

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140027.12 + phase: 0 /pseudo
(526 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
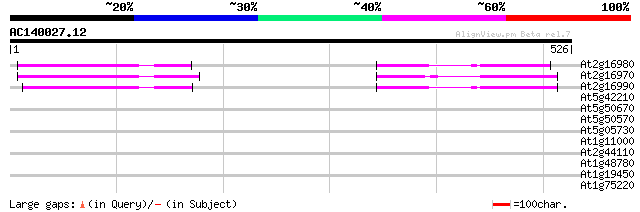
Score E
Sequences producing significant alignments: (bits) Value
At2g16980 putative tetracycline transporter protein 103 2e-22
At2g16970 putative tetracycline transporter protein 92 9e-19
At2g16990 putative tetracycline transporter protein 90 3e-18
At5g42210 putative protein 41 0.001
At5g50670 putative protein 31 1.5
At5g50570 putative protein 31 1.5
At5g05730 anthranilate synthase component I-1 precursor (sp|P32068) 30 3.3
At1g11000 membrane protein Mlo4 30 3.3
At2g44110 similar to Mlo proteins from H. vulgare 30 4.3
At1g48780 hypothetical protein 30 4.3
At1g19450 integral membrane protein, putative 30 4.3
At1g75220 integral membrane protein, putative 29 7.3
>At2g16980 putative tetracycline transporter protein
Length = 415
Score = 103 bits (258), Expect = 2e-22
Identities = 58/164 (35%), Positives = 94/164 (56%), Gaps = 15/164 (9%)
Query: 8 FELKPLFHLLLPLSIHWIAEAMTVSVLVDVTTTALCPQ-QSSCSKAIYINGLQQTITGIF 66
F L L HLL+ + + +AE + V+ DVT A+C SCS A+Y+ G+QQ G+
Sbjct: 4 FRLGELRHLLVTVFLSGLAEYLIRPVMTDVTVAAVCSGLDDSCSLAVYLTGVQQVTVGMG 63
Query: 67 KMAVLPLLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYYVLRTFSHIISQG 126
M ++P++G LSD +G K +L L + S++P A+L + + F YA+YV++T
Sbjct: 64 TMVMMPVIGNLSDRYGIKAMLTLPMCLSVLPPAILGYRRDTNFFYAFYVIKTLFD----- 118
Query: 127 SIFCISVAYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFL 170
+A VH +KR+++F + G+SS + V A++ ARFL
Sbjct: 119 ---------MAKNVHGTKRISMFGILAGVSSISGVCASLSARFL 153
Score = 77.0 bits (188), Expect = 2e-14
Identities = 46/163 (28%), Positives = 77/163 (47%), Gaps = 41/163 (25%)
Query: 345 QIVLLPILNPLVGEKVILCSALLASIAYAWLSGLAWAPWVSKIVTFYINLIQIYMFIWLA 404
Q+ +LP L+ +GE+ +L + LL A +AW+PWV +T +
Sbjct: 264 QLFILPTLSSTIGERKVLSTGLLMEFFNATCLSVAWSPWVPYAMTMLVP----------- 312
Query: 405 ENFITYISIMKVIEFNPLWQVPYLGGSFGIIYILEKPATYGIISKASSSTNQGKAQTFIA 464
G ++++ P+ GI S+ S+ QGK Q I+
Sbjct: 313 ----------------------------GAMFVM--PSVCGIASRQVGSSEQGKVQGCIS 342
Query: 465 GANSISGLLSPIVMSPLTSLFLSSDAPFECKGFSIICASVCMV 507
G + + +++P V SPLT+LFLS +APF GFSI+C ++ ++
Sbjct: 343 GVRAFAQVVAPFVYSPLTALFLSENAPFYFPGFSILCIAISLM 385
>At2g16970 putative tetracycline transporter protein
Length = 414
Score = 91.7 bits (226), Expect = 9e-19
Identities = 55/172 (31%), Positives = 89/172 (50%), Gaps = 15/172 (8%)
Query: 8 FELKPLFHLLLPLSIHWIAEAMTVSVLVDVTTTALCPQ-QSSCSKAIYINGLQQTITGIF 66
+ L L HLL + + +E + V+ DVT A+C +CS A+Y+ G++Q G+
Sbjct: 4 YRLGELRHLLTTVFLSGFSEFLVKPVMTDVTVAAVCSGLNETCSLAVYLTGVEQVTVGLG 63
Query: 67 KMAVLPLLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYYVLRTFSHIISQG 126
M ++P++G LSD +G K LL L + SI+P A+LA+ + F YA+Y+ +
Sbjct: 64 TMVMMPVIGNLSDRYGIKTLLTLPMCLSILPPAILAYRRDTNFFYAFYITKILFD----- 118
Query: 127 SIFCISVAYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFLPQNYIFVI 178
+A V KR+++F + G+ S + V A AR LP IF +
Sbjct: 119 ---------MAKNVCGRKRISMFGVLAGVRSISGVCATFSARLLPIASIFQV 161
Score = 73.9 bits (180), Expect = 2e-13
Identities = 49/169 (28%), Positives = 74/169 (42%), Gaps = 41/169 (24%)
Query: 345 QIVLLPILNPLVGEKVILCSALLASIAYAWLSGLAWAPWVSKIVTFYINLIQIYMFIWLA 404
Q+ +LP L +GE+ +L + LL A ++W+ WV T L+ + MF+
Sbjct: 264 QLFILPKLVSAIGERRVLSTGLLMDSVNAACLSVSWSAWVPYATTV---LVPVTMFVM-- 318
Query: 405 ENFITYISIMKVIEFNPLWQVPYLGGSFGIIYILEKPATYGIISKASSSTNQGKAQTFIA 464
P+ GI S+ QGK Q I+
Sbjct: 319 ------------------------------------PSVCGIASRQVGPGEQGKVQGCIS 342
Query: 465 GANSISGLLSPIVMSPLTSLFLSSDAPFECKGFSIICASVCMVSYLFLS 513
G S SG+++P + SPLT+LFLS APF GFS++C + ++ FLS
Sbjct: 343 GVKSFSGVVAPFIYSPLTALFLSEKAPFYFPGFSLLCVTFSLMIGFFLS 391
>At2g16990 putative tetracycline transporter protein
Length = 418
Score = 90.1 bits (222), Expect = 3e-18
Identities = 50/160 (31%), Positives = 83/160 (51%), Gaps = 15/160 (9%)
Query: 13 LFHLLLPLSIHWIAEAMTVSVLVDVTTTALCP-QQSSCSKAIYINGLQQTITGIFKMAVL 71
L H+L + + A M V V+ DVT A+C SCS A+Y+ G QQ G+ M ++
Sbjct: 8 LRHMLATVFLSAFAGFMVVPVITDVTVAAVCSGPDDSCSLAVYLTGFQQVAIGMGTMIMM 67
Query: 72 PLLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYYVLRTFSHIISQGSIFCI 131
P++G LSD +G K +L L + SI+P +L + + +F Y +Y+ + +
Sbjct: 68 PVIGNLSDRYGIKTILTLPMCLSIVPPVILGYRRDIKFFYVFYISKILTS---------- 117
Query: 132 SVAYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFLP 171
+A +H S R++ F + G+ + A + + ARFLP
Sbjct: 118 ----MAVNIHGSTRISAFGILAGIKTIAGLFGTLVARFLP 153
Score = 68.9 bits (167), Expect = 6e-12
Identities = 45/169 (26%), Positives = 72/169 (41%), Gaps = 41/169 (24%)
Query: 345 QIVLLPILNPLVGEKVILCSALLASIAYAWLSGLAWAPWVSKIVTFYINLIQIYMFIWLA 404
Q+ +LP +GE +L + L + ++WAPWV + T ++
Sbjct: 268 QLFVLPRFASAIGECKLLSTGLFMEFINMAIVSISWAPWVPYLTTVFVP----------- 316
Query: 405 ENFITYISIMKVIEFNPLWQVPYLGGSFGIIYILEKPATYGIISKASSSTNQGKAQTFIA 464
G ++++ P+ GI S+ QGK Q I+
Sbjct: 317 ----------------------------GALFVM--PSVCGIASRQVGPGEQGKVQGCIS 346
Query: 465 GANSISGLLSPIVMSPLTSLFLSSDAPFECKGFSIICASVCMVSYLFLS 513
G S +++P V SPLT+LFLS +APF GFS++C S+ + F S
Sbjct: 347 GVRSFGKVVAPFVFSPLTALFLSKNAPFYFPGFSLLCISLSSLIGFFQS 395
>At5g42210 putative protein
Length = 204
Score = 41.2 bits (95), Expect = 0.001
Identities = 50/174 (28%), Positives = 71/174 (40%), Gaps = 30/174 (17%)
Query: 132 SVAYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFLPQNYIFVILNYGLLLSGFNNT 191
S + AD V E +R + F +TG++S A V AN+ ARFL F +
Sbjct: 5 SADFQADNVPEGRRASAFGILTGITSCAFVCANLSARFLSIGVTFQVA----------AA 54
Query: 192 IDLLSTLY--AFLPS*NCETGSRKEPRVRLLY*I*GMPQK**S---SGCLSHVN*HQSFF 246
+ +LSTLY FLP + S P V + +K S C H N + F
Sbjct: 55 MGILSTLYMRLFLPD-SIRDNSLGAPIV--------INEKLSSPLLEDCPGHRN--RIFR 103
Query: 247 ELTCIHES*IF----PTLRGVALVSFFYKLGMTGIHSVLLQIFSLLCHYYLTNF 296
+ +HE L VA+VSFF L G+H+ + H+ F
Sbjct: 104 AIRLVHEMASLMRSSVPLFQVAMVSFFSSLAEAGLHASSMYYLKAKFHFNKDQF 157
>At5g50670 putative protein
Length = 359
Score = 31.2 bits (69), Expect = 1.5
Identities = 27/105 (25%), Positives = 53/105 (49%), Gaps = 13/105 (12%)
Query: 406 NFITYISIMKVIEFNPLWQVPYLGGS--FGIIYILEKPATYGIISKA--SSSTNQGKAQT 461
NF T K++EF+ GGS F +L ++S A ++ ++ G++Q+
Sbjct: 185 NFFTGFQGSKLLEFS--------GGSHVFPTTSVLNPSWGNSLVSVAVAANGSSYGQSQS 236
Query: 462 FIAGANSI-SGLLSPIVMSPLTSLFLSSDAPFECKGFSIICASVC 505
++ G++ +G++ PI SP ++ ++ PF + S AS+C
Sbjct: 237 YVVGSSPAKTGIMFPISSSPNSTRSIAKQFPFLQEEESSRTASLC 281
>At5g50570 putative protein
Length = 359
Score = 31.2 bits (69), Expect = 1.5
Identities = 27/105 (25%), Positives = 53/105 (49%), Gaps = 13/105 (12%)
Query: 406 NFITYISIMKVIEFNPLWQVPYLGGS--FGIIYILEKPATYGIISKA--SSSTNQGKAQT 461
NF T K++EF+ GGS F +L ++S A ++ ++ G++Q+
Sbjct: 185 NFFTGFQGSKLLEFS--------GGSHVFPTTSVLNPSWGNSLVSVAVAANGSSYGQSQS 236
Query: 462 FIAGANSI-SGLLSPIVMSPLTSLFLSSDAPFECKGFSIICASVC 505
++ G++ +G++ PI SP ++ ++ PF + S AS+C
Sbjct: 237 YVVGSSPAKTGIMFPISSSPNSTRSIAKQFPFLQEEESSRTASLC 281
>At5g05730 anthranilate synthase component I-1 precursor (sp|P32068)
Length = 595
Score = 30.0 bits (66), Expect = 3.3
Identities = 20/67 (29%), Positives = 33/67 (48%)
Query: 89 LTISTSIIPFALLAWNQSKEFVYAYYVLRTFSHIISQGSIFCISVAYVADVVHESKRVAV 148
LT S ++P + S Y R+ ++ S SI C+SV+ A +V ++K++A
Sbjct: 13 LTFSRRLLPSVASRYLSSSSVTVTGYSGRSSAYAPSFRSIKCVSVSPEASIVSDTKKLAD 72
Query: 149 FSWITGL 155
S T L
Sbjct: 73 ASKSTNL 79
>At1g11000 membrane protein Mlo4
Length = 573
Score = 30.0 bits (66), Expect = 3.3
Identities = 14/40 (35%), Positives = 23/40 (57%), Gaps = 2/40 (5%)
Query: 377 GLAWAPWVSKIVTFYINLIQIYMFIWLAENFITYISIMKV 416
G++W WV IV IN+ + M+ W+ +F+ I +M V
Sbjct: 277 GISWPLWVYAIVCICINVHGLNMYFWI--SFVPAILVMLV 314
>At2g44110 similar to Mlo proteins from H. vulgare
Length = 496
Score = 29.6 bits (65), Expect = 4.3
Identities = 12/40 (30%), Positives = 24/40 (60%), Gaps = 2/40 (5%)
Query: 377 GLAWAPWVSKIVTFYINLIQIYMFIWLAENFITYISIMKV 416
G++W WV ++ +N++ +++ WLA FI I ++ V
Sbjct: 275 GISWYLWVFVVLFLLLNIVAWHVYFWLA--FIPLILLLAV 312
>At1g48780 hypothetical protein
Length = 251
Score = 29.6 bits (65), Expect = 4.3
Identities = 13/33 (39%), Positives = 21/33 (63%)
Query: 429 GGSFGIIYILEKPATYGIISKASSSTNQGKAQT 461
GGSF +I +L P+T+G+ S S ++ K +T
Sbjct: 212 GGSFSVIPVLNGPSTFGLGSILRHSNSKDKTKT 244
>At1g19450 integral membrane protein, putative
Length = 488
Score = 29.6 bits (65), Expect = 4.3
Identities = 41/176 (23%), Positives = 75/176 (42%), Gaps = 22/176 (12%)
Query: 30 TVSVLVDVTTTALCPQQ-------SSCSKAIYINGLQQTIT---------GIFKMAVLPL 73
++SVL V AL P Q SS ++A L T++ + M
Sbjct: 46 SISVLACVLIVALGPIQFGFTCGYSSPTQAAITKDLGLTVSEYSVFGSLSNVGAMVGAIA 105
Query: 74 LGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYYVLRTFSHIISQGSIFCISV 133
GQ+++ GRK L++ +II + +++ + F+Y +L F G I
Sbjct: 106 SGQIAEYVGRKGSLMIAAIPNIIGWLSISFAKDTSFLYMGRLLEGF----GVGIISYTVP 161
Query: 134 AYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFLPQNYIFV--ILNYGLLLSG 187
Y+A++ ++ R A+ S + ++A + F+P + V +L LL+ G
Sbjct: 162 VYIAEIAPQTMRGALGSVNQLSVTIGIMLAYLLGLFVPWRILAVLGVLPCTLLIPG 217
>At1g75220 integral membrane protein, putative
Length = 487
Score = 28.9 bits (63), Expect = 7.3
Identities = 41/176 (23%), Positives = 74/176 (41%), Gaps = 22/176 (12%)
Query: 30 TVSVLVDVTTTALCPQQ-------SSCSKAIYINGLQQTIT---------GIFKMAVLPL 73
++SVL V AL P Q SS ++A L T++ + M
Sbjct: 45 SISVLACVLIVALGPIQFGFTCGYSSPTQAAITKDLGLTVSEYSVFGSLSNVGAMVGAIA 104
Query: 74 LGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYYVLRTFSHIISQGSIFCISV 133
GQ+++ GRK L++ +II + +++ + F+Y +L F G I
Sbjct: 105 SGQIAEYIGRKGSLMIAAIPNIIGWLCISFAKDTSFLYMGRLLEGF----GVGIISYTVP 160
Query: 134 AYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFLPQNYIFV--ILNYGLLLSG 187
Y+A++ ++ R + S + ++A + F+P + V IL LL+ G
Sbjct: 161 VYIAEIAPQNMRGGLGSVNQLSVTIGIMLAYLLGLFVPWRILAVLGILPCTLLIPG 216
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.339 0.147 0.482
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,796,039
Number of Sequences: 26719
Number of extensions: 416526
Number of successful extensions: 1339
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 1320
Number of HSP's gapped (non-prelim): 20
length of query: 526
length of database: 11,318,596
effective HSP length: 104
effective length of query: 422
effective length of database: 8,539,820
effective search space: 3603804040
effective search space used: 3603804040
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC140027.12


