

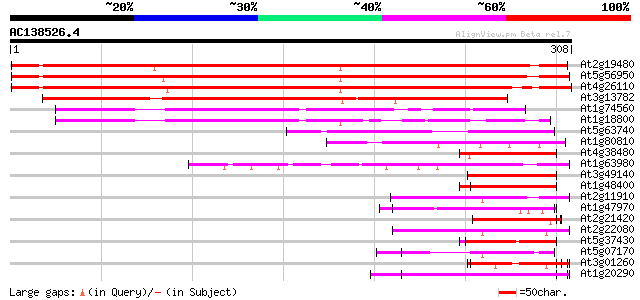
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138526.4 + phase: 0
(308 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g19480 putative nucleosome assembly protein 362 e-100
At5g56950 nucleosome assembly protein 358 2e-99
At4g26110 nucleosome assembly protein I-like protein 346 1e-95
At3g13782 nucleosome assembly protein, putative 261 3e-70
At1g74560 putative SET protein, phospatase 2A inhibitor 115 4e-26
At1g18800 unknown protein 114 7e-26
At5g63740 unknown protein 59 4e-09
At1g80810 unknown protein 53 2e-07
At4g38480 unknown protein 53 2e-07
At1g63980 unknown protein 53 2e-07
At3g49140 putative protein 52 3e-07
At1g48400 52 4e-07
At2g11910 unknown protein 52 5e-07
At1g47970 Unknown protein (T2J15.12) 52 5e-07
At2g21420 Mutator-like transposase 51 7e-07
At2g22080 En/Spm-like transposon protein 50 2e-06
At5g37430 contains similarity to unknown protein (pir||C71432) 50 2e-06
At5g07170 putative protein 49 3e-06
At3g01260 putative aldose 1-epimerase 49 3e-06
At1g20290 hypothetical protein 49 3e-06
>At2g19480 putative nucleosome assembly protein
Length = 379
Score = 362 bits (928), Expect = e-100
Identities = 181/321 (56%), Positives = 238/321 (73%), Gaps = 22/321 (6%)
Query: 2 IQALTQQRYKALQYLFASPIVEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPL 61
+Q L Q L+ L +P V KR+E L+EIQ ++D+++AKFFEERAALEAKYQ LYQPL
Sbjct: 33 LQNLAGQHSDVLENL--TPPVRKRVEFLREIQNQYDEMEAKFFEERAALEAKYQKLYQPL 90
Query: 62 YTKRYEIVNGLAEVEGA---------DIDEPEVKGVPYFWLVALQNNDEIADEITERDED 112
YTKRYEIVNG+ EVEGA + E KGVP FWL+AL+NN+ A+EITERDE
Sbjct: 91 YTKRYEIVNGVVEVEGAAEEVKSEQGEDKSAEEKGVPDFWLIALKNNEITAEEITERDEG 150
Query: 113 ALKYLKDIKYTRTTEPQGFKLEFFFDSNPYFSNTILTKTYHMVDEDEFIMEKAIGTEIEW 172
ALKYLKDIK++R EP+GFKLEFFFD NPYF NT+LTKTYHM+DEDE I+EKA+GTEIEW
Sbjct: 151 ALKYLKDIKWSRVEEPKGFKLEFFFDQNPYFKNTVLTKTYHMIDEDEPILEKALGTEIEW 210
Query: 173 LPGKSLTE-------KKDPNTAKQTIETEKFDSFFNFFNSLEIPKDGMGVDEEADEELQN 225
PGK LT+ KK K +TE +SFFNFF+ ++P D +D++ +ELQ
Sbjct: 211 YPGKCLTQKILKKKPKKGSKNTKPITKTEDCESFFNFFSPPQVPDDDEDLDDDMADELQG 270
Query: 226 EMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDEEEVEVEKEEED 285
+MEHDYDIGSTI++KII +AVSW+TGEA + + D++DDDD+ DE+DDEE+ E ++++E+
Sbjct: 271 QMEHDYDIGSTIKEKIISHAVSWFTGEAVEADDLDIEDDDDEIDEDDDEEDEEDDEDDEE 330
Query: 286 DNYGLDDDEEDDNEDVNNPKK 306
+ DD+++D+ E+ + KK
Sbjct: 331 E----DDEDDDEEEEADQGKK 347
>At5g56950 nucleosome assembly protein
Length = 374
Score = 358 bits (919), Expect = 2e-99
Identities = 184/322 (57%), Positives = 233/322 (72%), Gaps = 24/322 (7%)
Query: 2 IQALTQQRYKALQYLFASPIVEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPL 61
+Q L Q L+ L +P + +R+EVL+EIQ +HD+++ KF EERAALEAKYQ LYQPL
Sbjct: 33 LQNLAGQHSDVLENL--TPKIRRRVEVLREIQGKHDEIETKFREERAALEAKYQKLYQPL 90
Query: 62 YTKRYEIVNGLAEVEGADIDEP---------EVKGVPYFWLVALQNNDEIADEITERDED 112
Y KRYEIVNG EVEGA D E KGVP FWL AL+NND I++EITERDE
Sbjct: 91 YNKRYEIVNGATEVEGAPEDAKMDQGDEKTAEEKGVPSFWLTALKNNDVISEEITERDEG 150
Query: 113 ALKYLKDIKYTRTTEPQGFKLEFFFDSNPYFSNTILTKTYHMVDEDEFIMEKAIGTEIEW 172
AL YLKDIK+ + EP+GFKLEFFFD NPYF NT+LTK YHM+DEDE ++EKAIGTEI+W
Sbjct: 151 ALIYLKDIKWCKIEEPKGFKLEFFFDQNPYFKNTLLTKAYHMIDEDEPLLEKAIGTEIDW 210
Query: 173 LPGKSLTE-------KKDPNTAKQTIETEKFDSFFNFFNSLEIPKDGMGVDEEADEELQN 225
PGK LT+ KK AK +TE +SFFNFFN ++P D +DEE EELQN
Sbjct: 211 YPGKCLTQKILKKKPKKGAKNAKPITKTEDCESFFNFFNPPQVPDDDEDIDEERAEELQN 270
Query: 226 EMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDEEEVEVEKEEED 285
ME DYDIGSTIR+KIIP+AVSW+TGEA +GE ++D+DD+D+ +ED++E+ E E E+E
Sbjct: 271 LMEQDYDIGSTIREKIIPHAVSWFTGEAIEGEEFEIDNDDEDDIDEDEDEDEEDEDEDE- 329
Query: 286 DNYGLDDDEEDDNEDVNNPKKR 307
++D+ED+ E+V+ KK+
Sbjct: 330 -----EEDDEDEEEEVSKTKKK 346
>At4g26110 nucleosome assembly protein I-like protein
Length = 372
Score = 346 bits (887), Expect = 1e-95
Identities = 178/324 (54%), Positives = 229/324 (69%), Gaps = 27/324 (8%)
Query: 2 IQALTQQRYKALQYLFASPIVEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPL 61
+Q L QR L+ L +P V KR++ L++IQ +HD+L+AKF EERA LEAKYQ LYQPL
Sbjct: 33 LQNLAGQRSDVLENL--TPNVRKRVDALRDIQSQHDELEAKFREERAILEAKYQTLYQPL 90
Query: 62 YTKRYEIVNGLAEVEGADIDEPEV----------KGVPYFWLVALQNNDEIADEITERDE 111
Y KRYEIVNG EVE A D+ +V KGVP FWL AL+NND I++E+TERDE
Sbjct: 91 YVKRYEIVNGTTEVELAPEDDTKVDQGEEKTAEEKGVPSFWLTALKNNDVISEEVTERDE 150
Query: 112 DALKYLKDIKYTRTTEPQGFKLEFFFDSNPYFSNTILTKTYHMVDEDEFIMEKAIGTEIE 171
ALKYLKDIK+ + EP+GFKLEFFFD+NPYF NT+LTK+YHM+DEDE ++EKA+GTEI+
Sbjct: 151 GALKYLKDIKWCKIEEPKGFKLEFFFDTNPYFKNTVLTKSYHMIDEDEPLLEKAMGTEID 210
Query: 172 WLPGKSLTE-------KKDPNTAKQTIETEKFDSFFNFFNSLEIPKDGMGVDEEADEELQ 224
W PGK LT+ KK K + E +SFFNFF+ E+P + +DEE E+LQ
Sbjct: 211 WYPGKCLTQKILKKKPKKGSKNTKPITKLEDCESFFNFFSPPEVPDEDEDIDEERAEDLQ 270
Query: 225 NEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDEEEVEVEKEEE 284
N ME DYDIGSTIR+KIIP AVSW+TGEA + E ++DDD++D+ +ED++EE +EE
Sbjct: 271 NLMEQDYDIGSTIREKIIPRAVSWFTGEAMEAEDFEIDDDEEDDIDEDEDEE-----DEE 325
Query: 285 DDNYGLDDDEEDDNEDVNNPKKRV 308
D+ DDD+ED+ E K +
Sbjct: 326 DEE---DDDDEDEEESKTKKKPSI 346
>At3g13782 nucleosome assembly protein, putative
Length = 329
Score = 261 bits (668), Expect = 3e-70
Identities = 144/272 (52%), Positives = 179/272 (64%), Gaps = 24/272 (8%)
Query: 19 SPIVEKRLEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPLYTKRYEIVNGLAEVEGA 78
SP V KR+ LK+IQ HD+L+ KF E++ALEA Y LY+PL+ KRYEIVNG+ E E
Sbjct: 65 SPKVTKRVLFLKDIQVTHDELEEKFLAEKSALEATYDNLYKPLFAKRYEIVNGVVEAEA- 123
Query: 79 DIDEPEVKGVPYFWLVALQNNDEIADEITERDEDALKYLKDIKYTRTTEP-QGFKLEFFF 137
E +GVP FWL+A++ N+ +A+EITERDE ALKYLKDI+ R + + FKLEF F
Sbjct: 124 -----EKEGVPNFWLIAMKTNEMLANEITERDEAALKYLKDIRSCRVEDTSRNFKLEFLF 178
Query: 138 DSNPYFSNTILTKTYHMVDEDEFIMEKAIGTEIEWLPGKSLTEK-----------KDPNT 186
DSN YF N++L+KTYH+ DED ++EK IGT+IEW PGK LT K K N
Sbjct: 179 DSNLYFKNSVLSKTYHVNDEDGPVLEKVIGTDIEWFPGKCLTHKVVVKKKTKKGPKKVNN 238
Query: 187 AKQTIETEKFDSFFNFFNSLEIPK-----DGMGVDEEADEELQNEMEHDYDIGSTIRDKI 241
T +TE +SFFNFF EIP+ D D EELQN M+ DYDI TIRDK+
Sbjct: 239 IPMT-KTENCESFFNFFKPPEIPEIDEVDDYDDFDTIMTEELQNLMDQDYDIAVTIRDKL 297
Query: 242 IPNAVSWYTGEASDGESGDLDDDDDDEDEEDD 273
IP+AVSW+TGEA E D+DDDD DE+ D
Sbjct: 298 IPHAVSWFTGEALVDEDDSDDNDDDDNDEKSD 329
>At1g74560 putative SET protein, phospatase 2A inhibitor
Length = 256
Score = 115 bits (287), Expect = 4e-26
Identities = 73/259 (28%), Positives = 128/259 (49%), Gaps = 34/259 (13%)
Query: 26 LEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPLYTKRYEIVNGLAEVEGADIDEPEV 85
+E L+EIQ + + + K +E +E KY ++ +P+Y KR E++
Sbjct: 31 IEKLQEIQDDLEKINEKASDEVLEVEQKYNVIRKPVYDKRNEVI---------------- 74
Query: 86 KGVPYFWLVALQNNDEIADEITERDEDALKYLKDIKYTRTTEPQ-GFKLEFFFDSNPYFS 144
+ +P FW+ A ++ + D +TE D+ KYL ++ + + G+ + F F SNP+F
Sbjct: 75 QSIPGFWMTAFLSHPALGDLLTEEDQKIFKYLNSLEVEDAKDVKSGYSITFHFTSNPFFE 134
Query: 145 NTILTKTYHMVDEDEFIMEKAIGTEIEWLPGKSLTEKKDPNTAKQTIETEKFDSFFNFFN 204
+ LTKT+ ++E K T I+W GK L + + K +SFF +F
Sbjct: 135 DAKLTKTFTFLEEGT---TKITATPIKWKEGKGLPNGVNHDDKKGNKRALPEESFFTWFT 191
Query: 205 SLEIPKDGMGVDEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDD 264
+ +D A +E+ +E + I++ + N ++++ +A D E D DDD
Sbjct: 192 DAQHKED-------AGDEIHDE------VADIIKEDLWSNPLTYFNNDA-DEEDFDGDDD 237
Query: 265 DDDEDEEDDEEEVEVEKEE 283
D+E EEDD++E E + EE
Sbjct: 238 GDEEGEEDDDDEEEEDGEE 256
>At1g18800 unknown protein
Length = 256
Score = 114 bits (285), Expect = 7e-26
Identities = 78/275 (28%), Positives = 136/275 (49%), Gaps = 52/275 (18%)
Query: 26 LEVLKEIQKEHDDLKAKFFEERAALEAKYQLLYQPLYTKRYEIVNGLAEVEGADIDEPEV 85
+E L+EIQ + + + K +E +E KY ++ +P+Y KR EI+
Sbjct: 27 IEKLQEIQDDLEKINEKASDEVLEVEQKYNVIRKPVYDKRNEII---------------- 70
Query: 86 KGVPYFWLVALQNNDEIADEITERDEDALKYLKDIKYTRTTEPQ-GFKLEFFFDSNPYFS 144
K +P FWL A ++ + + +TE D+ KYL + + + G+ + F F+ NP+F
Sbjct: 71 KTIPDFWLTAFLSHPALGELLTEEDQKIFKYLSSLDVEDAKDVKSGYSITFSFNPNPFFE 130
Query: 145 NTILTKTYHMVDEDEFIMEKAIGTEIEWLPGKSLTE--KKDPNTAKQTIETEKFDSFFNF 202
+ LTKT+ ++E K T I+W GK L + N K+ + E SFF +
Sbjct: 131 DGKLTKTFTFLEEGT---TKITATPIKWKEGKGLANGVNHEKNGNKRALPEE---SFFTW 184
Query: 203 FNSLEIPKDGMGVDEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLD 262
F+ D + E++++EM+ D + I++ + PN ++++ +A D
Sbjct: 185 FS-----------DAQHKEDVEDEMQ-DEQVADIIKEDLWPNPLTYFNNDA--------D 224
Query: 263 DDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDD 297
++D D D++ DEEE E + +E DDDEED+
Sbjct: 225 EEDFDGDDDGDEEEKEGDSDE-------DDDEEDE 252
>At5g63740 unknown protein
Length = 226
Score = 58.5 bits (140), Expect = 4e-09
Identities = 45/148 (30%), Positives = 68/148 (45%), Gaps = 23/148 (15%)
Query: 153 HMVDEDEFIMEKAIGTEIEWLPGKSLTEKKDPNTAKQTIETEKFDSFFNFFNSLEIPKDG 212
++ DED+ MEK I T + +L +T E E + + N E DG
Sbjct: 9 YLRDEDKEEMEKRIRTVVI---DSNLVLSNSTCGICRTAEDEYIKVYDDHNNDGEGDGDG 65
Query: 213 MGV-DEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEE 271
G DE+ DE+ + + D D E D + D DDDDDD D+
Sbjct: 66 DGDGDEDEDEDADADEDEDED-------------------EDEDDDDDDDDDDDDDADDA 106
Query: 272 DDEEEVEVEKEEEDDNYGLDDDEEDDNE 299
DD+E+ + E ++ED++ DDD+E+D E
Sbjct: 107 DDDEDDDDEDDDEDEDDDDDDDDENDEE 134
>At1g80810 unknown protein
Length = 826
Score = 53.1 bits (126), Expect = 2e-07
Identities = 43/151 (28%), Positives = 72/151 (47%), Gaps = 28/151 (18%)
Query: 175 GKSLTEKKDPNT-AKQTIETEKFDSFFNFFNSLEIPKDGMGVDEEADEELQNEMEHDYDI 233
GK+L K+ N +T E ++ SLE D ++E +++ ++ E D
Sbjct: 683 GKNLRSLKELNAETDRTAEEQEV--------SLEAESDDRSEEQEYEDDCSDKKEQSQDK 734
Query: 234 G----STIRDKIIPNAVSWYTGEASDGE-------SGDLDDDDDDEDEEDD--EEEVEVE 280
G + +K PN+ GE S+ E + D++DD+++E+EE D E+E E E
Sbjct: 735 GVEAETKEEEKQYPNSEGESEGEDSESEEEPKWRETDDMEDDEEEEEEEIDHMEDEAEEE 794
Query: 281 KEEEDDNYG------LDDDEEDDNEDVNNPK 305
KEE DD ++ +EE++ ED K
Sbjct: 795 KEEVDDKEASANMSEIEKEEEEEEEDEEKRK 825
>At4g38480 unknown protein
Length = 471
Score = 52.8 bits (125), Expect = 2e-07
Identities = 25/59 (42%), Positives = 36/59 (60%), Gaps = 6/59 (10%)
Query: 248 WYTG------EASDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNED 300
W+ G + SD ES + DDDD EE++ EV+VE ++DDN DDD++DD +D
Sbjct: 404 WFDGYYVDDDDDSDDESSEESSDDDDSSEEEENGEVDVEITKDDDNDHGDDDDDDDEDD 462
Score = 35.4 bits (80), Expect = 0.039
Identities = 30/111 (27%), Positives = 49/111 (44%), Gaps = 17/111 (15%)
Query: 166 IGTEIE-WLPGKSLTEKK-DPNTAKQTIETEKFDSFFNFFNSLEIPKDGMGVDEEADEEL 223
I T+I+ W PG TEK P AKQ F ++ + D +E +D++
Sbjct: 372 IDTDIKIWTPGG--TEKPLSPGNAKQASGFGNPRWFDGYYVDDDDDSDDESSEESSDDDD 429
Query: 224 QNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDE 274
+E E + ++ I + D + GD DDDDD++D+ D +
Sbjct: 430 SSEEEENGEVDVEIT-------------KDDDNDHGDDDDDDDEDDDGDGD 467
Score = 28.9 bits (63), Expect = 3.7
Identities = 16/43 (37%), Positives = 23/43 (53%), Gaps = 8/43 (18%)
Query: 263 DDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDVNNPK 305
DDDDD D+E EE ++DD+ ++EE+ DV K
Sbjct: 411 DDDDDSDDESSEES-----SDDDDS---SEEEENGEVDVEITK 445
>At1g63980 unknown protein
Length = 391
Score = 52.8 bits (125), Expect = 2e-07
Identities = 66/235 (28%), Positives = 110/235 (46%), Gaps = 43/235 (18%)
Query: 99 NDEIADEITERDEDALKY----LKDIKYTRTTEPQGF--KLEFFFDSNPYFSNT---ILT 149
ND+ +D+ E ++DA+K LK + + T PQG + E N Y S IL
Sbjct: 90 NDDDSDKEDESEDDAVKSEPAKLKTV--AKVTRPQGRYKRREKGKLVNSYSSKDLEGILV 147
Query: 150 KTYHMVDEDEFIMEKAIGTEIEWLPGKSLTEKKDPNTAKQTIETEKFDSFFNFFNS---- 205
K D + + A +IE ++E +D + +Q IE E +++ F +
Sbjct: 148 KRTE--DPSPAVCDIADSMDIE-----IISEDQDASIKEQKIE-EPSSNWWGFKSGFVSG 199
Query: 206 -LEIPKDGMGVDEEADEEL--QNEMEHDYDI----------GSTIRDKIIPNAVSWYTGE 252
L K G + ++ ++ +N+ E+ Y++ G I+D+ A Y G+
Sbjct: 200 GLLGAKSGKKKLKASERKMFSENDQENLYNMVQDKATAGKQGLGIKDRPKKIAGVRYEGK 259
Query: 253 ASDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDVNNPKKR 307
+ ++ D DDDDDD+D+E+DEEE E E E DDD++D + + P KR
Sbjct: 260 KTSFDNSDDDDDDDDDDDEEDEEEDEDESE-------ADDDDKDSVIESSLPAKR 307
>At3g49140 putative protein
Length = 1229
Score = 52.4 bits (124), Expect = 3e-07
Identities = 24/49 (48%), Positives = 33/49 (66%)
Query: 252 EASDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNED 300
E D ESGD D +D EDE+++E +E E+EDD+ DDDE+DD+ D
Sbjct: 940 ETEDDESGDDDSEDTGEDEDEEEWVAILEDEDEDDDDDDDDDEDDDDSD 988
Score = 33.5 bits (75), Expect = 0.15
Identities = 13/41 (31%), Positives = 27/41 (65%)
Query: 260 DLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNED 300
D D + +D++ DD+ E E E+E++ + +DE++D++D
Sbjct: 936 DSDFETEDDESGDDDSEDTGEDEDEEEWVAILEDEDEDDDD 976
>At1g48400
Length = 513
Score = 52.0 bits (123), Expect = 4e-07
Identities = 19/47 (40%), Positives = 37/47 (78%)
Query: 254 SDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNED 300
+D + D DDDDDD+D++DD+++ + + +++DD+ DDD++DD++D
Sbjct: 279 NDYDYSDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDD 325
Score = 51.6 bits (122), Expect = 5e-07
Identities = 20/53 (37%), Positives = 36/53 (67%)
Query: 248 WYTGEASDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNED 300
W + D D DDDDDD+D++DD+++ + + +++DD+ DDD++DD+ D
Sbjct: 275 WVSTNDYDYSDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDGD 327
Score = 39.3 bits (90), Expect = 0.003
Identities = 13/34 (38%), Positives = 26/34 (76%)
Query: 255 DGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNY 288
D + D DDDDDD+D++DD+++ + + +++D +Y
Sbjct: 295 DDDDDDDDDDDDDDDDDDDDDDDDDDDDDDDGDY 328
Score = 32.0 bits (71), Expect = 0.43
Identities = 13/26 (50%), Positives = 18/26 (69%)
Query: 255 DGESGDLDDDDDDEDEEDDEEEVEVE 280
D + D DDDDDD+D++DD + VE
Sbjct: 307 DDDDDDDDDDDDDDDDDDDGDYYIVE 332
>At2g11910 unknown protein
Length = 168
Score = 51.6 bits (122), Expect = 5e-07
Identities = 34/112 (30%), Positives = 52/112 (46%), Gaps = 20/112 (17%)
Query: 210 KDGMGVDEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGES----------- 258
K G ++E D ++ + D D D+ N + GE +GE
Sbjct: 61 KRGSRIEENKDASDSDDDDDDEDADEDDDDEDDANDEDFSGGEGDEGEEEADPEDDPVTN 120
Query: 259 ---GDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDVNNPKKR 307
G D+DDDDE+ ++D+E+ + E EEE DDDEEDD++ P K+
Sbjct: 121 GGGGSDDEDDDDEEGDNDDEDEDNEDEEE------DDDEEDDDDVRQPPSKK 166
Score = 30.4 bits (67), Expect = 1.3
Identities = 24/88 (27%), Positives = 38/88 (42%), Gaps = 20/88 (22%)
Query: 216 DEEADEELQNE---MEHDYDIG--------STIRDKIIPNAVSWYTGEASDGESGDLDDD 264
DE+ADE+ +E + D+ G + D + N E D E GD DD+
Sbjct: 81 DEDADEDDDDEDDANDEDFSGGEGDEGEEEADPEDDPVTNGGGGSDDEDDDDEEGDNDDE 140
Query: 265 D---------DDEDEEDDEEEVEVEKEE 283
D DDE+++DD + +K +
Sbjct: 141 DEDNEDEEEDDDEEDDDDVRQPPSKKRK 168
>At1g47970 Unknown protein (T2J15.12)
Length = 198
Score = 51.6 bits (122), Expect = 5e-07
Identities = 35/119 (29%), Positives = 59/119 (49%), Gaps = 39/119 (32%)
Query: 204 NSLEIPKDGMGVDEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDD 263
N E P+ + D+++DEE D+G + D+ + DG GD DD
Sbjct: 11 NFKEEPEIQVLSDDDSDEEQVK------DVGEEVSDE-----------DDDDGSEGDDDD 53
Query: 264 DDDDEDEEDDEEEVEV--------------EKEE--------EDDNYGLDDDEEDDNED 300
D+++E+++DD+++V+V E EE +DD+ G DDD++DD+ED
Sbjct: 54 DEEEEEDDDDDDDVQVLQSLGGPPVQSAEDEDEEGDEDGNGDDDDDDGDDDDDDDDDED 112
Score = 47.4 bits (111), Expect = 1e-05
Identities = 29/98 (29%), Positives = 51/98 (51%), Gaps = 10/98 (10%)
Query: 211 DGMGVDEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDE 270
DG D++ DEE + + + D D+ ++ P S + E G+ DDDDDD D+
Sbjct: 45 DGSEGDDDDDEEEEEDDDDDDDV-QVLQSLGGPPVQSAEDEDEEGDEDGNGDDDDDDGDD 103
Query: 271 EDDEEEVEVEKEEEDDNYGLD---------DDEEDDNE 299
+DD+++ E E E++ + G + +DEED ++
Sbjct: 104 DDDDDDDEDEDVEDEGDLGTEYLVRPVGRAEDEEDASD 141
Score = 34.3 bits (77), Expect = 0.088
Identities = 25/97 (25%), Positives = 49/97 (49%), Gaps = 7/97 (7%)
Query: 216 DEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASD---GESGDLDDDDDDEDEED 272
D++ D++ ++E + D+G+ + + A +ASD E+G +D D+ ED+E+
Sbjct: 104 DDDDDDDEDEDVEDEGDLGTEYLVRPVGRAED--EEDASDFEPEENGVEEDIDEGEDDEN 161
Query: 273 DEEEVEVEKEEEDDNYGLDDDEEDD--NEDVNNPKKR 307
D + E +++E+D +ED + P KR
Sbjct: 162 DNSGGAGKSEAPPKRKRAPEEDEEDSGDEDDDRPPKR 198
Score = 34.3 bits (77), Expect = 0.088
Identities = 21/57 (36%), Positives = 32/57 (55%), Gaps = 17/57 (29%)
Query: 261 LDDDDDD-----------EDEEDDEEEV-----EVEKEEEDD-NYGLDDDEEDDNED 300
++DDDD D++ DEE+V EV E++DD + G DDD+E++ ED
Sbjct: 4 VNDDDDKNFKEEPEIQVLSDDDSDEEQVKDVGEEVSDEDDDDGSEGDDDDDEEEEED 60
Score = 32.7 bits (73), Expect = 0.25
Identities = 20/56 (35%), Positives = 30/56 (52%), Gaps = 13/56 (23%)
Query: 258 SGDLDDDDDDEDEE--------DDEEEVEV-----EKEEEDDNYGLDDDEEDDNED 300
+G DDDD + EE DD +E +V E +EDD+ G + D++DD E+
Sbjct: 2 TGVNDDDDKNFKEEPEIQVLSDDDSDEEQVKDVGEEVSDEDDDDGSEGDDDDDEEE 57
Score = 31.2 bits (69), Expect = 0.74
Identities = 19/48 (39%), Positives = 25/48 (51%), Gaps = 8/48 (16%)
Query: 257 ESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDVNNP 304
E D DDDD E ++DD+EE EEED DDD+ + + P
Sbjct: 37 EVSDEDDDDGSEGDDDDDEE-----EEED---DDDDDDVQVLQSLGGP 76
>At2g21420 Mutator-like transposase
Length = 468
Score = 51.2 bits (121), Expect = 7e-07
Identities = 21/49 (42%), Positives = 36/49 (72%), Gaps = 3/49 (6%)
Query: 255 DGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEE---DDNED 300
D + D DDDDDD+D++DD+++ + ++++EDD Y DD++ DDN+D
Sbjct: 392 DDDDDDDDDDDDDDDDDDDDDDDDDDEDDEDDGYIDSDDDDVDGDDNDD 440
Score = 48.1 bits (113), Expect = 6e-06
Identities = 18/49 (36%), Positives = 36/49 (72%)
Query: 255 DGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDVNN 303
D + D DDDDDD+D++DD+++ + + E+++D+ +D D++D + D N+
Sbjct: 391 DDDDDDDDDDDDDDDDDDDDDDDDDDDEDDEDDGYIDSDDDDVDGDDND 439
Score = 42.4 bits (98), Expect = 3e-04
Identities = 17/48 (35%), Positives = 33/48 (68%), Gaps = 2/48 (4%)
Query: 255 DGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDVN 302
D + D DDDDDD+D++D+++E + + +DD+ +D D+ DD +++
Sbjct: 400 DDDDDDDDDDDDDDDDDDEDDEDDGYIDSDDDD--VDGDDNDDGSELD 445
Score = 42.0 bits (97), Expect = 4e-04
Identities = 17/48 (35%), Positives = 31/48 (64%)
Query: 255 DGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDVN 302
D + D DDDDDD+D+++D+E+ ++DD G D+D+ + + +N
Sbjct: 401 DDDDDDDDDDDDDDDDDEDDEDDGYIDSDDDDVDGDDNDDGSELDGIN 448
Score = 39.3 bits (90), Expect = 0.003
Identities = 16/44 (36%), Positives = 31/44 (70%), Gaps = 2/44 (4%)
Query: 257 ESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNED 300
+ G L DD+D++DD+++ + + +++DD+ DDD+EDD +D
Sbjct: 382 QRGCLSPGCDDDDDDDDDDDDDDDDDDDDDDD--DDDDEDDEDD 423
Score = 33.9 bits (76), Expect = 0.11
Identities = 18/56 (32%), Positives = 30/56 (53%), Gaps = 5/56 (8%)
Query: 255 DGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDD-----NEDVNNPK 305
D + D DD+DD++D D ++ +V+ ++ DD LD +D +DVN K
Sbjct: 409 DDDDDDDDDEDDEDDGYIDSDDDDVDGDDNDDGSELDGINFEDFAVHLPKDVNEQK 464
Score = 29.6 bits (65), Expect = 2.2
Identities = 21/90 (23%), Positives = 40/90 (44%), Gaps = 11/90 (12%)
Query: 214 GVDEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDD 273
G D++ D++ ++ + D D D + D + G +D DDDD D +D+
Sbjct: 389 GCDDDDDDDDDDDDDDDDDDDDDDDDD----------DDEDDEDDGYIDSDDDDVDGDDN 438
Query: 274 EEEVEVEK-EEEDDNYGLDDDEEDDNEDVN 302
++ E++ ED L D + ++ N
Sbjct: 439 DDGSELDGINFEDFAVHLPKDVNEQKDESN 468
>At2g22080 En/Spm-like transposon protein
Length = 140
Score = 50.1 bits (118), Expect = 2e-06
Identities = 33/113 (29%), Positives = 56/113 (49%), Gaps = 16/113 (14%)
Query: 211 DGMGVDEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGES---GDLDDDDDD 267
+G G ++++ ++ + E D K P + GEA D + GD +DD+D
Sbjct: 26 EGNGNNDDSGDDGGEDEEGSADDAEGKETKKGPVSDPDLNGEAGDNDDEPEGDDGNDDED 85
Query: 268 EDEEDDEEEVEVEKEEEDDNYGLDDD-------------EEDDNEDVNNPKKR 307
+D ++++E E E E+E+D+ G +DD +EDD E + PKKR
Sbjct: 86 DDNHENDDEDEEEDEDENDDGGEEDDDEDAEVEEEEEEEDEDDEEALQPPKKR 138
Score = 33.1 bits (74), Expect = 0.20
Identities = 20/76 (26%), Positives = 39/76 (51%), Gaps = 11/76 (14%)
Query: 209 PKDGMGVDEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDE 268
P+ G D+E D+ +N+ E + + D GE D E ++++++++E
Sbjct: 75 PEGDDGNDDEDDDNHENDDEDEEEDEDENDDG----------GEEDDDEDAEVEEEEEEE 124
Query: 269 DEEDDEEEVEVEKEEE 284
D EDDEE ++ K+ +
Sbjct: 125 D-EDDEEALQPPKKRK 139
>At5g37430 contains similarity to unknown protein (pir||C71432)
Length = 607
Score = 49.7 bits (117), Expect = 2e-06
Identities = 20/50 (40%), Positives = 37/50 (74%), Gaps = 1/50 (2%)
Query: 251 GEASDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNED 300
G+ D + GD DDDD D+D +DD+++ + + +++DD+ G D D++DD++D
Sbjct: 557 GDDGDDDDGDDDDDDGDDDGDDDDDDGD-DGDDDDDDDGDDGDDDDDDDD 605
Score = 48.1 bits (113), Expect = 6e-06
Identities = 21/53 (39%), Positives = 32/53 (59%), Gaps = 5/53 (9%)
Query: 248 WYTGEASDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNED 300
W DG+ GD DD DDD+D+ DD+ + +++DD DDD++DD +D
Sbjct: 549 WLNLHDDDGDDGDDDDGDDDDDDGDDDGD-----DDDDDGDDGDDDDDDDGDD 596
Score = 34.7 bits (78), Expect = 0.067
Identities = 13/35 (37%), Positives = 24/35 (68%)
Query: 255 DGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYG 289
D + D DDD DD D++DD++ + + +++DD+ G
Sbjct: 573 DDDGDDDDDDGDDGDDDDDDDGDDGDDDDDDDDGG 607
>At5g07170 putative protein
Length = 542
Score = 49.3 bits (116), Expect = 3e-06
Identities = 26/84 (30%), Positives = 45/84 (52%), Gaps = 23/84 (27%)
Query: 216 DEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDEE 275
D +DEE +E H Y A+D ++ D DDD+DD+D++DD++
Sbjct: 89 DSSSDEEDDSESTHCY---------------------AADDDADDTDDDEDDDDDDDDDD 127
Query: 276 EVEVEKEEEDDNYGLDDDEEDDNE 299
+ + + +++DD+ DDDE D+E
Sbjct: 128 DDDDDDDDDDDDD--DDDESKDSE 149
Score = 46.6 bits (109), Expect = 2e-05
Identities = 31/105 (29%), Positives = 53/105 (49%), Gaps = 14/105 (13%)
Query: 202 FFNSLEIPKDGMGVDEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGES--- 258
F S+ + D+ + M+ I S+ D +I ++ S E D ES
Sbjct: 47 FVTSVSYDMGNISSDDHYCGDDDENMKIAAAIPSSQDDPLIDDSSS---DEEDDSESTHC 103
Query: 259 ----GDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNE 299
D DD DDDED++DD+++ + + +++DD DDD++DD+E
Sbjct: 104 YAADDDADDTDDDEDDDDDDDDDDDDDDDDDD----DDDDDDDDE 144
>At3g01260 putative aldose 1-epimerase
Length = 378
Score = 48.9 bits (115), Expect = 3e-06
Identities = 22/57 (38%), Positives = 35/57 (60%), Gaps = 2/57 (3%)
Query: 252 EASDGESGDLDDDD--DDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDVNNPKK 306
+ D GD DDDD DD D +DD + + + +D+N G DD ++D+N+D ++ KK
Sbjct: 43 DQDDDNDGDHDDDDHDDDNDHDDDNNDHDDDDNNDDNNDGDDDHDDDNNDDGDDEKK 99
Score = 43.5 bits (101), Expect = 1e-04
Identities = 21/53 (39%), Positives = 32/53 (59%), Gaps = 3/53 (5%)
Query: 254 SDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDD---EEDDNEDVNN 303
S ++ D D DDDD D++DD + + + +DDN DD+ ++DDN D NN
Sbjct: 28 SISKANDNDHDDDDHDQDDDNDGDHDDDDHDDDNDHDDDNNDHDDDDNNDDNN 80
Score = 42.7 bits (99), Expect = 2e-04
Identities = 20/55 (36%), Positives = 35/55 (63%), Gaps = 4/55 (7%)
Query: 254 SDGESGDLD-DDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDVNNPKKR 307
+DG+ D D DDD+D D+++++ + + + DDN DDD +DDN D + +K+
Sbjct: 48 NDGDHDDDDHDDDNDHDDDNNDHD---DDDNNDDNNDGDDDHDDDNNDDGDDEKK 99
Score = 42.4 bits (98), Expect = 3e-04
Identities = 17/47 (36%), Positives = 28/47 (59%)
Query: 254 SDGESGDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNED 300
+D + D D DDD++ + DD++ + ++D+N DDD DDN D
Sbjct: 35 NDHDDDDHDQDDDNDGDHDDDDHDDDNDHDDDNNDHDDDDNNDDNND 81
Score = 39.3 bits (90), Expect = 0.003
Identities = 17/57 (29%), Positives = 33/57 (57%), Gaps = 2/57 (3%)
Query: 249 YTGEASDGESGDLDDDDDDED--EEDDEEEVEVEKEEEDDNYGLDDDEEDDNEDVNN 303
+ G +D +D+D D+D ++DD+ + + + ++ DD+ DDD D ++D NN
Sbjct: 20 FKGGVTDHSISKANDNDHDDDDHDQDDDNDGDHDDDDHDDDNDHDDDNNDHDDDDNN 76
>At1g20290 hypothetical protein
Length = 497
Score = 48.9 bits (115), Expect = 3e-06
Identities = 24/91 (26%), Positives = 51/91 (55%)
Query: 216 DEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDEE 275
+EE +EE + E E + + ++ E + E + ++++++E+EE++EE
Sbjct: 393 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 452
Query: 276 EVEVEKEEEDDNYGLDDDEEDDNEDVNNPKK 306
E E E+EEE++ +++EE++ E+ KK
Sbjct: 453 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEKK 483
Score = 48.1 bits (113), Expect = 6e-06
Identities = 23/92 (25%), Positives = 52/92 (56%)
Query: 216 DEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDEE 275
+EE +EE + E E + + ++ E + E + ++++++E+EE++EE
Sbjct: 392 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 451
Query: 276 EVEVEKEEEDDNYGLDDDEEDDNEDVNNPKKR 307
E E E+EEE++ +++EE++ E+ +K+
Sbjct: 452 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEKK 483
Score = 46.2 bits (108), Expect = 2e-05
Identities = 26/102 (25%), Positives = 55/102 (53%), Gaps = 7/102 (6%)
Query: 199 FFNFFNSLEIPKDGMGVDEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGES 258
F+ F S+ +G V E +EE + E E + + ++ E + E
Sbjct: 345 FYKFTPSMVHVGEGTIVQAEEEEEEEEEEEEEEEEEEEEEEEEEEE-------EEEEEEE 397
Query: 259 GDLDDDDDDEDEEDDEEEVEVEKEEEDDNYGLDDDEEDDNED 300
+ ++++++E+EE++EEE E E+EEE++ +++EE++ E+
Sbjct: 398 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 439
Score = 45.1 bits (105), Expect = 5e-05
Identities = 23/92 (25%), Positives = 52/92 (56%)
Query: 216 DEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDEE 275
+EE +EE + E E + + ++ E + E + ++++++E+EE++EE
Sbjct: 400 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 459
Query: 276 EVEVEKEEEDDNYGLDDDEEDDNEDVNNPKKR 307
E E E+EEE++ +++EE++ + + KK+
Sbjct: 460 EEEEEEEEEEEEEEEEEEEEEEKKYMIVKKKK 491
Score = 45.1 bits (105), Expect = 5e-05
Identities = 23/92 (25%), Positives = 51/92 (55%)
Query: 216 DEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDEE 275
+EE +EE + E E + + ++ E + E + ++++++E+EE++EE
Sbjct: 399 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 458
Query: 276 EVEVEKEEEDDNYGLDDDEEDDNEDVNNPKKR 307
E E E+EEE++ +++EE++ + KK+
Sbjct: 459 EEEEEEEEEEEEEEEEEEEEEEEKKYMIVKKK 490
Score = 44.7 bits (104), Expect = 6e-05
Identities = 23/92 (25%), Positives = 50/92 (54%)
Query: 216 DEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDEE 275
+EE +EE + E E + + ++ E + E + ++++++E+EE++EE
Sbjct: 401 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 460
Query: 276 EVEVEKEEEDDNYGLDDDEEDDNEDVNNPKKR 307
E E E+EEE++ +++EE+ + KK+
Sbjct: 461 EEEEEEEEEEEEEEEEEEEEEKKYMIVKKKKK 492
Score = 44.3 bits (103), Expect = 8e-05
Identities = 24/92 (26%), Positives = 49/92 (53%)
Query: 216 DEEADEELQNEMEHDYDIGSTIRDKIIPNAVSWYTGEASDGESGDLDDDDDDEDEEDDEE 275
+EE +EE + E E + + ++ E + E + ++++++E+EE++EE
Sbjct: 402 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 461
Query: 276 EVEVEKEEEDDNYGLDDDEEDDNEDVNNPKKR 307
E E E+EEE++ +++EE V KK+
Sbjct: 462 EEEEEEEEEEEEEEEEEEEEKKYMIVKKKKKK 493
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.310 0.134 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,186,636
Number of Sequences: 26719
Number of extensions: 453470
Number of successful extensions: 17119
Number of sequences better than 10.0: 803
Number of HSP's better than 10.0 without gapping: 553
Number of HSP's successfully gapped in prelim test: 259
Number of HSP's that attempted gapping in prelim test: 4949
Number of HSP's gapped (non-prelim): 4655
length of query: 308
length of database: 11,318,596
effective HSP length: 99
effective length of query: 209
effective length of database: 8,673,415
effective search space: 1812743735
effective search space used: 1812743735
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC138526.4


