

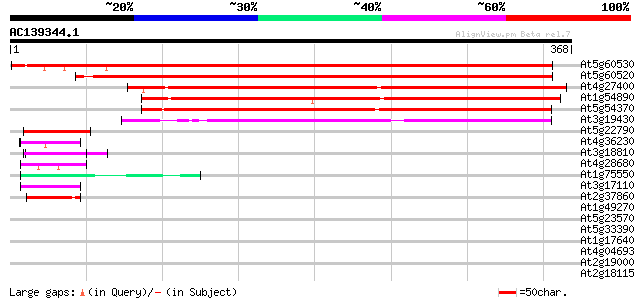
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139344.1 + phase: 0
(368 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g60530 late embryonic abundant protein - like 469 e-133
At5g60520 late embryonic abundant protein - like 457 e-129
At4g27400 unknown protein (At4g27400) 277 8e-75
At1g54890 hypothetical protein 263 1e-70
At5g54370 root cap protein 2-like protein 262 2e-70
At3g19430 putative late embryogenesis abundant protein 239 1e-63
At5g22790 putative protein 49 4e-06
At4g36230 putative glycine-rich cell wall protein 47 2e-05
At3g18810 protein kinase, putative 46 3e-05
At4g28680 aromatic amino-acid decarboxylase - like protein 45 8e-05
At1g75550 hypothetical protein 44 1e-04
At3g17110 hypothetical protein 42 7e-04
At2g37860 unknown protein 42 7e-04
At1g49270 hypothetical protein 41 0.001
At5g23570 unknown protein (At5g23570) 41 0.001
At5g33390 unknown protein (At5g33390) 40 0.002
At1g17640 RNA-binding protein, putative 40 0.002
At4g04693 putative protein 40 0.003
At2g19000 unknown protein 40 0.003
At2g18115 putative protein 39 0.003
>At5g60530 late embryonic abundant protein - like
Length = 439
Score = 469 bits (1208), Expect = e-133
Identities = 226/411 (54%), Positives = 289/411 (69%), Gaps = 57/411 (13%)
Query: 2 EAAIAQGQGNGNGNGNNGNG------KGNGNGNGNNGNG--------------------- 34
+ IAQ G G GNGNNG G KG G+ +NGNG
Sbjct: 20 DIVIAQSDG-GKGNGNNGKGNEVQVDKGKGDNGKSNGNGPKDKEQEKKDKEKAAKDKKEK 78
Query: 35 KGNDKEEGKEKGKEKEKAPKKKKAPKEE----------------------------SSNY 66
+ DKEE ++K KE+++ KK K KE+ ++ Y
Sbjct: 79 EKKDKEEKEKKDKERKEKEKKDKLEKEKKDKERKEKERKEKERKAKEKKDKEESEAAARY 138
Query: 67 QQLQTLPSGQERAFCKANNTCHFATLVCPAECKTRKPKKNKKDKGCFIDCSSKCEATCKF 126
+ L LP+GQE+A C+ +C++ TLVCP EC RKP KNK KGCFIDC++KCEATCK+
Sbjct: 139 RILSPLPTGQEQAMCQGRGSCYYKTLVCPGECPKRKPTKNKNTKGCFIDCTNKCEATCKW 198
Query: 127 RRANCDGYGSLCYDPRFVGGDGVMFYFHGAKGGNFAIVSDDEFQINAHFIGTRPQGRTRD 186
R+ NC+GYGSLCYDPRFVGGDGVMFYFHG+KGGNFAIVSD+ QINAHFIGTRP GRTRD
Sbjct: 199 RKTNCNGYGSLCYDPRFVGGDGVMFYFHGSKGGNFAIVSDNNLQINAHFIGTRPVGRTRD 258
Query: 187 YTWVQALSLMFDTHTLVIAANRVTQWNDNVDSLTVKWDGESVIIPTDDDAEWK-TNGDER 245
+TWVQAL++MF+ H LVI ANRV QW++ D+ T+++DGE + +P D+ +EW+ +G ++
Sbjct: 259 FTWVQALNVMFENHKLVITANRVNQWDETSDAFTIRYDGELITLPEDEQSEWREISGQKK 318
Query: 246 EVVVERTDDANSVRVTVSGLLEMDIRVRPIGEKENKAHNYQLPSDDAFAHLETQFKFKNP 305
++++ERTD+ NSVRV VS L++MDIRVRPIG++EN+ HNYQLP DDAFAHLETQFKF +
Sbjct: 319 DIIIERTDERNSVRVLVSDLVQMDIRVRPIGKEENRVHNYQLPQDDAFAHLETQFKFLDL 378
Query: 306 TDSIEGVLGQTYRPSYVSPVKRGVAMPMMGGEDKYQTPSLFSTTCKLCRFQ 356
++ +EGVLG+TYRP YVS K GV MP++GGEDKYQTPSLFS TC+LCRF+
Sbjct: 379 SELVEGVLGKTYRPDYVSSAKTGVPMPVLGGEDKYQTPSLFSPTCRLCRFK 429
>At5g60520 late embryonic abundant protein - like
Length = 338
Score = 457 bits (1177), Expect = e-129
Identities = 215/313 (68%), Positives = 252/313 (79%), Gaps = 5/313 (1%)
Query: 44 EKGKEKEKAPKKKKAPKEESSNYQQLQTLPSGQERAFCKANNTCHFATLVCPAECKTRKP 103
EKGK+ KK A S+NY L L SGQER C A +C+ L CP EC RKP
Sbjct: 21 EKGKKL-----KKGAYDAASTNYNVLYPLGSGQERVQCLARGSCNQKILTCPKECPERKP 75
Query: 104 KKNKKDKGCFIDCSSKCEATCKFRRANCDGYGSLCYDPRFVGGDGVMFYFHGAKGGNFAI 163
K NKK K CFIDCSSKCE TCK+R+ANC+GYGSLCYDPRFVGGDGVMFYFHG K GNFAI
Sbjct: 76 KMNKKKKACFIDCSSKCEVTCKWRKANCNGYGSLCYDPRFVGGDGVMFYFHGNKDGNFAI 135
Query: 164 VSDDEFQINAHFIGTRPQGRTRDYTWVQALSLMFDTHTLVIAANRVTQWNDNVDSLTVKW 223
VSD+ QINAHFIGTRP GRTRD+TWVQA S+MFD+H LVIAA +V W+D+VDSL V+W
Sbjct: 136 VSDENLQINAHFIGTRPAGRTRDFTWVQAFSVMFDSHNLVIAAKKVASWDDSVDSLVVRW 195
Query: 224 DGESVIIPTDDDAEWKTNGDEREVVVERTDDANSVRVTVSGLLEMDIRVRPIGEKENKAH 283
+GE V +PT+ +AEW+ + DEREV+VERTD+ N+VRVTVSG++++DI+VRPIG++E++ H
Sbjct: 196 NGEEVEVPTEGEAEWRIDLDEREVIVERTDERNNVRVTVSGIVQIDIQVRPIGKEEDRVH 255
Query: 284 NYQLPSDDAFAHLETQFKFKNPTDSIEGVLGQTYRPSYVSPVKRGVAMPMMGGEDKYQTP 343
YQLP DDAFAHLETQFKF N +D +EGVLG+TYRP YVSPVK GV MPMMGGEDKYQTP
Sbjct: 256 KYQLPKDDAFAHLETQFKFFNLSDLVEGVLGKTYRPGYVSPVKTGVPMPMMGGEDKYQTP 315
Query: 344 SLFSTTCKLCRFQ 356
SLFS C +CRFQ
Sbjct: 316 SLFSPLCNVCRFQ 328
>At4g27400 unknown protein (At4g27400)
Length = 341
Score = 277 bits (708), Expect = 8e-75
Identities = 127/292 (43%), Positives = 190/292 (64%), Gaps = 7/292 (2%)
Query: 78 RAFCKANNT--CHFATLVCPAECKTRKPKKNKKDKGCFIDCSSK-CEATCKFRRANCDGY 134
+ +C N T C + CP EC T N ++K C++DC CEA C+ + NC+ Y
Sbjct: 28 QTYCGGNATPRCQLRYIDCPEECPTEM-FPNSQNKICWVDCFKPLCEAVCRAVKPNCESY 86
Query: 135 GSLCYDPRFVGGDGVMFYFHGAKGGNFAIVSDDEFQINAHFIGTRPQGRTRDYTWVQALS 194
GS+C DPRF+GGDG++FYFHG +F+IVSD +FQINA F G RP GRTRD+TW+QAL
Sbjct: 87 GSICLDPRFIGGDGIVFYFHGKSNEHFSIVSDPDFQINARFTGHRPAGRTRDFTWIQALG 146
Query: 195 LMFDTHTLVIAANRVTQWNDNVDSLTVKWDGESVIIPTDDDAEWKTNGDEREVVVERTDD 254
+F++H + +V W+ N+D L DG+ +IIP + + W ++ ++++ +ER +
Sbjct: 147 FLFNSHKFSLETTKVATWDSNLDHLKFTIDGQDLIIPQETLSTWYSS--DKDIKIERLTE 204
Query: 255 ANSVRVTVSGLLEMDIRVRPIGEKENKAHNYQLPSDDAFAHLETQFKFKNPTDSIEGVLG 314
NSV VT+ E+ + V P+ +++++ HNY+LP DD FAH E QFKF N + ++G+LG
Sbjct: 205 KNSVIVTIKDKAEIMVNVVPVTKEDDRIHNYKLPVDDCFAHFEVQFKFINLSPKVDGILG 264
Query: 315 QTYRPSYVSPVKRGVAMPMMGGEDKYQTPSLFSTTCKLCRF-QRPSTSQGLI 365
+TYRP + +P K GV MP++GGED ++T SL S CK C F + P+ + G +
Sbjct: 265 RTYRPDFKNPAKPGVVMPVVGGEDSFRTSSLLSHVCKTCLFSEDPAVASGSV 316
>At1g54890 hypothetical protein
Length = 347
Score = 263 bits (672), Expect = 1e-70
Identities = 124/279 (44%), Positives = 181/279 (64%), Gaps = 7/279 (2%)
Query: 87 CHFATLVCPAECKTRKPKKNKKDKGCFIDCSSK-CEATCKFRRANCDGYGSLCYDPRFVG 145
C CP +C + P N K C IDC + C+ATC+ R+ NC+G GS C DPRF+G
Sbjct: 42 CFLKKQTCPKQCPSFSPP-NGSTKACVIDCFNPICKATCRNRKPNCNGKGSACLDPRFIG 100
Query: 146 GDGVMFYFHGAKGGNFAIVSDDEFQINAHFIGTRPQGRTRDYTWVQALSLMF--DTHTLV 203
GDG++FYFHG + +FA++SD +FQ+NA FIG RP GR RD+TW+Q+L L+F ++ T
Sbjct: 101 GDGIVFYFHGKRDEHFALISDVDFQVNARFIGLRPNGRARDFTWIQSLGLIFGPNSKTFS 160
Query: 204 IAANRVTQWNDNVDSLTVKWDGESVIIPTDDDAEWKTN-GDEREVVVERTDDANSVRVTV 262
+ A + +W+ VD L + ++G+ + +P D + W GD + +ERT D NSV VT+
Sbjct: 161 LEATKAEKWDHQVDHLRLSYEGKEISLPKGDTSVWSPPLGD--YIKIERTSDINSVLVTL 218
Query: 263 SGLLEMDIRVRPIGEKENKAHNYQLPSDDAFAHLETQFKFKNPTDSIEGVLGQTYRPSYV 322
+ E+ I V P+ ++++ H Y +P DD FAHLE QF+F + ++EGVLG+TY+ +
Sbjct: 219 QDIAEIWINVVPVTKEDDIIHKYGIPEDDCFAHLEVQFRFLKLSSNVEGVLGRTYKEDFK 278
Query: 323 SPVKRGVAMPMMGGEDKYQTPSLFSTTCKLCRFQRPSTS 361
+P K GVAMP++GGEDKY+T SL T+C C + S S
Sbjct: 279 NPAKPGVAMPVVGGEDKYRTASLLETSCNACVYSGGSRS 317
>At5g54370 root cap protein 2-like protein
Length = 337
Score = 262 bits (670), Expect = 2e-70
Identities = 116/270 (42%), Positives = 177/270 (64%), Gaps = 4/270 (1%)
Query: 87 CHFATLVCPAECKTRKPKKNKKDKGCFIDCSSK-CEATCKFRRANCDGYGSLCYDPRFVG 145
C+ + CP EC + K N K+K C+ DC C++ C+ R+ NC+ GS CYDPRF+G
Sbjct: 36 CYRKYIRCPEECPS-KTAMNSKNKVCYADCDRPTCKSQCRMRKPNCNRPGSACYDPRFIG 94
Query: 146 GDGVMFYFHGAKGGNFAIVSDDEFQINAHFIGTRPQGRTRDYTWVQALSLMFDTHTLVIA 205
GDG++FYFHG F++VSD + QIN FIG RP GR RD+TW+QAL +F+++ +
Sbjct: 95 GDGIVFYFHGKSNEEFSLVSDSDLQINGRFIGHRPAGRARDFTWIQALGFLFNSNKFSLE 154
Query: 206 ANRVTQWNDNVDSLTVKWDGESVIIPTDDDAEWKTNGDEREVVVERTDDANSVRVTVSGL 265
A + W++ +D L +DG+ + +P + + W + +++ +ER NSV VT+
Sbjct: 155 AAKTASWDNEIDHLKFSYDGQDLSVPEETLSTWYS--PNKDIKIERVSMRNSVIVTIKDK 212
Query: 266 LEMDIRVRPIGEKENKAHNYQLPSDDAFAHLETQFKFKNPTDSIEGVLGQTYRPSYVSPV 325
E+ I V P+ +++++ H+Y++PSDD FAHLE QF+F N + ++G+LG+TYRP + +P
Sbjct: 213 AEIMINVVPVTKEDDRIHSYKVPSDDCFAHLEVQFRFFNLSPKVDGILGRTYRPDFQNPA 272
Query: 326 KRGVAMPMMGGEDKYQTPSLFSTTCKLCRF 355
K GVAMP++GGED ++T SL S CK C F
Sbjct: 273 KPGVAMPVVGGEDSFKTSSLLSNDCKTCIF 302
>At3g19430 putative late embryogenesis abundant protein
Length = 550
Score = 239 bits (611), Expect = 1e-63
Identities = 121/284 (42%), Positives = 172/284 (59%), Gaps = 26/284 (9%)
Query: 74 SGQERAFCKANNT-CHFATLVCPAECKTRKPKKNKKDKGCFIDCSSKCEATCKFRRANCD 132
+G +R CK + C+ CPA+C + C +DC + C+ C NCD
Sbjct: 258 AGAKRVRCKKQRSPCYGVEYTCPADCP----------RSCQVDCVT-CKPVC-----NCD 301
Query: 133 GYGSLCYDPRFVGGDGVMFYFHGAKGGNFAIVSDDEFQINAHFIGTRPQGRTRDYTWVQA 192
GS+C DPRF+GGDG+ FYFHG K NF ++SD INAHFIG R G RD+TWVQ+
Sbjct: 302 KPGSVCQDPRFIGGDGLTFYFHGKKDSNFCLISDPNLHINAHFIGKRRAGMARDFTWVQS 361
Query: 193 LSLMFDTHTLVIAANRVTQWNDNVDSLTVKWDGESVIIPTDDDAEWKTN-GDEREVVVER 251
++++F TH L + A + W+D+VD + V +DG + +P D A W ++ G EV V+
Sbjct: 362 IAILFGTHRLYVGALKTATWDDSVDRIAVSFDGNVISLPQLDGARWTSSPGVYPEVSVK- 420
Query: 252 TDDANSVRVTVSGLLEMDIRVRPIGEKENKAHNYQLPSDDAFAHLETQFKFKNPTDSIEG 311
RV V GLL++ RV PI ++++ H Y + DD AHL+ FKF++ +D+++G
Sbjct: 421 -------RVEVEGLLKITARVVPITMEDSRIHGYDVKEDDCLAHLDLGFKFQDLSDNVDG 473
Query: 312 VLGQTYRPSYVSPVKRGVAMPMMGGEDKYQTPSLFSTTCKLCRF 355
VLGQTYR +YVS VK GV MP+MGG+ ++QT LF+ C RF
Sbjct: 474 VLGQTYRSNYVSRVKIGVHMPVMGGDREFQTTGLFAPDCSAARF 517
>At5g22790 putative protein
Length = 433
Score = 48.9 bits (115), Expect = 4e-06
Identities = 20/44 (45%), Positives = 29/44 (65%)
Query: 10 GNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAP 53
G+ NGN + G GNG+G G G+G+G+D E+ +K +EKE P
Sbjct: 104 GDENGNNDGGGNGGNGDGGGGGGDGEGDDGEDEADKAEEKEFGP 147
>At4g36230 putative glycine-rich cell wall protein
Length = 221
Score = 46.6 bits (109), Expect = 2e-05
Identities = 19/39 (48%), Positives = 20/39 (50%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G G G G G G G G G G GN GNGK N + K G
Sbjct: 93 GGGGGGGGGGGGQGSGGGGGEGNGGNGKDNSHKRNKSSG 131
Score = 42.0 bits (97), Expect = 5e-04
Identities = 21/51 (41%), Positives = 22/51 (42%), Gaps = 11/51 (21%)
Query: 7 QGQGNGNGNGNNGNGK-----------GNGNGNGNNGNGKGNDKEEGKEKG 46
QG G G G GN GNGK G G G G G G GN G+ G
Sbjct: 105 QGSGGGGGEGNGGNGKDNSHKRNKSSGGGGGGGGGGGGGSGNGSGRGRGGG 155
Score = 38.9 bits (89), Expect = 0.005
Identities = 16/36 (44%), Positives = 19/36 (52%)
Query: 7 QGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEG 42
+G G G G G G G G G G G G G G+D + G
Sbjct: 152 RGGGGGGGGGGGGGGGGGGGGGGGGGGGGGSDGKGG 187
Score = 38.5 bits (88), Expect = 0.006
Identities = 17/41 (41%), Positives = 19/41 (45%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKE 48
G GNG+G G G G G G G G G G G G G +
Sbjct: 143 GSGNGSGRGRGGGGGGGGGGGGGGGGGGGGGGGGGGGGGSD 183
Score = 38.1 bits (87), Expect = 0.008
Identities = 15/29 (51%), Positives = 16/29 (54%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKG 36
G G G G G +GNG G G G G G G G
Sbjct: 134 GGGGGGGGGGSGNGSGRGRGGGGGGGGGG 162
Score = 37.0 bits (84), Expect = 0.017
Identities = 17/40 (42%), Positives = 20/40 (49%), Gaps = 1/40 (2%)
Query: 7 QGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
+G+G G G G G G G G G G G G G +GK G
Sbjct: 150 RGRGGGGGGGGGGGGGGGGGGGGGGGGG-GGGGSDGKGGG 188
Score = 36.6 bits (83), Expect = 0.022
Identities = 14/29 (48%), Positives = 16/29 (54%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKG 36
G G G G NG+G+G G G G G G G
Sbjct: 136 GGGGGGGGSGNGSGRGRGGGGGGGGGGGG 164
Score = 36.6 bits (83), Expect = 0.022
Identities = 14/27 (51%), Positives = 15/27 (54%)
Query: 10 GNGNGNGNNGNGKGNGNGNGNNGNGKG 36
G G G G G G GNG+G G G G G
Sbjct: 132 GGGGGGGGGGGGSGNGSGRGRGGGGGG 158
Score = 36.2 bits (82), Expect = 0.029
Identities = 14/29 (48%), Positives = 16/29 (54%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKG 36
G G G G G G+G G+G G G G G G
Sbjct: 132 GGGGGGGGGGGGSGNGSGRGRGGGGGGGG 160
Score = 35.0 bits (79), Expect = 0.065
Identities = 14/41 (34%), Positives = 20/41 (48%)
Query: 12 GNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKA 52
G G G G G G G G G+ G G + GK+ ++ K+
Sbjct: 89 GGGGGGGGGGGGGGGGQGSGGGGGEGNGGNGKDNSHKRNKS 129
Score = 34.7 bits (78), Expect = 0.085
Identities = 14/29 (48%), Positives = 16/29 (54%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKG 36
G G+GNG+G G G G G G G G G
Sbjct: 141 GGGSGNGSGRGRGGGGGGGGGGGGGGGGG 169
Score = 34.7 bits (78), Expect = 0.085
Identities = 15/29 (51%), Positives = 18/29 (61%), Gaps = 1/29 (3%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKG 36
G G G+GNG +G G+G G G G G G G
Sbjct: 139 GGGGGSGNG-SGRGRGGGGGGGGGGGGGG 166
Score = 32.0 bits (71), Expect = 0.55
Identities = 13/29 (44%), Positives = 14/29 (47%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKG 36
G G G G G G G G G G+ G G G
Sbjct: 162 GGGGGGGGGGGGGGGGGGGGSDGKGGGWG 190
Score = 31.2 bits (69), Expect = 0.94
Identities = 14/31 (45%), Positives = 18/31 (57%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGND 38
G G G G G++G G G G G G G+G G +
Sbjct: 173 GGGGGGGGGSDGKGGGWGFGFGWGGDGPGGN 203
Score = 28.9 bits (63), Expect = 4.7
Identities = 13/29 (44%), Positives = 14/29 (47%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKG 36
G G G G G +GKG G G G G G
Sbjct: 171 GGGGGGGGGGGSDGKGGGWGFGFGWGGDG 199
>At3g18810 protein kinase, putative
Length = 700
Score = 46.2 bits (108), Expect = 3e-05
Identities = 19/54 (35%), Positives = 24/54 (44%)
Query: 11 NGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKKKAPKEESS 64
N NGN NNGN + NGNN NG ND G ++P + +S
Sbjct: 105 NNNGNNNNGNNNNGNDNNGNNNNGNNNDNNNQNNGGGSNNRSPPPPSRNSDRNS 158
Score = 41.6 bits (96), Expect = 7e-04
Identities = 20/43 (46%), Positives = 22/43 (50%), Gaps = 2/43 (4%)
Query: 10 GNGNGNGNNGNGK-GNGNGNGNNGN-GKGNDKEEGKEKGKEKE 50
GN N + NNGN K N NGN NNGN GND G +
Sbjct: 90 GNNNNDNNNGNNKDNNNNGNNNNGNNNNGNDNNGNNNNGNNND 132
Score = 40.4 bits (93), Expect = 0.002
Identities = 19/46 (41%), Positives = 22/46 (47%), Gaps = 8/46 (17%)
Query: 9 QGNGNGNGNNGNGK--------GNGNGNGNNGNGKGNDKEEGKEKG 46
QGN N +GNNGN GN N + NNGN K N+ G
Sbjct: 68 QGNNNNDGNNGNNNNDNNNNNNGNNNNDNNNGNNKDNNNNGNNNNG 113
Score = 39.7 bits (91), Expect = 0.003
Identities = 15/34 (44%), Positives = 17/34 (49%)
Query: 11 NGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKE 44
N NGN NN N GN N NNGN + G +
Sbjct: 87 NNNGNNNNDNNNGNNKDNNNNGNNNNGNNNNGND 120
Score = 38.9 bits (89), Expect = 0.005
Identities = 20/57 (35%), Positives = 27/57 (47%), Gaps = 4/57 (7%)
Query: 11 NGNGNGNNGNGK-GNGNGNGNNGNGKGNDKEEGKEKGKEKEK---APKKKKAPKEES 63
N NGN NNGN GN N N N NG G++ + ++ +P + AP S
Sbjct: 115 NNNGNDNNGNNNNGNNNDNNNQNNGGGSNNRSPPPPSRNSDRNSPSPPRALAPPRSS 171
Score = 32.3 bits (72), Expect = 0.42
Identities = 13/45 (28%), Positives = 18/45 (39%)
Query: 6 AQGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKE 50
+ Q + GNN N NGN N +N N + G K+
Sbjct: 59 SDSQSPPSPQGNNNNDGNNGNNNNDNNNNNNGNNNNDNNNGNNKD 103
>At4g28680 aromatic amino-acid decarboxylase - like protein
Length = 545
Score = 44.7 bits (104), Expect = 8e-05
Identities = 26/51 (50%), Positives = 28/51 (53%), Gaps = 8/51 (15%)
Query: 8 GQGNGNGNGN---NGNGKGNGNGNGN-----NGNGKGNDKEEGKEKGKEKE 50
G GNG NGN NGNG NGNGN N NGNGK N + K K + E
Sbjct: 13 GTGNGYSNGNGYTNGNGHTNGNGNYNGNGHVNGNGKANGAKVVKMKPMDSE 63
Score = 34.7 bits (78), Expect = 0.085
Identities = 22/53 (41%), Positives = 25/53 (46%), Gaps = 4/53 (7%)
Query: 12 GNGNG-NNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKKKAPKEES 63
G GNG +NGNG NGNG+ N G GN G G K K K +S
Sbjct: 13 GTGNGYSNGNGYTNGNGHTN---GNGNYNGNGHVNGNGKANGAKVVKMKPMDS 62
>At1g75550 hypothetical protein
Length = 167
Score = 43.9 bits (102), Expect = 1e-04
Identities = 34/118 (28%), Positives = 38/118 (31%), Gaps = 30/118 (25%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKKKAPKEESSNYQ 67
G G G G G G G G G G G G K GK KG+E K+
Sbjct: 79 GGGGGGGGGGGGGGWGWGGGGGGGGWYKWGCGGGGKGKGREGRGEFVKR----------- 127
Query: 68 QLQTLPSGQERAFCKANNTCHFATLVCPAECKTRKPKKNKKDKGCFIDCSSKCEATCK 125
E A CK C L CP C CF DC C+ C+
Sbjct: 128 ---------EYAECKGRGKCSGKRLECPQHCGGL----------CFYDCLFLCKPHCR 166
>At3g17110 hypothetical protein
Length = 175
Score = 41.6 bits (96), Expect = 7e-04
Identities = 18/39 (46%), Positives = 19/39 (48%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G NG G G G G G G GNG +GNG G G G
Sbjct: 87 GGNNGGGGGGGGRGGGGGGGNGGSGNGSGYGSGSGYGSG 125
Score = 40.8 bits (94), Expect = 0.001
Identities = 17/39 (43%), Positives = 19/39 (48%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G G+G GNG G G G G G G G G G+ G G
Sbjct: 40 GSGSGGGNGGGGGGGGGGGGGGGGGGGDGSGSGSGYGSG 78
Score = 40.4 bits (93), Expect = 0.002
Identities = 17/39 (43%), Positives = 19/39 (48%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G G GNG G G G G G G G G+G G+ G G
Sbjct: 42 GSGGGNGGGGGGGGGGGGGGGGGGGDGSGSGSGYGSGYG 80
Score = 40.4 bits (93), Expect = 0.002
Identities = 17/39 (43%), Positives = 20/39 (50%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G G+G+G GN G G G G G G G G G+ G G
Sbjct: 38 GYGSGSGGGNGGGGGGGGGGGGGGGGGGGDGSGSGSGYG 76
Score = 40.0 bits (92), Expect = 0.002
Identities = 17/39 (43%), Positives = 20/39 (50%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G GNG G G G G G G G G +G+G G+ G G
Sbjct: 44 GGGNGGGGGGGGGGGGGGGGGGGDGSGSGSGYGSGYGSG 82
Score = 40.0 bits (92), Expect = 0.002
Identities = 19/44 (43%), Positives = 23/44 (52%), Gaps = 5/44 (11%)
Query: 8 GQGNGNGNGNN-----GNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G G+G G GNN G G+G G G GN G+G G+ G G
Sbjct: 80 GSGSGYGGGNNGGGGGGGGRGGGGGGGNGGSGNGSGYGSGSGYG 123
Score = 38.5 bits (88), Expect = 0.006
Identities = 16/38 (42%), Positives = 19/38 (49%)
Query: 10 GNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGK 47
G G G G G G G GNG NG+G G+ G G+
Sbjct: 91 GGGGGGGGRGGGGGGGNGGSGNGSGYGSGSGYGSGAGR 128
Score = 37.4 bits (85), Expect = 0.013
Identities = 17/40 (42%), Positives = 21/40 (52%), Gaps = 5/40 (12%)
Query: 4 AIAQGQGNGNG-----NGNNGNGKGNGNGNGNNGNGKGND 38
A+ +G G+G+G G NG G G G G G G G G D
Sbjct: 28 ALGRGSGSGSGYGSGSGGGNGGGGGGGGGGGGGGGGGGGD 67
Score = 37.4 bits (85), Expect = 0.013
Identities = 16/31 (51%), Positives = 22/31 (70%), Gaps = 1/31 (3%)
Query: 7 QGQGNGNGNGNNGNGKGNGNGNG-NNGNGKG 36
+G G G GNG +GNG G G+G+G +G G+G
Sbjct: 99 RGGGGGGGNGGSGNGSGYGSGSGYGSGAGRG 129
Score = 36.2 bits (82), Expect = 0.029
Identities = 15/39 (38%), Positives = 19/39 (48%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G G+G G+G+ G G G G G G G G +G G
Sbjct: 34 GSGSGYGSGSGGGNGGGGGGGGGGGGGGGGGGGDGSGSG 72
Score = 35.8 bits (81), Expect = 0.038
Identities = 17/35 (48%), Positives = 21/35 (59%), Gaps = 1/35 (2%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEG 42
G G+G+G G +G G G+G G GNNG G G G
Sbjct: 68 GSGSGSGYG-SGYGSGSGYGGGNNGGGGGGGGRGG 101
Score = 35.4 bits (80), Expect = 0.050
Identities = 15/39 (38%), Positives = 17/39 (43%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G G G G G G G G G G +G+G G G G
Sbjct: 46 GNGGGGGGGGGGGGGGGGGGGDGSGSGSGYGSGYGSGSG 84
Score = 35.4 bits (80), Expect = 0.050
Identities = 17/39 (43%), Positives = 21/39 (53%), Gaps = 1/39 (2%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G+G G G G G G G G G+ NG+G G+ EG G
Sbjct: 127 GRGGGGGGGGGGGG-GGGGGSRGNGSGYGSGYGEGYGSG 164
Score = 35.0 bits (79), Expect = 0.065
Identities = 17/41 (41%), Positives = 20/41 (48%), Gaps = 1/41 (2%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNG-KGNDKEEGKEKGK 47
G G G+G G G G G G G G G G +GN G G+
Sbjct: 119 GSGYGSGAGRGGGGGGGGGGGGGGGGGSRGNGSGYGSGYGE 159
Score = 34.3 bits (77), Expect = 0.11
Identities = 16/39 (41%), Positives = 20/39 (51%), Gaps = 1/39 (2%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G G+G+G G +G G+G G G G G G G G G
Sbjct: 115 GYGSGSGYG-SGAGRGGGGGGGGGGGGGGGGGSRGNGSG 152
Score = 33.1 bits (74), Expect = 0.25
Identities = 13/29 (44%), Positives = 15/29 (50%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKG 36
G G G+G+G G G G G G G G G
Sbjct: 76 GSGYGSGSGYGGGNNGGGGGGGGRGGGGG 104
Score = 32.3 bits (72), Expect = 0.42
Identities = 14/35 (40%), Positives = 17/35 (48%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEG 42
G G+G G+G+ G NG G G G G G G
Sbjct: 74 GYGSGYGSGSGYGGGNNGGGGGGGGRGGGGGGGNG 108
Score = 32.3 bits (72), Expect = 0.42
Identities = 17/48 (35%), Positives = 21/48 (43%), Gaps = 1/48 (2%)
Query: 7 QGQGNGNGNGNNGNGKGNGNGNGNN-GNGKGNDKEEGKEKGKEKEKAP 53
+G G G G G G G G GNG+ G+G G G G +P
Sbjct: 128 RGGGGGGGGGGGGGGGGGSRGNGSGYGSGYGEGYGSGYGGGDNYGDSP 175
Score = 32.3 bits (72), Expect = 0.42
Identities = 14/39 (35%), Positives = 17/39 (42%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G G G G G G GNG +G G G+ G +G
Sbjct: 91 GGGGGGGGRGGGGGGGNGGSGNGSGYGSGSGYGSGAGRG 129
Score = 32.0 bits (71), Expect = 0.55
Identities = 14/39 (35%), Positives = 17/39 (42%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G G G G G G G G G +G+G G+ G G
Sbjct: 48 GGGGGGGGGGGGGGGGGGGDGSGSGSGYGSGYGSGSGYG 86
Score = 32.0 bits (71), Expect = 0.55
Identities = 16/35 (45%), Positives = 22/35 (62%), Gaps = 6/35 (17%)
Query: 8 GQGNGNGNG-NNGNGKGNGNGNGNN-----GNGKG 36
G G+G+G+G +G G G+G G GNN G G+G
Sbjct: 66 GDGSGSGSGYGSGYGSGSGYGGGNNGGGGGGGGRG 100
Score = 31.2 bits (69), Expect = 0.94
Identities = 16/41 (39%), Positives = 23/41 (56%), Gaps = 1/41 (2%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGN-NGNGKGNDKEEGKEKGK 47
G G G G +G+G G G+G G+ +G G GN+ G G+
Sbjct: 59 GGGGGGGGDGSGSGSGYGSGYGSGSGYGGGNNGGGGGGGGR 99
Score = 29.6 bits (65), Expect = 2.7
Identities = 17/40 (42%), Positives = 21/40 (52%), Gaps = 2/40 (5%)
Query: 8 GQGNGNGNG-NNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G G+G G+G +G+G G GN NG G G G G G
Sbjct: 70 GSGSGYGSGYGSGSGYGGGN-NGGGGGGGGRGGGGGGGNG 108
>At2g37860 unknown protein
Length = 432
Score = 41.6 bits (96), Expect = 7e-04
Identities = 21/36 (58%), Positives = 26/36 (71%), Gaps = 2/36 (5%)
Query: 12 GNG-NGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G+G NG++GNG G G GNG G+G+G D EE KE G
Sbjct: 112 GDGFNGSDGNGGGGGGGNGGEGDGEGEDYEE-KEFG 146
Score = 30.0 bits (66), Expect = 2.1
Identities = 22/63 (34%), Positives = 33/63 (51%), Gaps = 6/63 (9%)
Query: 15 NGNNGNGKGNGNGNGNNGNGKGNDKEEGK-EKGKEKEKAP--KKKKAPKEESSNYQQLQT 71
+GN G+G +GNG G G + +G+ E +EKE P K ++ KE + + T
Sbjct: 108 DGNVGDGFNGSDGNGGGGGGGNGGEGDGEGEDYEEKEFGPILKFEEVMKETEA---RGAT 164
Query: 72 LPS 74
LPS
Sbjct: 165 LPS 167
>At1g49270 hypothetical protein
Length = 699
Score = 41.2 bits (95), Expect = 0.001
Identities = 44/189 (23%), Positives = 58/189 (30%), Gaps = 30/189 (15%)
Query: 7 QGQGNGNGNGNNGNGKGN--GNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKKKAPKEES- 63
Q NGN NN N KGN +G+G N N K P+ P S
Sbjct: 103 QSDNNGNKGNNNENNKGNDGSSGDGGNKNMSHTPPPPSKTSDHSSHSQPRSLAPPTSNSG 162
Query: 64 SNYQQLQTLPSGQERAFCKANNTCHFATLVCPAECKTRKPKKNKKDKGCFIDCSSKCEAT 123
SN L G A ++ C +K KK+K D+ +
Sbjct: 163 SNSSSNDGLNIGAVIGLVAAAGILFIVMILLCVCCFRKKKKKSKLDQMPY---------- 212
Query: 124 CKFRRANCDGYGSLCYDPRFVGGDGVMFYFHGAKGGNFAIVSDDEFQINAHFIGTRPQGR 183
YGS Y GGD +Y N A + N H + P
Sbjct: 213 ----------YGSNAYPAGKTGGD--QYY-----NQNAATQQQQHYNQNDHIVNLPPPPG 255
Query: 184 TRDYTWVQA 192
+ WV +
Sbjct: 256 SMGTNWVSS 264
>At5g23570 unknown protein (At5g23570)
Length = 625
Score = 40.8 bits (94), Expect = 0.001
Identities = 22/62 (35%), Positives = 31/62 (49%), Gaps = 1/62 (1%)
Query: 7 QGQG-NGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKKKAPKEESSN 65
QG+G N +G GNN +G+GNGNG G N G + ++ P + P E N
Sbjct: 79 QGRGSNVSGRGNNVSGRGNGNGRGIQANISGRGRALSRKYDNNFVAPPPVSRPPLEGGWN 138
Query: 66 YQ 67
+Q
Sbjct: 139 WQ 140
>At5g33390 unknown protein (At5g33390)
Length = 118
Score = 40.0 bits (92), Expect = 0.002
Identities = 14/40 (35%), Positives = 22/40 (55%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGK 47
G G G+ G+ NG GNG G+G+G + +G+ G+
Sbjct: 29 GSGGSGSGGSGSGGRANGRGNGGRGSGRGGGRGDGRGDGR 68
Score = 36.2 bits (82), Expect = 0.029
Identities = 15/40 (37%), Positives = 24/40 (59%), Gaps = 1/40 (2%)
Query: 8 GQGNG-NGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G G+G +G+G NG+GNG G G+G+ + +G+ G
Sbjct: 32 GSGSGGSGSGGRANGRGNGGRGSGRGGGRGDGRGDGRGIG 71
Score = 34.7 bits (78), Expect = 0.085
Identities = 15/38 (39%), Positives = 21/38 (54%), Gaps = 6/38 (15%)
Query: 8 GQGNGNGNGNNGNGKGN------GNGNGNNGNGKGNDK 39
G+ NG GNG G+G+G G+G G G G+G +
Sbjct: 42 GRANGRGNGGRGSGRGGGRGDGRGDGRGIGGGGRGRGR 79
Score = 34.3 bits (77), Expect = 0.11
Identities = 15/34 (44%), Positives = 25/34 (73%), Gaps = 2/34 (5%)
Query: 4 AIAQGQGNGNGNGNNGNGKGNGNGNGNNGNGKGN 37
+++ G G+G G+G+ G+G G G+G+G NG+GN
Sbjct: 18 SVSGGSGSG-GSGSGGSGSG-GSGSGGRANGRGN 49
Score = 34.3 bits (77), Expect = 0.11
Identities = 17/40 (42%), Positives = 23/40 (57%), Gaps = 2/40 (5%)
Query: 8 GQGNG-NGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G G+G +G+G +G+G G NG GN G G G G +G
Sbjct: 27 GSGSGGSGSGGSGSG-GRANGRGNGGRGSGRGGGRGDGRG 65
Score = 32.3 bits (72), Expect = 0.42
Identities = 14/44 (31%), Positives = 23/44 (51%), Gaps = 7/44 (15%)
Query: 11 NGNGNGNNGNGKGNG-------NGNGNNGNGKGNDKEEGKEKGK 47
NG G+G++G+G G G+G+ G+G G G+ G+
Sbjct: 4 NGGGSGSSGSGSGGSVSGGSGSGGSGSGGSGSGGSGSGGRANGR 47
Score = 31.2 bits (69), Expect = 0.94
Identities = 15/41 (36%), Positives = 22/41 (53%), Gaps = 1/41 (2%)
Query: 8 GQGNGNGNGN-NGNGKGNGNGNGNNGNGKGNDKEEGKEKGK 47
G+G+G G G +G G G G G G G G+ G+ +G+
Sbjct: 51 GRGSGRGGGRGDGRGDGRGIGGGGRGRGRIPAAFMGRGRGR 91
>At1g17640 RNA-binding protein, putative
Length = 369
Score = 40.0 bits (92), Expect = 0.002
Identities = 20/43 (46%), Positives = 24/43 (55%), Gaps = 4/43 (9%)
Query: 3 AAIAQGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEK 45
AA A G G +GNGNNG+ G GNG NG G D+ +K
Sbjct: 331 AAAAYGGGKSHGNGNNGSSSGKGNGT----NGSGPDRYHPYQK 369
>At4g04693 putative protein
Length = 187
Score = 39.7 bits (91), Expect = 0.003
Identities = 25/73 (34%), Positives = 34/73 (46%), Gaps = 14/73 (19%)
Query: 10 GNGNGNG-NNGNGKGNGNGNGNNGNGKG-------------NDKEEGKEKGKEKEKAPKK 55
GNG GNG ++G G G GN GN GNG ++ ++G E +E E A +
Sbjct: 112 GNGVGNGGDDGGGNGEGNVGGNGGNGNQEAVEIQDVGDFGLDNDDDGDEDDEENEAAADE 171
Query: 56 KKAPKEESSNYQQ 68
+K N QQ
Sbjct: 172 QKQAIVVEENKQQ 184
Score = 36.6 bits (83), Expect = 0.022
Identities = 15/32 (46%), Positives = 19/32 (58%)
Query: 16 GNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGK 47
G+NG G G G GN GNG GN ++G G+
Sbjct: 96 GDNGGGNGGDGGCGNGGNGVGNGGDDGGGNGE 127
Score = 34.7 bits (78), Expect = 0.085
Identities = 22/44 (50%), Positives = 26/44 (59%), Gaps = 5/44 (11%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNN---GNGKGNDKEEGKEKGKE 48
G GNG G+G GNG GNG GNG + GNG+GN G +E
Sbjct: 99 GGGNG-GDGGCGNG-GNGVGNGGDDGGGNGEGNVGGNGGNGNQE 140
Score = 34.3 bits (77), Expect = 0.11
Identities = 17/34 (50%), Positives = 19/34 (55%), Gaps = 3/34 (8%)
Query: 13 NGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
NG GN G+G G GNG NG G G D G +G
Sbjct: 98 NGGGNGGDG---GCGNGGNGVGNGGDDGGGNGEG 128
Score = 31.2 bits (69), Expect = 0.94
Identities = 16/36 (44%), Positives = 17/36 (46%), Gaps = 1/36 (2%)
Query: 11 NGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
NG GNG +G G GNG NG G EG G
Sbjct: 98 NGGGNGGDG-GCGNGGNGVGNGGDDGGGNGEGNVGG 132
>At2g19000 unknown protein
Length = 125
Score = 39.7 bits (91), Expect = 0.003
Identities = 18/50 (36%), Positives = 21/50 (42%), Gaps = 6/50 (12%)
Query: 81 CKANNTCHFATLVCPAECKTRKP------KKNKKDKGCFIDCSSKCEATC 124
C A C L CP EC K + GC DC++KC ATC
Sbjct: 74 CSAQGACSGKKLTCPEECYKSTNVNKDGYKSTSRSGGCSFDCTTKCAATC 123
>At2g18115 putative protein
Length = 296
Score = 39.3 bits (90), Expect = 0.003
Identities = 19/48 (39%), Positives = 25/48 (51%)
Query: 10 GNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKKK 57
G G +G+ G G G G GNGKG+ K +G + GKE +K K
Sbjct: 133 GAGGSSGDGGAGGGGSGRVGRRGNGKGDGKGQGDDGGKEDCNDHEKNK 180
Score = 37.4 bits (85), Expect = 0.013
Identities = 16/40 (40%), Positives = 19/40 (47%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGK 47
G G G G G G+G G G G G G G G +G G+
Sbjct: 209 GGGGGGGGGGGGSGPGGGGGGGGGGGGGGGGVGDGYGYGR 248
Score = 36.6 bits (83), Expect = 0.022
Identities = 18/41 (43%), Positives = 26/41 (62%), Gaps = 2/41 (4%)
Query: 10 GNGNGN-GNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEK 49
G G+G G GNGKG+G G G++G GK + + K KG ++
Sbjct: 145 GGGSGRVGRRGNGKGDGKGQGDDG-GKEDCNDHEKNKGDDR 184
Score = 35.4 bits (80), Expect = 0.050
Identities = 15/35 (42%), Positives = 16/35 (44%)
Query: 2 EAAIAQGQGNGNGNGNNGNGKGNGNGNGNNGNGKG 36
E + G G G G G G G G G G G G G G
Sbjct: 201 EPSPGSGGGGGGGGGGGGGGSGPGGGGGGGGGGGG 235
Score = 34.7 bits (78), Expect = 0.085
Identities = 18/46 (39%), Positives = 23/46 (49%), Gaps = 1/46 (2%)
Query: 6 AQGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEK 51
A G G G G+G+ GNG G+GKG + GKE + EK
Sbjct: 134 AGGSSGDGGAGGGGSGRVGRRGNGK-GDGKGQGDDGGKEDCNDHEK 178
Score = 34.7 bits (78), Expect = 0.085
Identities = 16/39 (41%), Positives = 18/39 (46%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G G G G G +G G G G G G G G G G +G
Sbjct: 211 GGGGGGGGGGSGPGGGGGGGGGGGGGGGGVGDGYGYGRG 249
Score = 32.7 bits (73), Expect = 0.32
Identities = 15/39 (38%), Positives = 17/39 (43%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G G G G G G G G G G G G G+ G+ G
Sbjct: 213 GGGGGGGGSGPGGGGGGGGGGGGGGGGVGDGYGYGRGWG 251
Score = 32.3 bits (72), Expect = 0.42
Identities = 18/59 (30%), Positives = 31/59 (52%), Gaps = 3/59 (5%)
Query: 9 QGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKE--KEKAPKKKKAPKEESSN 65
+G G++G+G G G+G G +GN K +GK +G + KE +K ++ +N
Sbjct: 129 EGCNGAGGSSGDGGAGGGGSGRVGR-RGNGKGDGKGQGDDGGKEDCNDHEKNKGDDRNN 186
Score = 28.9 bits (63), Expect = 4.7
Identities = 12/27 (44%), Positives = 13/27 (47%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNG 34
G G G G G G G G+G G G G
Sbjct: 226 GGGGGGGGGGGGGGVGDGYGYGRGWGG 252
Score = 28.5 bits (62), Expect = 6.1
Identities = 13/29 (44%), Positives = 14/29 (47%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKG 36
G G G G +G G G G G G G G G
Sbjct: 232 GGGGGGGGVGDGYGYGRGWGGGYGGGGGG 260
Score = 28.1 bits (61), Expect = 8.0
Identities = 13/29 (44%), Positives = 14/29 (47%)
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKG 36
G G G G G G+G G G G G G G
Sbjct: 230 GGGGGGGGGGVGDGYGYGRGWGGGYGGGG 258
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.133 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,992,167
Number of Sequences: 26719
Number of extensions: 521671
Number of successful extensions: 9885
Number of sequences better than 10.0: 294
Number of HSP's better than 10.0 without gapping: 194
Number of HSP's successfully gapped in prelim test: 100
Number of HSP's that attempted gapping in prelim test: 4330
Number of HSP's gapped (non-prelim): 2687
length of query: 368
length of database: 11,318,596
effective HSP length: 101
effective length of query: 267
effective length of database: 8,619,977
effective search space: 2301533859
effective search space used: 2301533859
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC139344.1


