05
Identities = 27/51 (52%), Positives = 28/51 (53%), Gaps = 11/51 (21%)
Query: 3 ERGGDRGGFGSGFG-GRGGDRG----------GRGGRGDRGRGGRRRAGGR 42
+RGG RGG G G G G GGDRG GRGG GDRG GR GR
Sbjct: 2 DRGGYRGGRGDGRGRGGGGDRGRGYSGRGDGRGRGGDGDRGYSGRGDGHGR 52
Score = 40.4 bits (93), Expect = 0.001
Identities = 28/56 (50%), Positives = 28/56 (50%), Gaps = 19/56 (33%)
Query: 5 GGDRG------GFGSGFGGRG-------GDRGGRGGRGDRGRG------GRRRAGG 41
GGDRG G G G GG G GD GRGG GDRGRG GR R GG
Sbjct: 19 GGDRGRGYSGRGDGRGRGGDGDRGYSGRGDGHGRGGGGDRGRGYSGRGDGRGRGGG 74
Score = 40.0 bits (92), Expect = 0.001
Identities = 31/63 (49%), Positives = 34/63 (53%), Gaps = 18/63 (28%)
Query: 5 GGDRG----GFGSGFG-GRGGDRG----------GRGGRGDRGRG--GRRRAGGRREEEE 47
GGDRG G G G G G GGDRG GRGG GDRGRG GR R G ++ +
Sbjct: 55 GGDRGRGYSGRGDGRGRGGGGDRGRGYSGRGDGHGRGGGGDRGRGYSGRGR-GFVQDRDG 113
Query: 48 KWV 50
WV
Sbjct: 114 GWV 116
Score = 38.9 bits (89), Expect = 0.003
Identities = 26/49 (53%), Positives = 26/49 (53%), Gaps = 13/49 (26%)
Query: 6 GDRG--GFGSGFG-GRGGDRG----------GRGGRGDRGRGGRRRAGG 41
GDRG G G G G G GGDRG GRGG GDRGRG R G
Sbjct: 39 GDRGYSGRGDGHGRGGGGDRGRGYSGRGDGRGRGGGGDRGRGYSGRGDG 87
>At4g38680 glycine-rich protein 2 (GRP2)
Length = 203
Score = 43.1 bits (100), Expect = 2e-04
Identities = 20/34 (58%), Positives = 22/34 (63%)
Query: 2 AERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGG 35
+E GG GG G G+GG GG GG GG G GRGG
Sbjct: 145 SEGGGGYGGGGGGYGGGGGYGGGGGGYGGGGRGG 178
Score = 40.4 bits (93), Expect = 0.001
Identities = 21/31 (67%), Positives = 21/31 (67%), Gaps = 1/31 (3%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGG 35
GG GG GSG GG GG GG GGRG GRGG
Sbjct: 99 GGRGGGRGSG-GGYGGGGGGYGGRGGGGRGG 128
Score = 40.4 bits (93), Expect = 0.001
Identities = 22/38 (57%), Positives = 22/38 (57%), Gaps = 3/38 (7%)
Query: 6 GDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRR 43
G RGGFG GGRGG RG GG G G G R GG R
Sbjct: 92 GGRGGFG---GGRGGGRGSGGGYGGGGGGYGGRGGGGR 126
Score = 40.0 bits (92), Expect = 0.001
Identities = 20/33 (60%), Positives = 20/33 (60%)
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGR 36
RGG GG G G G GG GG GG G RG GGR
Sbjct: 94 RGGFGGGRGGGRGSGGGYGGGGGGYGGRGGGGR 126
Score = 37.7 bits (86), Expect = 0.007
Identities = 18/33 (54%), Positives = 19/33 (57%)
Query: 9 GGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG+G G GG GG G GG G G GGR GG
Sbjct: 149 GGYGGGGGGYGGGGGYGGGGGGYGGGGRGGGGG 181
Score = 35.8 bits (81), Expect = 0.026
Identities = 19/33 (57%), Positives = 21/33 (63%), Gaps = 3/33 (9%)
Query: 5 GGDRGGFGSG--FGGRGGDRGGRGGRGDRGRGG 35
GG GG+G G +GG GG GG GGRG G GG
Sbjct: 152 GGGGGGYGGGGGYGGGGGGYGG-GGRGGGGGGG 183
Score = 33.5 bits (75), Expect = 0.13
Identities = 18/39 (46%), Positives = 19/39 (48%)
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGR 42
+G GG G GG GG RGG G G GG GGR
Sbjct: 83 QGNSGGGSSGGRGGFGGGRGGGRGSGGGYGGGGGGYGGR 121
Score = 31.6 bits (70), Expect = 0.49
Identities = 16/30 (53%), Positives = 16/30 (53%)
Query: 14 GFGGRGGDRGGRGGRGDRGRGGRRRAGGRR 43
G GG GG GG GG G G GG GG R
Sbjct: 147 GGGGYGGGGGGYGGGGGYGGGGGGYGGGGR 176
Score = 31.2 bits (69), Expect = 0.64
Identities = 19/29 (65%), Positives = 20/29 (68%), Gaps = 5/29 (17%)
Query: 4 RGGDRG---GFGSGFGGRGGDRGGRGGRG 29
RGG RG G+G G GG GG RGG GGRG
Sbjct: 101 RGGGRGSGGGYGGGGGGYGG-RGG-GGRG 127
Score = 27.7 bits (60), Expect = 7.1
Identities = 15/39 (38%), Positives = 17/39 (43%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRR 43
GG GG+G G G GG G G+ G R G R
Sbjct: 165 GGGGGGYGGGGRGGGGGGGSCYSCGESGHFARDCTSGGR 203
>At5g59630 glycine-rich protein - like
Length = 253
Score = 41.6 bits (96), Expect = 5e-04
Identities = 24/71 (33%), Positives = 34/71 (47%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVKEGKI 64
GG GG G G GG GG GG GG G G GG GG + + + + + +K+ +
Sbjct: 80 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGKLQLLGLKPYSKKMKKKLV 139
Query: 65 RSLEQIYLHSL 75
EQ+ + L
Sbjct: 140 NGTEQVAIKIL 150
Score = 40.4 bits (93), Expect = 0.001
Identities = 20/37 (54%), Positives = 20/37 (54%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G GG GG GG GG G G GG GG
Sbjct: 68 GGSSGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 104
Score = 39.7 bits (91), Expect = 0.002
Identities = 20/37 (54%), Positives = 20/37 (54%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G GG GG GG GG G G GG GG
Sbjct: 73 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 109
Score = 39.7 bits (91), Expect = 0.002
Identities = 20/37 (54%), Positives = 20/37 (54%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G GG GG GG GG G G GG GG
Sbjct: 72 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 108
Score = 39.7 bits (91), Expect = 0.002
Identities = 20/37 (54%), Positives = 20/37 (54%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G GG GG GG GG G G GG GG
Sbjct: 74 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 110
Score = 39.7 bits (91), Expect = 0.002
Identities = 20/37 (54%), Positives = 20/37 (54%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G GG GG GG GG G G GG GG
Sbjct: 78 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 114
Score = 39.7 bits (91), Expect = 0.002
Identities = 20/37 (54%), Positives = 20/37 (54%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G GG GG GG GG G G GG GG
Sbjct: 76 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 112
Score = 39.7 bits (91), Expect = 0.002
Identities = 20/37 (54%), Positives = 20/37 (54%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G GG GG GG GG G G GG GG
Sbjct: 77 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 113
Score = 39.7 bits (91), Expect = 0.002
Identities = 20/37 (54%), Positives = 20/37 (54%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G GG GG GG GG G G GG GG
Sbjct: 79 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 115
Score = 39.7 bits (91), Expect = 0.002
Identities = 20/37 (54%), Positives = 20/37 (54%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G GG GG GG GG G G GG GG
Sbjct: 75 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 111
Score = 37.7 bits (86), Expect = 0.007
Identities = 19/37 (51%), Positives = 19/37 (51%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
G GG G G GG GG GG GG G G GG GG
Sbjct: 69 GSSGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 105
Score = 33.5 bits (75), Expect = 0.13
Identities = 17/39 (43%), Positives = 18/39 (45%)
Query: 3 ERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
+ G D G GG GG GG GG G G GG GG
Sbjct: 59 DNGLDSSNSGGSSGGGGGGGGGGGGGGGGGGGGGGGGGG 97
>At5g02530 unknown protein
Length = 292
Score = 41.6 bits (96), Expect = 5e-04
Identities = 24/43 (55%), Positives = 25/43 (57%), Gaps = 1/43 (2%)
Query: 4 RGGDRGGF-GSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREE 45
RG RGGF G GG G RGGRG RGRGGR G R+E
Sbjct: 228 RGRGRGGFMGRPRGGGFGGGNFRGGRGARGRGGRGSGGRGRDE 270
Score = 28.5 bits (62), Expect = 4.2
Identities = 15/37 (40%), Positives = 15/37 (40%)
Query: 6 GDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGR 42
G RG G G G G RGG G R R G R
Sbjct: 21 GSRGRGGIGGGNNTGGRGGSGSNSGPSRRFANRVGAR 57
>At3g23450 hypothetical protein
Length = 452
Score = 41.6 bits (96), Expect = 5e-04
Identities = 21/37 (56%), Positives = 21/37 (56%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G GFGG GG GG GG G G GG GG
Sbjct: 66 GGFGGGAGGGFGGGGGGGGGGGGGGGGGFGGGGGFGG 102
Score = 39.3 bits (90), Expect = 0.002
Identities = 20/37 (54%), Positives = 20/37 (54%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GGFG G GG G GG GG G G GG GG
Sbjct: 62 GGAGGGFGGGAGGGFGGGGGGGGGGGGGGGGGFGGGG 98
Score = 37.7 bits (86), Expect = 0.007
Identities = 19/36 (52%), Positives = 19/36 (52%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAG 40
GG GG G GFGG GG GG GG G GG G
Sbjct: 84 GGGGGGGGGGFGGGGGFGGGHGGGVGGGVGGGHGGG 119
Score = 37.7 bits (86), Expect = 0.007
Identities = 21/39 (53%), Positives = 21/39 (53%), Gaps = 2/39 (5%)
Query: 5 GGDRGGFGSGFGG--RGGDRGGRGGRGDRGRGGRRRAGG 41
GG GGFG GFGG GG GG GG G GG GG
Sbjct: 42 GGKGGGFGGGFGGGAGGGVGGGAGGGFGGGAGGGFGGGG 80
Score = 36.6 bits (83), Expect = 0.015
Identities = 19/36 (52%), Positives = 19/36 (52%)
Query: 6 GDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
G GG G GFG GG GG GG G G GG GG
Sbjct: 359 GKGGGIGGGFGKGGGIGGGIGGGGGFGGGGGFGKGG 394
Score = 36.2 bits (82), Expect = 0.020
Identities = 20/37 (54%), Positives = 20/37 (54%), Gaps = 2/37 (5%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G GFGG G GG GG G G GG GG
Sbjct: 58 GGVGGGAGGGFGGGAG--GGFGGGGGGGGGGGGGGGG 92
Score = 35.0 bits (79), Expect = 0.045
Identities = 19/37 (51%), Positives = 19/37 (51%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G GG GG GG GG G G G GG
Sbjct: 74 GGFGGGGGGGGGGGGGGGGGFGGGGGFGGGHGGGVGG 110
Score = 35.0 bits (79), Expect = 0.045
Identities = 21/37 (56%), Positives = 22/37 (58%), Gaps = 3/37 (8%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GGFG G GG GG GG GG G G GG + GG
Sbjct: 362 GGIGGGFGKG-GGIGGGIGGGGGFG--GGGGFGKGGG 395
Score = 35.0 bits (79), Expect = 0.045
Identities = 20/38 (52%), Positives = 20/38 (52%), Gaps = 1/38 (2%)
Query: 5 GGDRGGFGSGFGGRGGD-RGGRGGRGDRGRGGRRRAGG 41
GG GGFG G GG GG GG GG G G G GG
Sbjct: 70 GGAGGGFGGGGGGGGGGGGGGGGGFGGGGGFGGGHGGG 107
Score = 34.7 bits (78), Expect = 0.058
Identities = 17/30 (56%), Positives = 17/30 (56%)
Query: 6 GDRGGFGSGFGGRGGDRGGRGGRGDRGRGG 35
G GGFG G G GG GG GG G G GG
Sbjct: 419 GGGGGFGKGGGFGGGGFGGGGGGGGGGGGG 448
Score = 34.3 bits (77), Expect = 0.076
Identities = 20/39 (51%), Positives = 20/39 (51%), Gaps = 2/39 (5%)
Query: 5 GGDRGGFGSGFGG--RGGDRGGRGGRGDRGRGGRRRAGG 41
GG GGFG G GG GG GG GG G GG GG
Sbjct: 46 GGFGGGFGGGAGGGVGGGAGGGFGGGAGGGFGGGGGGGG 84
Score = 33.9 bits (76), Expect = 0.099
Identities = 18/37 (48%), Positives = 19/37 (50%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG G G G GG G GG GG G G+GG GG
Sbjct: 386 GGGGFGKGGGIGGGIGKGGGFGGGGGFGKGGGIGGGG 422
Score = 33.9 bits (76), Expect = 0.099
Identities = 21/41 (51%), Positives = 22/41 (53%), Gaps = 4/41 (9%)
Query: 5 GGDRGGFGSG--FGGRGG--DRGGRGGRGDRGRGGRRRAGG 41
GG GG G G FGG GG GG GG G G+GG GG
Sbjct: 394 GGIGGGIGKGGGFGGGGGFGKGGGIGGGGGFGKGGGFGGGG 434
Score = 33.5 bits (75), Expect = 0.13
Identities = 18/37 (48%), Positives = 21/37 (56%), Gaps = 1/37 (2%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G GFG +GG GG G+G GG + GG
Sbjct: 114 GGHGGGVGGGFG-KGGGIGGGIGKGGGVGGGIGKGGG 149
Score = 33.1 bits (74), Expect = 0.17
Identities = 19/42 (45%), Positives = 22/42 (52%), Gaps = 5/42 (11%)
Query: 5 GGDRGGFGSGFG-----GRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GGFG G G G+GG GG G+G GG + GG
Sbjct: 208 GGVGGGFGKGGGVGGGIGKGGGVGGGFGKGGGVGGGIGKGGG 249
Score = 33.1 bits (74), Expect = 0.17
Identities = 18/37 (48%), Positives = 20/37 (53%), Gaps = 1/37 (2%)
Query: 6 GDRGGFGSGFGGRGGDRGGRG-GRGDRGRGGRRRAGG 41
G GG G G GG GG GG G G+G GG + GG
Sbjct: 369 GKGGGIGGGIGGGGGFGGGGGFGKGGGIGGGIGKGGG 405
Score = 32.7 bits (73), Expect = 0.22
Identities = 18/37 (48%), Positives = 18/37 (48%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G GG GG GG G G G G GG
Sbjct: 78 GGGGGGGGGGGGGGGGFGGGGGFGGGHGGGVGGGVGG 114
Score = 32.7 bits (73), Expect = 0.22
Identities = 19/37 (51%), Positives = 20/37 (53%), Gaps = 1/37 (2%)
Query: 6 GDRGGFGSGFG-GRGGDRGGRGGRGDRGRGGRRRAGG 41
G GGFG G G G+GG GG GG G G G GG
Sbjct: 401 GKGGGFGGGGGFGKGGGIGGGGGFGKGGGFGGGGFGG 437
Score = 32.3 bits (72), Expect = 0.29
Identities = 19/37 (51%), Positives = 20/37 (53%), Gaps = 1/37 (2%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G GFGG GG G G G G+GG GG
Sbjct: 375 GGGIGG-GGGFGGGGGFGKGGGIGGGIGKGGGFGGGG 410
Score = 32.3 bits (72), Expect = 0.29
Identities = 19/42 (45%), Positives = 22/42 (52%), Gaps = 5/42 (11%)
Query: 5 GGDRGGFGSGFG-----GRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GGFG G G G+GG GG G+G GG + GG
Sbjct: 118 GGVGGGFGKGGGIGGGIGKGGGVGGGIGKGGGIGGGIGKGGG 159
Score = 32.3 bits (72), Expect = 0.29
Identities = 19/37 (51%), Positives = 19/37 (51%), Gaps = 1/37 (2%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GGFG G GG GG GG G G G G GG
Sbjct: 88 GGGGGGFGGG-GGFGGGHGGGVGGGVGGGHGGGVGGG 123
Score = 32.3 bits (72), Expect = 0.29
Identities = 19/42 (45%), Positives = 22/42 (52%), Gaps = 5/42 (11%)
Query: 5 GGDRGGFGSGFG-----GRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GGFG G G G+GG GG G+G GG + GG
Sbjct: 228 GGVGGGFGKGGGVGGGIGKGGGIGGGIGKGGGIGGGIGKGGG 269
Score = 32.0 bits (71), Expect = 0.38
Identities = 19/37 (51%), Positives = 19/37 (51%), Gaps = 1/37 (2%)
Query: 6 GDRGGFGSGFG-GRGGDRGGRGGRGDRGRGGRRRAGG 41
G GGFG G G G GG G GG G G GG GG
Sbjct: 407 GGGGGFGKGGGIGGGGGFGKGGGFGGGGFGGGGGGGG 443
Score = 32.0 bits (71), Expect = 0.38
Identities = 16/31 (51%), Positives = 17/31 (54%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGG 35
GG G G G GG G GG GG G G+GG
Sbjct: 322 GGGGFGKGGGIGGGIGKGGGIGGGGGFGKGG 352
Score = 31.6 bits (70), Expect = 0.49
Identities = 19/38 (50%), Positives = 20/38 (52%), Gaps = 1/38 (2%)
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
+GG GG G GFG GG GG G G G GG GG
Sbjct: 316 KGGGIGG-GGGFGKGGGIGGGIGKGGGIGGGGGFGKGG 352
Score = 31.2 bits (69), Expect = 0.64
Identities = 18/36 (50%), Positives = 19/36 (52%), Gaps = 1/36 (2%)
Query: 6 GDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
G GG G GFG GG GG G G G GG + GG
Sbjct: 225 GKGGGVGGGFGKGGGVGGGIGKGGGIG-GGIGKGGG 259
Score = 31.2 bits (69), Expect = 0.64
Identities = 19/39 (48%), Positives = 19/39 (48%), Gaps = 2/39 (5%)
Query: 5 GGDRGGFGSGFGGRG--GDRGGRGGRGDRGRGGRRRAGG 41
GG G G G GG G G GG GG G G GG GG
Sbjct: 408 GGGGFGKGGGIGGGGGFGKGGGFGGGGFGGGGGGGGGGG 446
Score = 31.2 bits (69), Expect = 0.64
Identities = 17/36 (47%), Positives = 17/36 (47%)
Query: 6 GDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
G GG G G G GG GG G G G GG GG
Sbjct: 295 GKGGGIGGGIGKGGGIGGGIGKGGGIGGGGGFGKGG 330
Score = 31.2 bits (69), Expect = 0.64
Identities = 17/36 (47%), Positives = 20/36 (55%), Gaps = 1/36 (2%)
Query: 6 GDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
G GG G GF G+GG GG G+G GG + GG
Sbjct: 205 GKGGGVGGGF-GKGGGVGGGIGKGGGVGGGFGKGGG 239
Score = 31.2 bits (69), Expect = 0.64
Identities = 18/37 (48%), Positives = 18/37 (48%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG G G GFG GG GG G G G GG GG
Sbjct: 380 GGGGFGGGGGFGKGGGIGGGIGKGGGFGGGGGFGKGG 416
Score = 30.8 bits (68), Expect = 0.84
Identities = 19/37 (51%), Positives = 20/37 (53%), Gaps = 2/37 (5%)
Query: 6 GDRGGFGSGFG-GRGGDRGGRGGRGDRGRGGRRRAGG 41
G GG G G G G+GG GG GG G G GG GG
Sbjct: 413 GKGGGIGGGGGFGKGGGFGG-GGFGGGGGGGGGGGGG 448
Score = 30.8 bits (68), Expect = 0.84
Identities = 17/33 (51%), Positives = 18/33 (54%), Gaps = 2/33 (6%)
Query: 5 GGDRGGF--GSGFGGRGGDRGGRGGRGDRGRGG 35
GG GG G G GG G GG GG G G+GG
Sbjct: 298 GGIGGGIGKGGGIGGGIGKGGGIGGGGGFGKGG 330
Score = 30.8 bits (68), Expect = 0.84
Identities = 18/42 (42%), Positives = 21/42 (49%), Gaps = 5/42 (11%)
Query: 5 GGDRGGFGSGFG-----GRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G G G+GG GG G+G GG + GG
Sbjct: 188 GGIGGGIGKGGGIGGGIGKGGGVGGGFGKGGGVGGGIGKGGG 229
Score = 30.4 bits (67), Expect = 1.1
Identities = 18/42 (42%), Positives = 21/42 (49%), Gaps = 5/42 (11%)
Query: 5 GGDRGGFGSGFG-----GRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G G G+GG GG G+G GG + GG
Sbjct: 128 GGIGGGIGKGGGVGGGIGKGGGIGGGIGKGGGVGGGIGKGGG 169
Score = 30.4 bits (67), Expect = 1.1
Identities = 17/30 (56%), Positives = 17/30 (56%), Gaps = 1/30 (3%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRG 34
GG G G GFGG GG GG GG G G G
Sbjct: 420 GGGGFGKGGGFGG-GGFGGGGGGGGGGGGG 448
Score = 30.4 bits (67), Expect = 1.1
Identities = 18/42 (42%), Positives = 21/42 (49%), Gaps = 5/42 (11%)
Query: 5 GGDRGGFGSGFG-----GRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G G G+GG GG G+G GG + GG
Sbjct: 178 GGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGVGGGFGKGGG 219
Score = 30.4 bits (67), Expect = 1.1
Identities = 18/34 (52%), Positives = 19/34 (54%), Gaps = 4/34 (11%)
Query: 6 GDRGGFGSGFGG--RGGDRGGRGG--RGDRGRGG 35
G GGFG G GG GG GG GG G G+GG
Sbjct: 95 GGGGGFGGGHGGGVGGGVGGGHGGGVGGGFGKGG 128
Score = 30.0 bits (66), Expect = 1.4
Identities = 18/42 (42%), Positives = 21/42 (49%), Gaps = 5/42 (11%)
Query: 5 GGDRGGFGSGFG-----GRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G G G+GG GG G+G GG + GG
Sbjct: 138 GGVGGGIGKGGGIGGGIGKGGGVGGGIGKGGGIGGGIGKGGG 179
Score = 30.0 bits (66), Expect = 1.4
Identities = 18/42 (42%), Positives = 21/42 (49%), Gaps = 5/42 (11%)
Query: 5 GGDRGGFGSGFG-----GRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G G G+GG GG G+G GG + GG
Sbjct: 168 GGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGG 209
Score = 30.0 bits (66), Expect = 1.4
Identities = 18/42 (42%), Positives = 21/42 (49%), Gaps = 5/42 (11%)
Query: 5 GGDRGGFGSGFG-----GRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G G G+GG GG G+G GG + GG
Sbjct: 258 GGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGG 299
Score = 30.0 bits (66), Expect = 1.4
Identities = 18/42 (42%), Positives = 21/42 (49%), Gaps = 5/42 (11%)
Query: 5 GGDRGGFGSGFG-----GRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G G G+GG GG G+G GG + GG
Sbjct: 238 GGVGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGG 279
Score = 30.0 bits (66), Expect = 1.4
Identities = 18/42 (42%), Positives = 21/42 (49%), Gaps = 5/42 (11%)
Query: 5 GGDRGGFGSGFG-----GRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G G G+GG GG G+G GG + GG
Sbjct: 148 GGIGGGIGKGGGVGGGIGKGGGIGGGIGKGGGIGGGIGKGGG 189
Score = 30.0 bits (66), Expect = 1.4
Identities = 17/37 (45%), Positives = 19/37 (50%), Gaps = 1/37 (2%)
Query: 6 GDRGGFGSGFGGRGGDRGGRG-GRGDRGRGGRRRAGG 41
G GG G G G GG GG G G+G GG + GG
Sbjct: 327 GKGGGIGGGIGKGGGIGGGGGFGKGGGIGGGIGKGGG 363
Score = 30.0 bits (66), Expect = 1.4
Identities = 18/42 (42%), Positives = 21/42 (49%), Gaps = 5/42 (11%)
Query: 5 GGDRGGFGSGFG-----GRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G G G+GG GG G+G GG + GG
Sbjct: 268 GGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGG 309
Score = 30.0 bits (66), Expect = 1.4
Identities = 18/42 (42%), Positives = 21/42 (49%), Gaps = 5/42 (11%)
Query: 5 GGDRGGFGSGFG-----GRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G G G+GG GG G+G GG + GG
Sbjct: 158 GGVGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGG 199
Score = 30.0 bits (66), Expect = 1.4
Identities = 18/42 (42%), Positives = 21/42 (49%), Gaps = 5/42 (11%)
Query: 5 GGDRGGFGSGFG-----GRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G G G+GG GG G+G GG + GG
Sbjct: 278 GGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGG 319
Score = 30.0 bits (66), Expect = 1.4
Identities = 18/42 (42%), Positives = 20/42 (46%), Gaps = 5/42 (11%)
Query: 5 GGDRGGFGSGF-----GGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G GG GG GG G+G GG + GG
Sbjct: 98 GGFGGGHGGGVGGGVGGGHGGGVGGGFGKGGGIGGGIGKGGG 139
Score = 30.0 bits (66), Expect = 1.4
Identities = 18/42 (42%), Positives = 21/42 (49%), Gaps = 5/42 (11%)
Query: 5 GGDRGGFGSGFG-----GRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G G G+GG GG G+G GG + GG
Sbjct: 248 GGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGG 289
Score = 30.0 bits (66), Expect = 1.4
Identities = 17/37 (45%), Positives = 19/37 (50%), Gaps = 1/37 (2%)
Query: 6 GDRGGFGSGFGGRGGDRGGRG-GRGDRGRGGRRRAGG 41
G GG G G G GG GG G G+G GG + GG
Sbjct: 305 GKGGGIGGGIGKGGGIGGGGGFGKGGGIGGGIGKGGG 341
Score = 29.6 bits (65), Expect = 1.9
Identities = 18/41 (43%), Positives = 20/41 (47%), Gaps = 5/41 (12%)
Query: 6 GDRGGFGSGFG-----GRGGDRGGRGGRGDRGRGGRRRAGG 41
G GGFG G G G+GG GG G+G GG GG
Sbjct: 343 GGGGGFGKGGGIGGGIGKGGGIGGGFGKGGGIGGGIGGGGG 383
Score = 29.3 bits (64), Expect = 2.4
Identities = 17/28 (60%), Positives = 17/28 (60%), Gaps = 3/28 (10%)
Query: 6 GDRGGFGSG-FGGRGGDRGGRGGRGDRG 32
G GGFG G FGG GG GG GG G G
Sbjct: 425 GKGGGFGGGGFGGGGG--GGGGGGGGIG 450
Score = 29.3 bits (64), Expect = 2.4
Identities = 17/37 (45%), Positives = 18/37 (47%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G G GG G G G G+GG GG
Sbjct: 288 GGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGG 324
Score = 29.3 bits (64), Expect = 2.4
Identities = 18/44 (40%), Positives = 21/44 (46%), Gaps = 7/44 (15%)
Query: 5 GGDRGGFGSGFG-------GRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G G G+GG GG G+G GG + GG
Sbjct: 330 GGIGGGIGKGGGIGGGGGFGKGGGIGGGIGKGGGIGGGFGKGGG 373
Score = 29.3 bits (64), Expect = 2.4
Identities = 18/39 (46%), Positives = 19/39 (48%), Gaps = 2/39 (5%)
Query: 5 GGDRGGFGSG--FGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G GG GG G G G G+GG GG
Sbjct: 308 GGIGGGIGKGGGIGGGGGFGKGGGIGGGIGKGGGIGGGG 346
Score = 28.1 bits (61), Expect = 5.5
Identities = 13/23 (56%), Positives = 14/23 (60%)
Query: 4 RGGDRGGFGSGFGGRGGDRGGRG 26
+GG GG G G GG GG GG G
Sbjct: 426 KGGGFGGGGFGGGGGGGGGGGGG 448
Score = 28.1 bits (61), Expect = 5.5
Identities = 18/37 (48%), Positives = 18/37 (48%), Gaps = 1/37 (2%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G GG GG G GG G GG GG
Sbjct: 352 GGIGGGIGKG-GGIGGGFGKGGGIGGGIGGGGGFGGG 387
>At2g05580 unknown protein
Length = 302
Score = 41.6 bits (96), Expect = 5e-04
Identities = 18/40 (45%), Positives = 26/40 (65%)
Query: 3 ERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGR 42
++GG GG G GG GG +GG+ G G G+GG++ GG+
Sbjct: 129 QKGGGGGGQGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGQ 168
Score = 41.6 bits (96), Expect = 5e-04
Identities = 20/47 (42%), Positives = 30/47 (63%), Gaps = 2/47 (4%)
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGR--GDRGRGGRRRAGGRREEEEK 48
+GG +GG G G GG+ G GG+GG+ G G+GG++ GG + +K
Sbjct: 95 QGGQKGGGGGGQGGQKGGGGGQGGQKGGGGGQGGQKGGGGGGQGGQK 141
Score = 41.2 bits (95), Expect = 6e-04
Identities = 20/41 (48%), Positives = 29/41 (69%), Gaps = 2/41 (4%)
Query: 4 RGGDRGGFGSGFGG-RGGDRGGRGG-RGDRGRGGRRRAGGR 42
+GG +GG G G GG +GG GG+GG +G G+GG++ GG+
Sbjct: 137 QGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGQGGQKGGGGQ 177
Score = 40.8 bits (94), Expect = 8e-04
Identities = 23/43 (53%), Positives = 25/43 (57%), Gaps = 5/43 (11%)
Query: 4 RGGDRGGFGSGF---GGRGGDRGGRGGRGDR--GRGGRRRAGG 41
+GG +GG G G GGRGG GGRGG G G GG R GG
Sbjct: 255 QGGHKGGGGGGHVGGGGRGGGGGGRGGGGSGGGGGGGSGRGGG 297
Score = 40.8 bits (94), Expect = 8e-04
Identities = 18/38 (47%), Positives = 24/38 (62%)
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
+GG +GG G G GG +GG+ G G RG+GG + GG
Sbjct: 168 QGGQKGGGGQGGQKGGGGQGGQKGGGGRGQGGMKGGGG 205
Score = 40.0 bits (92), Expect = 0.001
Identities = 20/41 (48%), Positives = 28/41 (67%), Gaps = 3/41 (7%)
Query: 4 RGGDRGGFGSGFGG-RGGDRGGRGGR--GDRGRGGRRRAGG 41
+GG +GG G G GG +GG GG+GG+ G G+GG++ GG
Sbjct: 84 QGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGGQGGQKGGGG 124
Score = 39.7 bits (91), Expect = 0.002
Identities = 17/39 (43%), Positives = 25/39 (63%)
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGR 42
+GG +GG G G GG+ G G G +G G+GG++ GG+
Sbjct: 148 QGGQKGGGGGGQGGQKGGGGQGGQKGGGGQGGQKGGGGQ 186
Score = 39.7 bits (91), Expect = 0.002
Identities = 19/41 (46%), Positives = 25/41 (60%), Gaps = 4/41 (9%)
Query: 4 RGGDRGGFGSGF----GGRGGDRGGRGGRGDRGRGGRRRAG 40
+GG +GG G G GG+GG +GG G G +G GGR + G
Sbjct: 159 QGGQKGGGGQGGQKGGGGQGGQKGGGGQGGQKGGGGRGQGG 199
Score = 39.3 bits (90), Expect = 0.002
Identities = 19/37 (51%), Positives = 21/37 (56%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG +GG G G +GG GG G G RG GG R GG
Sbjct: 246 GGHKGGGGGQGGHKGGGGGGHVGGGGRGGGGGGRGGG 282
Score = 38.9 bits (89), Expect = 0.003
Identities = 19/44 (43%), Positives = 27/44 (61%), Gaps = 4/44 (9%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEK 48
GG +GG G GG+GG +GG GG G+GG++ GG + +K
Sbjct: 113 GGGQGGQKGGGGGQGGQKGGGGG----GQGGQKGGGGGGQGGQK 152
Score = 38.5 bits (88), Expect = 0.004
Identities = 19/48 (39%), Positives = 29/48 (59%), Gaps = 3/48 (6%)
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGR---GDRGRGGRRRAGGRREEEEK 48
+GG +GG G G +GG GG+GG+ G G+GG++ GG + +K
Sbjct: 116 QGGQKGGGGGQGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGGGQGGQK 163
Score = 38.5 bits (88), Expect = 0.004
Identities = 20/45 (44%), Positives = 28/45 (61%), Gaps = 5/45 (11%)
Query: 3 ERGGDRGGFGS--GFGGRGGDRGGRGGRGDRG---RGGRRRAGGR 42
++GG GG G G GG+GG +GG G G +G +GG++ GGR
Sbjct: 151 QKGGGGGGQGGQKGGGGQGGQKGGGGQGGQKGGGGQGGQKGGGGR 195
Score = 38.1 bits (87), Expect = 0.005
Identities = 22/41 (53%), Positives = 25/41 (60%), Gaps = 3/41 (7%)
Query: 5 GGDRGGFG--SGFGGRGGDRGGRGGRGDRGRGGRRRAGGRR 43
GG +GG G G GG+GG +GG GG G G GGR GG R
Sbjct: 240 GGGQGGGGHKGGGGGQGGHKGGGGG-GHVGGGGRGGGGGGR 279
Score = 38.1 bits (87), Expect = 0.005
Identities = 18/37 (48%), Positives = 21/37 (56%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG RG G GG GG G +GG G G+GG + GG
Sbjct: 192 GGGRGQGGMKGGGGGGQGGHKGGGGGGGQGGHKGGGG 228
Score = 38.1 bits (87), Expect = 0.005
Identities = 20/39 (51%), Positives = 25/39 (63%), Gaps = 2/39 (5%)
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGR-GDRGRGGRRRAGG 41
+GG +GG G G GG GG +GG GG+ G +G GG GG
Sbjct: 232 QGGHKGGGGGGQGG-GGHKGGGGGQGGHKGGGGGGHVGG 269
Score = 37.7 bits (86), Expect = 0.007
Identities = 18/39 (46%), Positives = 24/39 (61%), Gaps = 2/39 (5%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGR--RRAGG 41
GG G G G GG+GG +GG G G +G GG+ ++ GG
Sbjct: 146 GGQGGQKGGGGGGQGGQKGGGGQGGQKGGGGQGGQKGGG 184
Score = 37.7 bits (86), Expect = 0.007
Identities = 18/31 (58%), Positives = 18/31 (58%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGG 35
GG RGG G G GG G GG GG G G GG
Sbjct: 269 GGGRGGGGGGRGGGGSGGGGGGGSGRGGGGG 299
Score = 37.4 bits (85), Expect = 0.009
Identities = 19/44 (43%), Positives = 24/44 (54%), Gaps = 6/44 (13%)
Query: 4 RGGDRGGFGSGFGGR------GGDRGGRGGRGDRGRGGRRRAGG 41
+GG +GG G G GG GG G +GG G G+GG + GG
Sbjct: 197 QGGMKGGGGGGQGGHKGGGGGGGQGGHKGGGGGGGQGGHKGGGG 240
Score = 37.0 bits (84), Expect = 0.012
Identities = 18/43 (41%), Positives = 26/43 (59%), Gaps = 4/43 (9%)
Query: 3 ERGGDRGGFGSGFGGR----GGDRGGRGGRGDRGRGGRRRAGG 41
E ++GG G G GG+ GG +GG+ G G G+GG++ GG
Sbjct: 72 ESKNNQGGIGRGQGGQKGGGGGGQGGQKGGGGGGQGGQKGGGG 114
Score = 36.6 bits (83), Expect = 0.015
Identities = 20/45 (44%), Positives = 25/45 (55%), Gaps = 6/45 (13%)
Query: 5 GGDRGGF--GSGFGGRGGDRGGRGGRGDRGR----GGRRRAGGRR 43
GG +GG G G GG+GG +GG GG G G GG + GG +
Sbjct: 205 GGGQGGHKGGGGGGGQGGHKGGGGGGGQGGHKGGGGGGQGGGGHK 249
Score = 36.2 bits (82), Expect = 0.020
Identities = 19/39 (48%), Positives = 24/39 (60%), Gaps = 4/39 (10%)
Query: 5 GGDRGGF--GSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG +GG G G GG+GG +GG GG +G GG + GG
Sbjct: 217 GGGQGGHKGGGGGGGQGGHKGGGGG--GQGGGGHKGGGG 253
Score = 35.8 bits (81), Expect = 0.026
Identities = 18/40 (45%), Positives = 23/40 (57%), Gaps = 2/40 (5%)
Query: 4 RGGDRGGFGSGF--GGRGGDRGGRGGRGDRGRGGRRRAGG 41
+GG +GG G G GG G +GG G G G+GG + GG
Sbjct: 177 QGGQKGGGGQGGQKGGGGRGQGGMKGGGGGGQGGHKGGGG 216
Score = 35.8 bits (81), Expect = 0.026
Identities = 19/47 (40%), Positives = 24/47 (50%), Gaps = 8/47 (17%)
Query: 4 RGGDRGGFG--------SGFGGRGGDRGGRGGRGDRGRGGRRRAGGR 42
+GG +GG G G GG+GG +GG GG G G G GG+
Sbjct: 186 QGGQKGGGGRGQGGMKGGGGGGQGGHKGGGGGGGQGGHKGGGGGGGQ 232
Score = 35.4 bits (80), Expect = 0.034
Identities = 19/44 (43%), Positives = 23/44 (52%), Gaps = 6/44 (13%)
Query: 4 RGGDRGGFGSGF------GGRGGDRGGRGGRGDRGRGGRRRAGG 41
+GG +GG G G GG GG +GG G G G+GG GG
Sbjct: 208 QGGHKGGGGGGGQGGHKGGGGGGGQGGHKGGGGGGQGGGGHKGG 251
Score = 35.4 bits (80), Expect = 0.034
Identities = 17/37 (45%), Positives = 20/37 (53%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G GG GG +GG G +G G G + GG
Sbjct: 226 GGGGGGQGGHKGGGGGGQGGGGHKGGGGGQGGHKGGG 262
Score = 35.4 bits (80), Expect = 0.034
Identities = 17/34 (50%), Positives = 22/34 (64%), Gaps = 2/34 (5%)
Query: 4 RGGDRGGFGSG--FGGRGGDRGGRGGRGDRGRGG 35
+GG +GG G G G +GG GG+GG G +G GG
Sbjct: 220 QGGHKGGGGGGGQGGHKGGGGGGQGGGGHKGGGG 253
Score = 33.9 bits (76), Expect = 0.099
Identities = 19/37 (51%), Positives = 20/37 (53%), Gaps = 1/37 (2%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG +GG G GG GGRGG G GRGG GG
Sbjct: 252 GGGQGGHKGGGGGGHVGGGGRGG-GGGGRGGGGSGGG 287
Score = 33.9 bits (76), Expect = 0.099
Identities = 18/32 (56%), Positives = 18/32 (56%), Gaps = 2/32 (6%)
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGG 35
RGG GG G GG GG GG GRG G GG
Sbjct: 272 RGGGGGGRGG--GGSGGGGGGGSGRGGGGGGG 301
Score = 33.1 bits (74), Expect = 0.17
Identities = 19/37 (51%), Positives = 19/37 (51%), Gaps = 1/37 (2%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G GG GG GG G G G GR GG
Sbjct: 264 GGHVGGGGRG-GGGGGRGGGGSGGGGGGGSGRGGGGG 299
Score = 31.2 bits (69), Expect = 0.64
Identities = 17/40 (42%), Positives = 20/40 (49%), Gaps = 2/40 (5%)
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGR--GDRGRGGRRRAGG 41
+GG GG G G GG G GG G G+GG + GG
Sbjct: 224 KGGGGGGGQGGHKGGGGGGQGGGGHKGGGGGQGGHKGGGG 263
>At1g77030 hypothetical protein
Length = 349
Score = 41.6 bits (96), Expect = 5e-04
Identities = 27/51 (52%), Positives = 30/51 (57%), Gaps = 11/51 (21%)
Query: 2 AERGGDRGGFGSGFGGRGGD-RGGRGGRGDRGRGGR---------RRAGGR 42
++RGG RGG G GGRGG RGGRGG D G GGR R+GGR
Sbjct: 202 SKRGG-RGGRGGARGGRGGGARGGRGGSRDFGGGGRDFGSSSDRGGRSGGR 251
Score = 33.9 bits (76), Expect = 0.099
Identities = 28/70 (40%), Positives = 30/70 (42%), Gaps = 29/70 (41%)
Query: 4 RGGDRGGFGSGF-GGRGG---------------DRGGRGGRGDRG----------RGGRR 37
RGG RGG G G GGRGG DRGGR G D G RGG+R
Sbjct: 210 RGGARGGRGGGARGGRGGSRDFGGGGRDFGSSSDRGGRSGGRDFGGRRGGASTSSRGGKR 269
Query: 38 ---RAGGRRE 44
R GG +
Sbjct: 270 GGGRGGGESD 279
Score = 31.6 bits (70), Expect = 0.49
Identities = 16/26 (61%), Positives = 17/26 (64%), Gaps = 1/26 (3%)
Query: 20 GDRGGRGG-RGDRGRGGRRRAGGRRE 44
G RGGRGG RG RG G R GG R+
Sbjct: 205 GGRGGRGGARGGRGGGARGGRGGSRD 230
Score = 30.0 bits (66), Expect = 1.4
Identities = 17/36 (47%), Positives = 20/36 (55%), Gaps = 4/36 (11%)
Query: 2 AERGGDRGGFGSGFGGR--GGDRGGRGGRGDRGRGG 35
++RGG GG FGGR G RGG+ GRGG
Sbjct: 242 SDRGGRSGG--RDFGGRRGGASTSSRGGKRGGGRGG 275
>At1g09175 putative GPI-anchored protein
Length = 150
Score = 41.6 bits (96), Expect = 5e-04
Identities = 19/36 (52%), Positives = 21/36 (57%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAG 40
GG RGG G G GRGG+ G G RG GGR +G
Sbjct: 71 GGGRGGGGGGSSGRGGNSGKGGSRGSSNSGGRGSSG 106
Score = 35.8 bits (81), Expect = 0.026
Identities = 20/39 (51%), Positives = 23/39 (58%), Gaps = 4/39 (10%)
Query: 8 RGGFGSGFGGRGGDRGGRGGR-GDRGRGGRR---RAGGR 42
R G G GGRGG GG GR G+ G+GG R +GGR
Sbjct: 64 RSSGGGGGGGRGGGGGGSSGRGGNSGKGGSRGSSNSGGR 102
Score = 34.3 bits (77), Expect = 0.076
Identities = 18/31 (58%), Positives = 18/31 (58%), Gaps = 1/31 (3%)
Query: 13 SGFGGRGGDRGGRGGRGDRGRGGRRRAGGRR 43
S GG GG RGG GG G GRGG GG R
Sbjct: 65 SSGGGGGGGRGGGGG-GSSGRGGNSGKGGSR 94
Score = 28.1 bits (61), Expect = 5.5
Identities = 15/33 (45%), Positives = 16/33 (48%), Gaps = 1/33 (3%)
Query: 4 RGGDRGGFGS-GFGGRGGDRGGRGGRGDRGRGG 35
+GG RG S G G G GG GG G GG
Sbjct: 90 KGGSRGSSNSGGRGSSGNGNGGTGGGHSVGSGG 122
>At5g58470 RNA/ssDNA-binding protein - like
Length = 422
Score = 41.2 bits (95), Expect = 6e-04
Identities = 23/39 (58%), Positives = 25/39 (63%), Gaps = 2/39 (5%)
Query: 6 GDRGGFGSG-FGGRGGDRGGRGGRGDRGRGGRRRAGGRR 43
G RG G G +GGRGG GG GGRG+RG GG GG R
Sbjct: 37 GGRGASGGGSYGGRGG-YGGGGGRGNRGGGGGGYQGGDR 74
Score = 40.8 bits (94), Expect = 8e-04
Identities = 21/39 (53%), Positives = 23/39 (58%), Gaps = 1/39 (2%)
Query: 6 GDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRRE 44
G RGG+G G GGRG GG GG RGGR GG R+
Sbjct: 48 GGRGGYGGG-GGRGNRGGGGGGYQGGDRGGRGSGGGGRD 85
Score = 40.0 bits (92), Expect = 0.001
Identities = 21/41 (51%), Positives = 23/41 (55%), Gaps = 4/41 (9%)
Query: 5 GGDRGGFGSGFGGRG----GDRGGRGGRGDRGRGGRRRAGG 41
GG GG +G+GGRG G GGRGG G G G R GG
Sbjct: 26 GGGYGGGDAGYGGRGASGGGSYGGRGGYGGGGGRGNRGGGG 66
Score = 37.0 bits (84), Expect = 0.012
Identities = 24/65 (36%), Positives = 25/65 (37%), Gaps = 27/65 (41%)
Query: 4 RGGDRGGFGSGFGGRGGD---------------------------RGGRGGRGDRGRGGR 36
+GGDRGG GSG GGR GD G G DRG GG
Sbjct: 70 QGGDRGGRGSGGGGRDGDWRCPNPSCGNVNFARRVECNKCGALAPSGTSSGANDRGGGGY 129
Query: 37 RRAGG 41
R GG
Sbjct: 130 SRGGG 134
Score = 36.6 bits (83), Expect = 0.015
Identities = 18/37 (48%), Positives = 19/37 (50%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GGD G G G+GG GGRG G GGR GG
Sbjct: 19 GGDGYGGGGGYGGGDAGYGGRGASGGGSYGGRGGYGG 55
Score = 36.2 bits (82), Expect = 0.020
Identities = 20/32 (62%), Positives = 21/32 (65%), Gaps = 2/32 (6%)
Query: 5 GGDRGGFGSGFGG-RGGDRGGRGGRGDRGRGG 35
GG RG G G GG +GGDRGGRG G GR G
Sbjct: 56 GGGRGNRGGGGGGYQGGDRGGRGS-GGGGRDG 86
Score = 36.2 bits (82), Expect = 0.020
Identities = 21/43 (48%), Positives = 23/43 (52%), Gaps = 7/43 (16%)
Query: 3 ERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGG--RRRAGGRR 43
+RGG RGG G G+GG GGDR R D G R GG R
Sbjct: 381 QRGGGRGGGGGGYGGGGGDR-----RRDNYSSGPDRNHHGGNR 418
Score = 34.3 bits (77), Expect = 0.076
Identities = 16/31 (51%), Positives = 17/31 (54%)
Query: 12 GSGFGGRGGDRGGRGGRGDRGRGGRRRAGGR 42
G G+GG GG GG G G RG G GGR
Sbjct: 20 GDGYGGGGGYGGGDAGYGGRGASGGGSYGGR 50
Score = 33.9 bits (76), Expect = 0.099
Identities = 18/34 (52%), Positives = 20/34 (57%), Gaps = 1/34 (2%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRR 38
GG RGG+GS GG RGGR G D G RR+
Sbjct: 222 GGPRGGYGSDAPSTGG-RGGRSGGYDGGSAPRRQ 254
Score = 31.2 bits (69), Expect = 0.64
Identities = 17/41 (41%), Positives = 20/41 (48%)
Query: 1 MAERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
M + G G +GG G GG G GD G GGR +GG
Sbjct: 4 MYNQDGGGGAPIPSYGGDGYGGGGGYGGGDAGYGGRGASGG 44
Score = 30.0 bits (66), Expect = 1.4
Identities = 18/40 (45%), Positives = 18/40 (45%), Gaps = 4/40 (10%)
Query: 7 DRGGFGSGFGG----RGGDRGGRGGRGDRGRGGRRRAGGR 42
DRGG G GG RGG RGGR G R G R
Sbjct: 123 DRGGGGYSRGGGDSDRGGGRGGRNDSGRSYESSRYDGGSR 162
Score = 29.6 bits (65), Expect = 1.9
Identities = 18/38 (47%), Positives = 19/38 (49%), Gaps = 3/38 (7%)
Query: 1 MAERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRR 38
MAE+ R F RGG RGG GG G G RRR
Sbjct: 368 MAEKSAPRA---PTFDQRGGGRGGGGGGYGGGGGDRRR 402
Score = 28.9 bits (63), Expect = 3.2
Identities = 18/35 (51%), Positives = 19/35 (53%), Gaps = 3/35 (8%)
Query: 10 GFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRRE 44
G SG RGG RGG GD RGG R GGR +
Sbjct: 116 GTSSGANDRGGGGYSRGG-GDSDRGGGR--GGRND 147
>At5g04290 glycine-rich protein
Length = 1493
Score = 41.2 bits (95), Expect = 6e-04
Identities = 20/36 (55%), Positives = 21/36 (57%)
Query: 7 DRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGR 42
+ G G GFG R G G RGGR GRGGR GGR
Sbjct: 1327 EHSGGGRGFGERRGGGGFRGGRNQSGRGGRSFDGGR 1362
Score = 33.1 bits (74), Expect = 0.17
Identities = 22/48 (45%), Positives = 26/48 (53%), Gaps = 6/48 (12%)
Query: 2 AERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKW 49
+E+ DRG G GFGGR G G RGGR GRG + G E+ W
Sbjct: 1082 SEQTFDRG--GRGFGGRRGG-GRRGGRDQFGRGS---SFGNSEDPAPW 1123
Score = 30.4 bits (67), Expect = 1.1
Identities = 15/41 (36%), Positives = 21/41 (50%), Gaps = 1/41 (2%)
Query: 9 GGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKW 49
GG G G GG G+ G GG+G+ RGG+ + E +
Sbjct: 398 GGKGEGSGGGKGEGSG-GGKGEGSRGGKGEGSSDFKSESSY 437
Score = 28.1 bits (61), Expect = 5.5
Identities = 19/44 (43%), Positives = 22/44 (49%), Gaps = 9/44 (20%)
Query: 6 GDRGGFGSGFGGRGGDRGG-------RGGRGDRGRGGRRRAGGR 42
G + GS +G + D GG RGGRG GR G R GGR
Sbjct: 1064 GKKDDGGSSWGKK--DEGGYSEQTFDRGGRGFGGRRGGGRRGGR 1105
Score = 27.7 bits (60), Expect = 7.1
Identities = 13/25 (52%), Positives = 16/25 (64%), Gaps = 1/25 (4%)
Query: 6 GDRGGFGSGFGGRGGDRGGRGGRGD 30
G GG G G GG G+ G RGG+G+
Sbjct: 403 GSGGGKGEGSGGGKGE-GSRGGKGE 426
Score = 27.3 bits (59), Expect = 9.3
Identities = 13/30 (43%), Positives = 14/30 (46%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRG 34
GG G G G G G G G RG +G G
Sbjct: 398 GGKGEGSGGGKGEGSGGGKGEGSRGGKGEG 427
>At3g20470 glycine-rich protein, putative
Length = 174
Score = 40.4 bits (93), Expect = 0.001
Identities = 20/37 (54%), Positives = 20/37 (54%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G GFGG G GG GG G G GG GG
Sbjct: 115 GGAGGGSGGGFGGGAGAGGGLGGGGGAGGGGGFGGGG 151
Score = 39.7 bits (91), Expect = 0.002
Identities = 20/37 (54%), Positives = 20/37 (54%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G GFGG G GG GG G G G AGG
Sbjct: 83 GGAGGGAGGGFGGGAGSGGGLGGGGGAGGGFGGGAGG 119
Score = 38.1 bits (87), Expect = 0.005
Identities = 20/37 (54%), Positives = 20/37 (54%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG G G GFGG GG GG GG G G G AGG
Sbjct: 55 GGSGLGGGGGFGGGGGLGGGAGGGGGLGGGAGGGAGG 91
Score = 36.6 bits (83), Expect = 0.015
Identities = 20/39 (51%), Positives = 20/39 (51%), Gaps = 2/39 (5%)
Query: 5 GGDRGGFGSGFGGRG--GDRGGRGGRGDRGRGGRRRAGG 41
GG GGFG G G G G GG GG G G GG GG
Sbjct: 119 GGSGGGFGGGAGAGGGLGGGGGAGGGGGFGGGGGSGIGG 157
Score = 36.2 bits (82), Expect = 0.020
Identities = 18/36 (50%), Positives = 19/36 (52%)
Query: 6 GDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
G GG G G GG GG GG GG G GG +GG
Sbjct: 66 GGGGGLGGGAGGGGGLGGGAGGGAGGGFGGGAGSGG 101
Score = 35.4 bits (80), Expect = 0.034
Identities = 18/36 (50%), Positives = 18/36 (50%)
Query: 6 GDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
G GG G G G GG GG GG G GG AGG
Sbjct: 98 GSGGGLGGGGGAGGGFGGGAGGGSGGGFGGGAGAGG 133
Score = 35.0 bits (79), Expect = 0.045
Identities = 18/37 (48%), Positives = 19/37 (50%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GGFG G G GG GG G G G G +GG
Sbjct: 87 GGAGGGFGGGAGSGGGLGGGGGAGGGFGGGAGGGSGG 123
Score = 33.9 bits (76), Expect = 0.099
Identities = 17/36 (47%), Positives = 17/36 (47%)
Query: 6 GDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
G GG G GFGG G G G G G GG GG
Sbjct: 104 GGGGGAGGGFGGGAGGGSGGGFGGGAGAGGGLGGGG 139
Score = 32.3 bits (72), Expect = 0.29
Identities = 16/31 (51%), Positives = 16/31 (51%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGG 35
GG G G GFGG GG G G G G GG
Sbjct: 137 GGGGAGGGGGFGGGGGSGIGGGFGGGAGAGG 167
Score = 32.0 bits (71), Expect = 0.38
Identities = 20/37 (54%), Positives = 20/37 (54%), Gaps = 2/37 (5%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG GSG GG GG GG GG G GG GG
Sbjct: 50 GGGIGG-GSGLGG-GGGFGGGGGLGGGAGGGGGLGGG 84
Score = 31.2 bits (69), Expect = 0.64
Identities = 20/39 (51%), Positives = 20/39 (51%), Gaps = 4/39 (10%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDR--GRGGRRRAGG 41
GG GGFG G GG G GG GG G GG AGG
Sbjct: 107 GGAGGGFGGGAGG--GSGGGFGGGAGAGGGLGGGGGAGG 143
Score = 30.8 bits (68), Expect = 0.84
Identities = 18/40 (45%), Positives = 18/40 (45%), Gaps = 3/40 (7%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGG---RGDRGRGGRRRAGG 41
GG GG G GG GG GG G G G GG GG
Sbjct: 68 GGGLGGGAGGGGGLGGGAGGGAGGGFGGGAGSGGGLGGGG 107
Score = 30.8 bits (68), Expect = 0.84
Identities = 18/37 (48%), Positives = 18/37 (48%), Gaps = 1/37 (2%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G GG G GG G G G GG GG
Sbjct: 44 GGGLGG-GGGIGGGSGLGGGGGFGGGGGLGGGAGGGG 79
Score = 30.8 bits (68), Expect = 0.84
Identities = 17/37 (45%), Positives = 17/37 (45%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G GG GG GG G G GG GG
Sbjct: 72 GGGAGGGGGLGGGAGGGAGGGFGGGAGSGGGLGGGGG 108
Score = 30.0 bits (66), Expect = 1.4
Identities = 19/37 (51%), Positives = 19/37 (51%), Gaps = 2/37 (5%)
Query: 6 GDRGGFGSGFGG-RGGDRGGRGGRGDRGRGGRRRAGG 41
G GG G G GG GG GG G G G GG AGG
Sbjct: 76 GGGGGLGGGAGGGAGGGFGGGAGSGG-GLGGGGGAGG 111
Score = 30.0 bits (66), Expect = 1.4
Identities = 13/23 (56%), Positives = 13/23 (56%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGG 27
GG G G GFGG G GG GG
Sbjct: 149 GGGGSGIGGGFGGGAGAGGGFGG 171
Score = 29.6 bits (65), Expect = 1.9
Identities = 19/37 (51%), Positives = 20/37 (53%), Gaps = 2/37 (5%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G+G GG G GG G G G GG AGG
Sbjct: 133 GGLGGGGGAGGGGGFGGGGGSGIGG--GFGGGAGAGG 167
Score = 29.6 bits (65), Expect = 1.9
Identities = 17/36 (47%), Positives = 18/36 (49%), Gaps = 1/36 (2%)
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAG 40
GG GG G+G G GG GG G G G GG G
Sbjct: 78 GGGLGG-GAGGGAGGGFGGGAGSGGGLGGGGGAGGG 112
Score = 27.7 bits (60), Expect = 7.1
Identities = 16/31 (51%), Positives = 16/31 (51%), Gaps = 1/31 (3%)
Query: 6 GDRGGFGSGFG-GRGGDRGGRGGRGDRGRGG 35
G GGFG G G G GG GG G G GG
Sbjct: 142 GGGGGFGGGGGSGIGGGFGGGAGAGGGFGGG 172
>At2g35920 ATP-dependent RNA helicase A like protein
Length = 995
Score = 40.4 bits (93), Expect = 0.001
Identities = 21/43 (48%), Positives = 23/43 (52%)
Query: 16 GGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRL 58
GGR G G RG RG GGR GG R E+ W PV + RL
Sbjct: 10 GGRRGGGHSSGRRGGRGGGGRGGGGGGRGEQRWWDPVWRAERL 52
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.140 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,614,296
Number of Sequences: 26719
Number of extensions: 311863
Number of successful extensions: 6929
Number of sequences better than 10.0: 234
Number of HSP's better than 10.0 without gapping: 192
Number of HSP's successfully gapped in prelim test: 42
Number of HSP's that attempted gapping in prelim test: 1526
Number of HSP's gapped (non-prelim): 2633
length of query: 279
length of database: 11,318,596
effective HSP length: 98
effective length of query: 181
effective length of database: 8,700,134
effective search space: 1574724254
effective search space used: 1574724254
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC140104.12


