

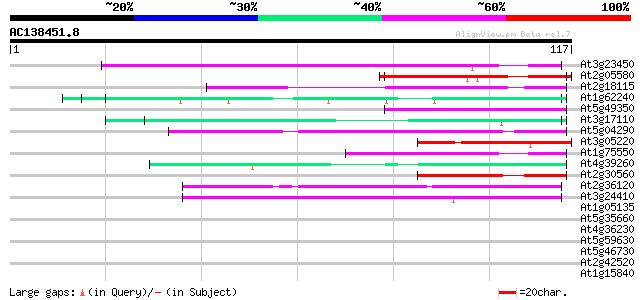
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138451.8 - phase: 0
(117 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g23450 hypothetical protein 45 6e-06
At2g05580 unknown protein 42 4e-05
At2g18115 putative protein 42 5e-05
At1g62240 unknown protein (At1g62240) 41 8e-05
At5g49350 unknown protein 41 1e-04
At3g17110 hypothetical protein 40 2e-04
At5g04290 glycine-rich protein 40 2e-04
At3g05220 unknown protein 40 2e-04
At1g75550 hypothetical protein 40 2e-04
At4g39260 glycine-rich protein (clone AtGRP8) 39 3e-04
At2g30560 putative glycine-rich protein 39 4e-04
At2g36120 unknown protein 39 5e-04
At3g24410 hypothetical protein 38 7e-04
At1g05135 unknown protein 38 0.001
At5g35660 unknown protein 37 0.001
At4g36230 putative glycine-rich cell wall protein 37 0.001
At5g59630 glycine-rich protein - like 37 0.002
At5g46730 unknown protein 37 0.002
At2g42520 putative ATP-dependent RNA helicase 37 0.002
At1g15840 hypothetical protein 37 0.002
>At3g23450 hypothetical protein
Length = 452
Score = 45.1 bits (105), Expect = 6e-06
Identities = 38/96 (39%), Positives = 41/96 (42%), Gaps = 6/96 (6%)
Query: 20 LGKGTLLQEGDRNTNECGGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKG 79
+GKG + G GG I KG G G GI KG G K I G
Sbjct: 264 IGKGGGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGG 323
Query: 80 TGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
G+ G GGG GKGG GGGG G GKGGG
Sbjct: 324 GGFGKGGGIGGGIGKGGGIGGGG------GFGKGGG 353
Score = 38.5 bits (88), Expect = 5e-04
Identities = 38/103 (36%), Positives = 43/103 (40%), Gaps = 14/103 (13%)
Query: 20 LGKGTLLQEGDRNTNECGGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKG 79
+GKG + G GG I KG G G GI KG G K + G
Sbjct: 154 IGKGGGVGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGV--G 211
Query: 80 TGWQTTGRKGGGSGKG-------GKGGGGGYRIPIPGVGKGGG 115
G+ G GGG GKG GKGGG G G+GKGGG
Sbjct: 212 GGFGKGGGVGGGIGKGGGVGGGFGKGGGVG-----GGIGKGGG 249
Score = 37.7 bits (86), Expect = 0.001
Identities = 37/103 (35%), Positives = 43/103 (40%), Gaps = 14/103 (13%)
Query: 20 LGKGTLLQEGDRNTNECGGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKG 79
+GKG + G GG I KG G G G+ KG G K + G
Sbjct: 174 IGKGGGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGVGGGFGKGGGVGGGIGKGGGV--G 231
Query: 80 TGWQTTGRKGGGSGKG-------GKGGGGGYRIPIPGVGKGGG 115
G+ G GGG GKG GKGGG G G+GKGGG
Sbjct: 232 GGFGKGGGVGGGIGKGGGIGGGIGKGGGIG-----GGIGKGGG 269
Score = 37.7 bits (86), Expect = 0.001
Identities = 38/99 (38%), Positives = 41/99 (41%), Gaps = 14/99 (14%)
Query: 20 LGKGTLLQEGDRNTNECGGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKG 79
+GKG + G GG I KG G G GI KG G K I G
Sbjct: 254 IGKGGGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGG 313
Query: 80 TGWQTTGRKGGGSGKG---GKGGGGGYRIPIPGVGKGGG 115
G KGGG G G GKGGG G G+GKGGG
Sbjct: 314 IG------KGGGIGGGGGFGKGGGIG-----GGIGKGGG 341
Score = 37.0 bits (84), Expect = 0.002
Identities = 38/98 (38%), Positives = 42/98 (42%), Gaps = 10/98 (10%)
Query: 20 LGKGTLLQEGDRNTNECGGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKG 79
+GKG + G GG I KG G G G G + G I K I G
Sbjct: 294 IGKGGGIGGGIGKGGGIGGGIGKGGGIGGGGGFGKG-------GGIGGGI-GKGGGIGGG 345
Query: 80 TGWQTTGRKGGGSGKGGK-GGGGGYRIPIPGVGKGGGG 116
G+ G GGG GKGG GGG G I G G GGGG
Sbjct: 346 GGFGKGGGIGGGIGKGGGIGGGFGKGGGIGG-GIGGGG 382
Score = 36.6 bits (83), Expect = 0.002
Identities = 37/96 (38%), Positives = 40/96 (41%), Gaps = 10/96 (10%)
Query: 20 LGKGTLLQEGDRNTNECGGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKG 79
+GKG + G GG I KG G G GI KG G K I G
Sbjct: 144 IGKGGGIGGGIGKGGGVGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGG 203
Query: 80 TGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
G KGGG G GG G GGG G+GKGGG
Sbjct: 204 IG------KGGGVG-GGFGKGGGVG---GGIGKGGG 229
Score = 36.2 bits (82), Expect = 0.003
Identities = 36/96 (37%), Positives = 40/96 (41%), Gaps = 10/96 (10%)
Query: 20 LGKGTLLQEGDRNTNECGGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKG 79
+GKG + G GG I KG G G G+ KG G K I G
Sbjct: 204 IGKGGGVGGGFGKGGGVGGGIGKGGGVGGGFGKGGGVGGGIGKGGGIGGGIGKGGGIGGG 263
Query: 80 TGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
G KGGG G GG G GGG G+GKGGG
Sbjct: 264 IG------KGGGIG-GGIGKGGGIG---GGIGKGGG 289
Score = 36.2 bits (82), Expect = 0.003
Identities = 35/99 (35%), Positives = 40/99 (40%), Gaps = 10/99 (10%)
Query: 20 LGKGTLLQEGDRNTNECGGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKG 79
+GKG + G GG I G G G G G + G I K G
Sbjct: 358 IGKGGGIGGGFGKGGGIGGGIGGGGGFGGGGGFGKG-------GGIGGGI-GKGGGFGGG 409
Query: 80 TGWQTTGRKGGGSG--KGGKGGGGGYRIPIPGVGKGGGG 116
G+ G GGG G KGG GGGG+ G G GGGG
Sbjct: 410 GGFGKGGGIGGGGGFGKGGGFGGGGFGGGGGGGGGGGGG 448
Score = 36.2 bits (82), Expect = 0.003
Identities = 37/96 (38%), Positives = 40/96 (41%), Gaps = 10/96 (10%)
Query: 20 LGKGTLLQEGDRNTNECGGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKG 79
+GKG + G GG I KG G G GI KG G K I G
Sbjct: 224 IGKGGGVGGGFGKGGGVGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGG 283
Query: 80 TGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
G KGGG G GG G GGG G+GKGGG
Sbjct: 284 IG------KGGGIG-GGIGKGGGIG---GGIGKGGG 309
Score = 35.4 bits (80), Expect = 0.005
Identities = 36/95 (37%), Positives = 39/95 (40%), Gaps = 10/95 (10%)
Query: 21 GKGTLLQEGDRNTNECGGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGT 80
GKG + G GG I KG G G G+ KG G K I G
Sbjct: 125 GKGGGIGGGIGKGGGVGGGIGKGGGIGGGIGKGGGVGGGIGKGGGIGGGIGKGGGIGGGI 184
Query: 81 GWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
G KGGG G GG G GGG G+GKGGG
Sbjct: 185 G------KGGGIG-GGIGKGGGIG---GGIGKGGG 209
Score = 35.4 bits (80), Expect = 0.005
Identities = 20/38 (52%), Positives = 20/38 (52%), Gaps = 6/38 (15%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 67 GFGGGAGGGFGGGGGGGGGGGGGG------GGGFGGGG 98
Score = 34.7 bits (78), Expect = 0.008
Identities = 32/81 (39%), Positives = 34/81 (41%), Gaps = 10/81 (12%)
Query: 37 GGAILKGNE--GKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGK 94
GG I KG G G G GI KG G + K I G G GGG GK
Sbjct: 333 GGGIGKGGGIGGGGGFGKGGGIGGGIGKGGGIGGGFGKGGGIGGGIGGGGGFGGGGGFGK 392
Query: 95 GGKGGGGGYRIPIPGVGKGGG 115
GG GG G+GKGGG
Sbjct: 393 GGGIGG--------GIGKGGG 405
Score = 34.3 bits (77), Expect = 0.010
Identities = 19/37 (51%), Positives = 19/37 (51%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
G G G GGG G GG GGGGG G G GGG
Sbjct: 63 GAGGGFGGGAGGGFGGGGGGGGGGGGGGGGGFGGGGG 99
Score = 34.3 bits (77), Expect = 0.010
Identities = 37/96 (38%), Positives = 39/96 (40%), Gaps = 12/96 (12%)
Query: 21 GKGTLLQEGDRNTNECGGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGT 80
GKG + G GG I KG G G GI KG G K I G
Sbjct: 235 GKGGGVGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGIGKGGGIGGGI 294
Query: 81 GWQTTGRKGGG-SGKGGKGGGGGYRIPIPGVGKGGG 115
G KGGG G GKGGG G G+GKGGG
Sbjct: 295 G------KGGGIGGGIGKGGGIG-----GGIGKGGG 319
Score = 33.1 bits (74), Expect = 0.023
Identities = 30/86 (34%), Positives = 33/86 (37%), Gaps = 16/86 (18%)
Query: 37 GGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKG- 95
GG G G G G G G+ G + G G G GGG GKG
Sbjct: 73 GGGFGGGGGGGGGGGGGGGGGFGGGGGFGGG----HGGGVGGGVGGGHGGGVGGGFGKGG 128
Query: 96 ------GKGGGGGYRIPIPGVGKGGG 115
GKGGG G G+GKGGG
Sbjct: 129 GIGGGIGKGGGVG-----GGIGKGGG 149
Score = 32.7 bits (73), Expect = 0.030
Identities = 35/89 (39%), Positives = 39/89 (43%), Gaps = 18/89 (20%)
Query: 37 GGAILKGNEGKSNDGCGDGIEIHC---RKGYVDGTIWDKDISITKGTGWQTTGRKGGGSG 93
GG + G+ G G G G I + G V G I K I G G G GGG G
Sbjct: 109 GGGVGGGHGGGVGGGFGKGGGIGGGIGKGGGVGGGI-GKGGGIGGGIG--KGGGVGGGIG 165
Query: 94 KG-------GKGGGGGYRIPIPGVGKGGG 115
KG GKGGG G G+GKGGG
Sbjct: 166 KGGGIGGGIGKGGGIG-----GGIGKGGG 189
Score = 32.0 bits (71), Expect = 0.051
Identities = 19/39 (48%), Positives = 21/39 (53%), Gaps = 1/39 (2%)
Query: 79 GTGWQTTGRKGGGSGKG-GKGGGGGYRIPIPGVGKGGGG 116
G G G GGG+G G G G GGG+ G G GGGG
Sbjct: 51 GFGGGAGGGVGGGAGGGFGGGAGGGFGGGGGGGGGGGGG 89
Score = 30.8 bits (68), Expect = 0.11
Identities = 24/67 (35%), Positives = 27/67 (39%), Gaps = 12/67 (17%)
Query: 37 GGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGG 96
GG I KG G G G I G+ KG G+ G GGG G GG
Sbjct: 397 GGGIGKGGGFGGGGGFGKGGGIGGGGGF------------GKGGGFGGGGFGGGGGGGGG 444
Query: 97 KGGGGGY 103
GGG G+
Sbjct: 445 GGGGIGH 451
Score = 30.4 bits (67), Expect = 0.15
Identities = 29/87 (33%), Positives = 33/87 (37%), Gaps = 20/87 (22%)
Query: 37 GGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGG 96
GG G G G G G+ G+ G G G+ G GGG G GG
Sbjct: 41 GGGKGGGFGGGFGGGAGGGVGGGAGGGFGGGA----------GGGFGGGGGGGGGGGGGG 90
Query: 97 KGG-------GGGYRIPIPGVGKGGGG 116
GG GGG+ GVG G GG
Sbjct: 91 GGGFGGGGGFGGGHG---GGVGGGVGG 114
Score = 30.4 bits (67), Expect = 0.15
Identities = 19/46 (41%), Positives = 22/46 (47%)
Query: 71 DKDISITKGTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
D+ + G G G GGG+G G GG GG G G GGGG
Sbjct: 35 DEKTLVGGGKGGGFGGGFGGGAGGGVGGGAGGGFGGGAGGGFGGGG 80
Score = 27.7 bits (60), Expect = 0.96
Identities = 18/38 (47%), Positives = 18/38 (47%), Gaps = 5/38 (13%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG GG GGG G GVG G GG
Sbjct: 86 GGGGGGGGFGGGGGFGGGHGGGVG-----GGVGGGHGG 118
Score = 25.8 bits (55), Expect = 3.6
Identities = 17/45 (37%), Positives = 21/45 (45%), Gaps = 4/45 (8%)
Query: 72 KDISITKGTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
+D+S + +T G G G GG GGG GVG G GG
Sbjct: 26 RDVSSGRDEDEKTLVGGGKGGGFGGGFGGGAG----GGVGGGAGG 66
>At2g05580 unknown protein
Length = 302
Score = 42.4 bits (98), Expect = 4e-05
Identities = 22/40 (55%), Positives = 25/40 (62%), Gaps = 1/40 (2%)
Query: 79 GTGWQTTGRKGGGSGKGG-KGGGGGYRIPIPGVGKGGGGK 117
G G G KGGG G+GG KGGGGG + G G GGGG+
Sbjct: 240 GGGQGGGGHKGGGGGQGGHKGGGGGGHVGGGGRGGGGGGR 279
Score = 40.4 bits (93), Expect = 1e-04
Identities = 23/40 (57%), Positives = 26/40 (64%), Gaps = 5/40 (12%)
Query: 78 KGTGWQTTGRKGGGSGKGG-KGGGGGYRIPIPGVGKGGGG 116
KG G G+KGGG G+GG KGGGGG + G KGGGG
Sbjct: 110 KGGGGGQGGQKGGGGGQGGQKGGGGGGQ----GGQKGGGG 145
Score = 38.9 bits (89), Expect = 4e-04
Identities = 23/43 (53%), Positives = 24/43 (55%), Gaps = 7/43 (16%)
Query: 78 KGTGWQTTGRKGGGSGK----GGKGGGGGYRIPIPGVGKGGGG 116
KG G G KGGG G GG+GGGGG R G G GGGG
Sbjct: 249 KGGGGGQGGHKGGGGGGHVGGGGRGGGGGGR---GGGGSGGGG 288
Score = 38.5 bits (88), Expect = 5e-04
Identities = 22/39 (56%), Positives = 24/39 (61%), Gaps = 6/39 (15%)
Query: 79 GTGWQTTGRKGGGSGKGG-KGGGGGYRIPIPGVGKGGGG 116
G G G+KGGG G+GG KGGGGG G KGGGG
Sbjct: 101 GGGGGQGGQKGGGGGQGGQKGGGGG-----QGGQKGGGG 134
Score = 37.4 bits (85), Expect = 0.001
Identities = 30/80 (37%), Positives = 33/80 (40%), Gaps = 9/80 (11%)
Query: 37 GGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGG 96
GG G++G G G G KG G K G G GGG G GG
Sbjct: 228 GGGGQGGHKGGGGGGQGGG----GHKGGGGGQGGHKGGGGGGHVGGGGRGGGGGGRGGGG 283
Query: 97 KGGGGGYRIPIPGVGKGGGG 116
GGGGG G G+GGGG
Sbjct: 284 SGGGGG-----GGSGRGGGG 298
Score = 35.8 bits (81), Expect = 0.004
Identities = 19/39 (48%), Positives = 23/39 (58%), Gaps = 5/39 (12%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGGK 117
G G Q + GGG G+GG+ GGGG G KGGGG+
Sbjct: 144 GGGGQGGQKGGGGGGQGGQKGGGG-----QGGQKGGGGQ 177
Score = 35.8 bits (81), Expect = 0.004
Identities = 22/41 (53%), Positives = 24/41 (57%), Gaps = 6/41 (14%)
Query: 78 KGTGWQTTGRKGGGSGK--GGKGGGGGYRIPIPGVGKGGGG 116
KG G G+KGGG G G KGGGGG + G KGGGG
Sbjct: 120 KGGGGGQGGQKGGGGGGQGGQKGGGGGGQ----GGQKGGGG 156
Score = 35.4 bits (80), Expect = 0.005
Identities = 21/39 (53%), Positives = 23/39 (58%), Gaps = 6/39 (15%)
Query: 79 GTGWQTTGRKGGGSGKGG-KGGGGGYRIPIPGVGKGGGG 116
G G Q + GGG G+GG KGGGGG G KGGGG
Sbjct: 91 GGGGQGGQKGGGGGGQGGQKGGGGG-----QGGQKGGGG 124
Score = 35.0 bits (79), Expect = 0.006
Identities = 19/40 (47%), Positives = 23/40 (57%), Gaps = 5/40 (12%)
Query: 78 KGTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGGK 117
KG G G + GG G+GG+ GGGG G KGGGG+
Sbjct: 152 KGGGGGGQGGQKGGGGQGGQKGGGG-----QGGQKGGGGQ 186
Score = 35.0 bits (79), Expect = 0.006
Identities = 22/40 (55%), Positives = 26/40 (65%), Gaps = 7/40 (17%)
Query: 78 KGTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGGK 117
KG G Q G+KGGG G+GG+ GGGG G KGGGG+
Sbjct: 163 KGGGGQG-GQKGGG-GQGGQKGGGG-----QGGQKGGGGR 195
Score = 34.3 bits (77), Expect = 0.010
Identities = 23/54 (42%), Positives = 29/54 (53%), Gaps = 6/54 (11%)
Query: 65 VDGTIWDKDISITKGTGWQTTGRKGGGSGK--GGKGGGGGYRIPIPGVGKGGGG 116
+DG ++ + G G G+KGGG G G KGGGGG + G KGGGG
Sbjct: 65 LDGEEEEESKNNQGGIGRGQGGQKGGGGGGQGGQKGGGGGGQ----GGQKGGGG 114
Score = 33.9 bits (76), Expect = 0.013
Identities = 21/41 (51%), Positives = 23/41 (55%), Gaps = 7/41 (17%)
Query: 78 KGTGWQTTGRKGGGSGKGG--KGGGGGYRIPIPGVGKGGGG 116
KG G Q + GGG G+GG GGGGG G KGGGG
Sbjct: 181 KGGGGQGGQKGGGGRGQGGMKGGGGGG-----QGGHKGGGG 216
Score = 33.9 bits (76), Expect = 0.013
Identities = 20/39 (51%), Positives = 23/39 (58%), Gaps = 1/39 (2%)
Query: 78 KGTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
KG G Q G+KGGG G KGGGG + + G G GG G
Sbjct: 172 KGGGGQG-GQKGGGGQGGQKGGGGRGQGGMKGGGGGGQG 209
Score = 31.6 bits (70), Expect = 0.066
Identities = 18/39 (46%), Positives = 19/39 (48%), Gaps = 5/39 (12%)
Query: 78 KGTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
+G G G GG G G GGGGG G KGGGG
Sbjct: 195 RGQGGMKGGGGGGQGGHKGGGGGGG-----QGGHKGGGG 228
Score = 31.2 bits (69), Expect = 0.087
Identities = 16/31 (51%), Positives = 20/31 (63%), Gaps = 1/31 (3%)
Query: 86 GRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G +GG G GG GG GG++ G G+GGGG
Sbjct: 218 GGQGGHKGGGGGGGQGGHK-GGGGGGQGGGG 247
Score = 30.4 bits (67), Expect = 0.15
Identities = 13/21 (61%), Positives = 15/21 (70%)
Query: 81 GWQTTGRKGGGSGKGGKGGGG 101
G + G GGGSG+GG GGGG
Sbjct: 281 GGGSGGGGGGGSGRGGGGGGG 301
Score = 30.0 bits (66), Expect = 0.19
Identities = 15/31 (48%), Positives = 18/31 (57%), Gaps = 6/31 (19%)
Query: 86 GRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G +GG G GG GG GG++ G GGGG
Sbjct: 206 GGQGGHKGGGGGGGQGGHK------GGGGGG 230
Score = 29.3 bits (64), Expect = 0.33
Identities = 13/24 (54%), Positives = 14/24 (58%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGG 102
G G G GGG G G+GGGGG
Sbjct: 276 GGGRGGGGSGGGGGGGSGRGGGGG 299
Score = 28.1 bits (61), Expect = 0.73
Identities = 15/30 (50%), Positives = 16/30 (53%)
Query: 86 GRKGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
G GGG G GGGGG + G G GGG
Sbjct: 202 GGGGGGQGGHKGGGGGGGQGGHKGGGGGGG 231
Score = 26.2 bits (56), Expect = 2.8
Identities = 12/24 (50%), Positives = 13/24 (54%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGG 102
G G +G GGG G GGGGG
Sbjct: 278 GRGGGGSGGGGGGGSGRGGGGGGG 301
>At2g18115 putative protein
Length = 296
Score = 42.0 bits (97), Expect = 5e-05
Identities = 28/75 (37%), Positives = 33/75 (43%), Gaps = 23/75 (30%)
Query: 42 KGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGGKGGGG 101
KG++ + DG G GI I G G + + GGG G GG GGGG
Sbjct: 180 KGDDRNNGDGVGIGIGI--------------------GIGGEPSPGSGGGGGGGGGGGGG 219
Query: 102 GYRIPIPGVGKGGGG 116
G PG G GGGG
Sbjct: 220 GSG---PGGGGGGGG 231
Score = 35.0 bits (79), Expect = 0.006
Identities = 16/27 (59%), Positives = 17/27 (62%)
Query: 89 GGGSGKGGKGGGGGYRIPIPGVGKGGG 115
GG SG GG GGGG R+ G GKG G
Sbjct: 135 GGSSGDGGAGGGGSGRVGRRGNGKGDG 161
Score = 32.0 bits (71), Expect = 0.051
Identities = 18/38 (47%), Positives = 20/38 (52%), Gaps = 5/38 (13%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G+G G GGG G GG G G GY G G+G GG
Sbjct: 220 GSGPGGGGGGGGGGGGGGGGVGDGY-----GYGRGWGG 252
Score = 31.6 bits (70), Expect = 0.066
Identities = 20/39 (51%), Positives = 20/39 (51%), Gaps = 1/39 (2%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGY-RIPIPGVGKGGGG 116
G G G GGG G GG G G GY R G G GGGG
Sbjct: 222 GPGGGGGGGGGGGGGGGGVGDGYGYGRGWGGGYGGGGGG 260
Score = 30.8 bits (68), Expect = 0.11
Identities = 27/79 (34%), Positives = 28/79 (35%), Gaps = 23/79 (29%)
Query: 37 GGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGG 96
GG G G G GDG GY G GW G GG G GG
Sbjct: 225 GGGGGGGGGGGGGGGVGDGY------GY--------------GRGW---GGGYGGGGGGG 261
Query: 97 KGGGGGYRIPIPGVGKGGG 115
G GG + G G GGG
Sbjct: 262 WGWGGDSNGGVWGPGNGGG 280
Score = 28.1 bits (61), Expect = 0.73
Identities = 31/94 (32%), Positives = 39/94 (40%), Gaps = 22/94 (23%)
Query: 22 KGTLLQEGDRNTNECGGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTG 81
K +QEGD E G I +G E + GC G +G +G + G G
Sbjct: 94 KARRVQEGD--VEEGGCEICRGRE-RDCGGCTGG------EGSCEGC---NGAGGSSGDG 141
Query: 82 WQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
G GGGSG+ G+ G G G GKG G
Sbjct: 142 ----GAGGGGSGRVGRRGNG------KGDGKGQG 165
>At1g62240 unknown protein (At1g62240)
Length = 227
Score = 41.2 bits (95), Expect = 8e-05
Identities = 34/112 (30%), Positives = 44/112 (38%), Gaps = 12/112 (10%)
Query: 12 GVHEYANWLGKGTLLQEGDRNTN---ECGGAILKGNEGKSNDGCGDGIEIHCRKGYV--- 65
G+ + AN G G G+ N + +C G I+ G G G G G G
Sbjct: 65 GIDDNANGYGDGN----GNGNAHGRADCPGGIVVGGGGGGGGGGGGGGGSGGSNGSFFNG 120
Query: 66 --DGTIWDKDISITKGTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
GT + +G + GGGSG+G GGGGG G G G G
Sbjct: 121 SGSGTGYGSGDGRVSSSGEYSASAGGGGSGEGSGGGGGGDGSSGSGSGSGSG 172
Score = 39.7 bits (91), Expect = 2e-04
Identities = 32/102 (31%), Positives = 41/102 (39%), Gaps = 17/102 (16%)
Query: 21 GKGTLLQEGDRNTNECGGAILKGN------EGKSNDGCGDGIEIHCRKGYVDGTIWDKDI 74
G G + G+ + + GG +G+ +G S G G G G GT D+
Sbjct: 130 GDGRVSSSGEYSASAGGGGSGEGSGGGGGGDGSSGSGSGSG----SGSGSGTGTASGPDV 185
Query: 75 SITKGTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
+ G GGG G GG GGGGG G G G GG
Sbjct: 186 YMHVEGG-------GGGGGGGGGGGGGGVDGSGSGSGSGSGG 220
Score = 39.7 bits (91), Expect = 2e-04
Identities = 34/108 (31%), Positives = 42/108 (38%), Gaps = 15/108 (13%)
Query: 16 YANWLGKGTLLQEGDRNTNECGGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDIS 75
+ N G GT GD + G G S +G G G G DG+ S
Sbjct: 117 FFNGSGSGTGYGSGDGRVSSSGEYSASAGGGGSGEGSGGG-------GGGDGSSGSGSGS 169
Query: 76 IT-KGTGWQTTGR-------KGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
+ G+G T +GGG G GG GGGGG + G G G G
Sbjct: 170 GSGSGSGTGTASGPDVYMHVEGGGGGGGGGGGGGGGGVDGSGSGSGSG 217
>At5g49350 unknown protein
Length = 302
Score = 40.8 bits (94), Expect = 1e-04
Identities = 20/38 (52%), Positives = 21/38 (54%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G TG GGG G GG GGGG + G G GGGG
Sbjct: 58 GVGGGGTGFGGGGLGAGGSGGGGAGGLGSGGTGVGGGG 95
Score = 35.4 bits (80), Expect = 0.005
Identities = 16/28 (57%), Positives = 17/28 (60%)
Query: 89 GGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
GGG G GG GGGG + G G GGGG
Sbjct: 94 GGGLGAGGSGGGGAGGLGGGGTGVGGGG 121
Score = 33.1 bits (74), Expect = 0.023
Identities = 32/94 (34%), Positives = 36/94 (38%), Gaps = 17/94 (18%)
Query: 24 TLLQEGDRNTNECGGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQ 83
T +E D GG I G G G G G G+ G + G G
Sbjct: 29 TYSEEDDSEVGGEGGGIGGGGTGFGGGGTGVG---GGGTGFGGGGL---------GAGGS 76
Query: 84 TTGRKGG-GSGKGGKGGGGGYRIPIPGVGKGGGG 116
G GG GSG G GGGGG + G GGGG
Sbjct: 77 GGGGAGGLGSGGTGVGGGGG----LGAGGSGGGG 106
Score = 33.1 bits (74), Expect = 0.023
Identities = 20/38 (52%), Positives = 21/38 (54%), Gaps = 1/38 (2%)
Query: 79 GTGWQTTGRKG-GGSGKGGKGGGGGYRIPIPGVGKGGG 115
GTG G G GGSG GG GG GG + G G GGG
Sbjct: 88 GTGVGGGGGLGAGGSGGGGAGGLGGGGTGVGGGGTGGG 125
Score = 32.7 bits (73), Expect = 0.030
Identities = 18/36 (50%), Positives = 19/36 (52%), Gaps = 5/36 (13%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGG 114
GTG G GGG G GG GGGG + G G GG
Sbjct: 144 GTGGGGAGGLGGGGGNGGFGGGG-----LGGGGLGG 174
Score = 30.4 bits (67), Expect = 0.15
Identities = 18/41 (43%), Positives = 20/41 (47%), Gaps = 3/41 (7%)
Query: 79 GTGWQTTGRKGGGS---GKGGKGGGGGYRIPIPGVGKGGGG 116
G+G G GGG G GG GGG G+ G G GGG
Sbjct: 101 GSGGGGAGGLGGGGTGVGGGGTGGGTGFNGGGTGFGPFGGG 141
Score = 29.6 bits (65), Expect = 0.25
Identities = 19/40 (47%), Positives = 21/40 (52%), Gaps = 2/40 (5%)
Query: 79 GTGWQTTGRKGGGSGKGG--KGGGGGYRIPIPGVGKGGGG 116
GTG+ G G G+G GG GGGG G G GGGG
Sbjct: 132 GTGFGPFGGGGLGTGGGGAGGLGGGGGNGGFGGGGLGGGG 171
Score = 29.6 bits (65), Expect = 0.25
Identities = 18/39 (46%), Positives = 20/39 (51%), Gaps = 1/39 (2%)
Query: 79 GTGWQTTGRKGG-GSGKGGKGGGGGYRIPIPGVGKGGGG 116
G+G G GG G+G G GG GG GVG GG G
Sbjct: 85 GSGGTGVGGGGGLGAGGSGGGGAGGLGGGGTGVGGGGTG 123
Score = 28.5 bits (62), Expect = 0.56
Identities = 17/37 (45%), Positives = 19/37 (50%), Gaps = 3/37 (8%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
GTG+ G G G GG G GGG G+G GGG
Sbjct: 125 GTGFNGGGTGFGPFGGGGLGTGGG---GAGGLGGGGG 158
Score = 28.1 bits (61), Expect = 0.73
Identities = 30/97 (30%), Positives = 36/97 (36%), Gaps = 9/97 (9%)
Query: 21 GKGTLLQEGDRNTNECGGAILKG-NEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKG 79
G GT G GG G G + G G G+ G G + + G
Sbjct: 61 GGGTGFGGGGLGAGGSGGGGAGGLGSGGTGVGGGGGLGAGGSGGGGAGGLGGGGTGVGGG 120
Query: 80 TGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
TG GGG+G G GGG G+G GGGG
Sbjct: 121 GTGGGTGFNGGGTGFG-PFGGG-------GLGTGGGG 149
Score = 24.6 bits (52), Expect = 8.1
Identities = 13/26 (50%), Positives = 13/26 (50%)
Query: 77 TKGTGWQTTGRKGGGSGKGGKGGGGG 102
T G G G GG G GG G GGG
Sbjct: 145 TGGGGAGGLGGGGGNGGFGGGGLGGG 170
>At3g17110 hypothetical protein
Length = 175
Score = 40.0 bits (92), Expect = 2e-04
Identities = 30/87 (34%), Positives = 31/87 (35%), Gaps = 15/87 (17%)
Query: 29 GDRNTNECGGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRK 88
G N GG G G N G G+G GY G G
Sbjct: 87 GGNNGGGGGGGGRGGGGGGGNGGSGNGSGYGSGSGYGSGA---------------GRGGG 131
Query: 89 GGGSGKGGKGGGGGYRIPIPGVGKGGG 115
GGG G GG GGGGG R G G G G
Sbjct: 132 GGGGGGGGGGGGGGSRGNGSGYGSGYG 158
Score = 39.7 bits (91), Expect = 2e-04
Identities = 34/110 (30%), Positives = 39/110 (34%), Gaps = 25/110 (22%)
Query: 21 GKGTLLQEGDRNTNECGGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGT 80
G G+ G N GG G G G GDG G+ + G
Sbjct: 34 GSGSGYGSGSGGGNGGGGGGGGGGGGGGGGGGGDGSG--------SGSGYGSGYGSGSGY 85
Query: 81 GWQTTGRKGGGSGKGGKGGGG--------------GYRIPIPGVGKGGGG 116
G G GGG G+GG GGGG GY G G+GGGG
Sbjct: 86 GGGNNGGGGGGGGRGGGGGGGNGGSGNGSGYGSGSGYG---SGAGRGGGG 132
Score = 34.3 bits (77), Expect = 0.010
Identities = 18/42 (42%), Positives = 23/42 (53%), Gaps = 6/42 (14%)
Query: 75 SITKGTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
++ +G+G + G G G GG GGGGG G G GGGG
Sbjct: 28 ALGRGSGSGSGYGSGSGGGNGGGGGGGG------GGGGGGGG 63
Score = 31.2 bits (69), Expect = 0.087
Identities = 18/39 (46%), Positives = 19/39 (48%), Gaps = 6/39 (15%)
Query: 78 KGTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
K G + G GSG GG GGGG G G GGGG
Sbjct: 27 KALGRGSGSGSGYGSGSGGGNGGGG------GGGGGGGG 59
Score = 30.8 bits (68), Expect = 0.11
Identities = 19/42 (45%), Positives = 20/42 (47%), Gaps = 3/42 (7%)
Query: 79 GTGWQTTGRKGGGSGKG---GKGGGGGYRIPIPGVGKGGGGK 117
G G G G GSG G G G G GY G G GGGG+
Sbjct: 58 GGGGGGGGGDGSGSGSGYGSGYGSGSGYGGGNNGGGGGGGGR 99
Score = 30.4 bits (67), Expect = 0.15
Identities = 34/112 (30%), Positives = 39/112 (34%), Gaps = 22/112 (19%)
Query: 21 GKGTLLQEGDRNTNECGGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGT 80
G G G N GG G G G G G GY G+ + + G
Sbjct: 36 GSGYGSGSGGGNGGGGGGGGGGGGGGGGGGGDGSGSGSGYGSGYGSGSGYGGGNNGGGGG 95
Query: 81 GWQTTGRKGGGSG---------KG-------GKGGGGGYRIPIPGVGKGGGG 116
G G GGG+G G G+GGGGG G G GGGG
Sbjct: 96 GGGRGGGGGGGNGGSGNGSGYGSGSGYGSGAGRGGGGG------GGGGGGGG 141
>At5g04290 glycine-rich protein
Length = 1493
Score = 39.7 bits (91), Expect = 2e-04
Identities = 27/83 (32%), Positives = 35/83 (41%), Gaps = 5/83 (6%)
Query: 34 NECGGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSG 93
N+ GG G + DG G + G G+ W K + + G+ W GGGS
Sbjct: 1184 NDAGGGSSWGKQDSGGDGSSWGKQ---DGGGDSGSAWGKQNNTSGGSSWGKQSDAGGGSS 1240
Query: 94 KGGKGGGGGYRIPIPGVGKGGGG 116
G + GGGG G GGGG
Sbjct: 1241 WGKQDGGGGG--SSWGKQDGGGG 1261
Score = 35.8 bits (81), Expect = 0.004
Identities = 21/60 (35%), Positives = 26/60 (43%), Gaps = 7/60 (11%)
Query: 43 GNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGGKGGGGG 102
G G S+ G DG G G+ W K + G+ W GGGS G + GGGG
Sbjct: 1246 GGGGGSSWGKQDG-------GGGSGSAWGKQNETSNGSSWGKQNDSGGGSSWGKQDGGGG 1298
Score = 35.8 bits (81), Expect = 0.004
Identities = 26/78 (33%), Positives = 32/78 (40%), Gaps = 10/78 (12%)
Query: 46 GKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTG---------RKGGGSGKGG 96
GK ND G G + G G+ W K G+ W G GGG G G
Sbjct: 1279 GKQNDS-GGGSSWGKQDGGGGGSSWGKQNDGGGGSSWGKQGDGGSKPWNEHSGGGRGFGE 1337
Query: 97 KGGGGGYRIPIPGVGKGG 114
+ GGGG+R G+GG
Sbjct: 1338 RRGGGGFRGGRNQSGRGG 1355
Score = 32.3 bits (72), Expect = 0.039
Identities = 20/58 (34%), Positives = 25/58 (42%), Gaps = 2/58 (3%)
Query: 46 GKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGS--GKGGKGGGG 101
GK + G G G + +G+ W K G+ W GGGS GK GGGG
Sbjct: 1254 GKQDGGGGSGSAWGKQNETSNGSSWGKQNDSGGGSSWGKQDGGGGGSSWGKQNDGGGG 1311
Score = 30.8 bits (68), Expect = 0.11
Identities = 22/76 (28%), Positives = 27/76 (34%), Gaps = 20/76 (26%)
Query: 34 NECGGAILKGNE--------GKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTT 85
N+ GG G + GK NDG G G + W K G+ W
Sbjct: 1148 NDAGGGSSWGKQDNGVGSSWGKQNDGSGGG------------SSWGKQNDAGGGSSWGKQ 1195
Query: 86 GRKGGGSGKGGKGGGG 101
G GS G + GGG
Sbjct: 1196 DSGGDGSSWGKQDGGG 1211
Score = 28.5 bits (62), Expect = 0.56
Identities = 16/49 (32%), Positives = 19/49 (38%)
Query: 67 GTIWDKDISITKGTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
G+ W K G+ W GGGS G + G G G GGG
Sbjct: 1129 GSSWGKQDGDGGGSSWGKENDAGGGSSWGKQDNGVGSSWGKQNDGSGGG 1177
Score = 26.2 bits (56), Expect = 2.8
Identities = 24/66 (36%), Positives = 32/66 (48%), Gaps = 5/66 (7%)
Query: 37 GGAIL-KGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKG 95
GG++ K ++G+S+ G D E K G+ W K G QT R GG G G
Sbjct: 1039 GGSLWGKKDDGESSWGKKDDGESSWGKKDDGGSSWGKKDE--GGYSEQTFDR--GGRGFG 1094
Query: 96 GKGGGG 101
G+ GGG
Sbjct: 1095 GRRGGG 1100
Score = 25.8 bits (55), Expect = 3.6
Identities = 16/31 (51%), Positives = 17/31 (54%), Gaps = 6/31 (19%)
Query: 86 GRKGGGSGKG-GKGGGGGYRIPIPGVGKGGG 115
G KG GSG G G+G GGG G G GG
Sbjct: 398 GGKGEGSGGGKGEGSGGG-----KGEGSRGG 423
Score = 24.6 bits (52), Expect = 8.1
Identities = 10/24 (41%), Positives = 14/24 (57%)
Query: 78 KGTGWQTTGRKGGGSGKGGKGGGG 101
+G+G GGG G+G +GG G
Sbjct: 402 EGSGGGKGEGSGGGKGEGSRGGKG 425
>At3g05220 unknown protein
Length = 541
Score = 39.7 bits (91), Expect = 2e-04
Identities = 20/34 (58%), Positives = 24/34 (69%), Gaps = 3/34 (8%)
Query: 86 GRKGGGSGKGGKGGGGGYRIPI--PGVGKGGGGK 117
G KGGG G G KGGGGG+ IP+ G+G+G GK
Sbjct: 229 GFKGGGGG-GKKGGGGGFEIPVQMKGMGEGKNGK 261
Score = 31.6 bits (70), Expect = 0.066
Identities = 25/74 (33%), Positives = 32/74 (42%), Gaps = 2/74 (2%)
Query: 45 EGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGGK--GGGGG 102
EGK N G G + + G + K G ++ G+K G G K GGGGG
Sbjct: 275 EGKENKGGGKTGKTDAKSGGGGLLGFFKKGKSGNGDEKKSAGKKDGHGGNKVKSHGGGGG 334
Query: 103 YRIPIPGVGKGGGG 116
+ G KGGGG
Sbjct: 335 VQHYDSGPKKGGGG 348
Score = 28.5 bits (62), Expect = 0.56
Identities = 25/79 (31%), Positives = 31/79 (38%), Gaps = 4/79 (5%)
Query: 42 KGNEGKSNDGCGDGIEIHCR-KGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGGK--- 97
KG G G G G EI + KG +G G + G++ G GK GK
Sbjct: 231 KGGGGGGKKGGGGGFEIPVQMKGMGEGKNGKDGKKGKGGEKGKKEGKENKGGGKTGKTDA 290
Query: 98 GGGGGYRIPIPGVGKGGGG 116
GGG + GK G G
Sbjct: 291 KSGGGGLLGFFKKGKSGNG 309
Score = 28.1 bits (61), Expect = 0.73
Identities = 32/84 (38%), Positives = 38/84 (45%), Gaps = 9/84 (10%)
Query: 37 GGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQ---TTGRKGGGSG 93
GG L G K G GD + +K DG +K S G G Q + +KGGG
Sbjct: 293 GGGGLLGFFKKGKSGNGDEKKSAGKK---DGHGGNKVKSHGGGGGVQHYDSGPKKGGGGT 349
Query: 94 KGGKGGGGGYRI-PIPGVGKGGGG 116
KG GG GG I + KGGGG
Sbjct: 350 KG--GGHGGLDIDELMKHSKGGGG 371
Score = 26.9 bits (58), Expect = 1.6
Identities = 15/35 (42%), Positives = 17/35 (47%)
Query: 83 QTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGGK 117
Q G G +GK GK G GG + G GGGK
Sbjct: 250 QMKGMGEGKNGKDGKKGKGGEKGKKEGKENKGGGK 284
Score = 26.2 bits (56), Expect = 2.8
Identities = 20/59 (33%), Positives = 26/59 (43%), Gaps = 5/59 (8%)
Query: 63 GYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGGKG----GGGGYRIPIPGVGKGGGGK 117
G +D + K +S G + G GGG G GGKG G + + G K GGK
Sbjct: 17 GNIDPALLVKKLS-KSGKHAEILGGGGGGGGGGGKGFPNLNGQFANLNMGGNNKPKGGK 74
>At1g75550 hypothetical protein
Length = 167
Score = 39.7 bits (91), Expect = 2e-04
Identities = 23/46 (50%), Positives = 24/46 (52%), Gaps = 6/46 (13%)
Query: 71 DKDISITKGTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
DK S G+G G GGG G GG GGGGG G G GGGG
Sbjct: 53 DKKRSYNGGSGSYRWGWGGGGGGGGGGGGGGG------GGGGGGGG 92
Score = 33.9 bits (76), Expect = 0.013
Identities = 18/31 (58%), Positives = 18/31 (58%), Gaps = 5/31 (16%)
Query: 86 GRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G GGG G GG GGGGG G G GGGG
Sbjct: 74 GGGGGGGGGGGGGGGGG-----GGWGWGGGG 99
Score = 33.5 bits (75), Expect = 0.017
Identities = 19/38 (50%), Positives = 20/38 (52%), Gaps = 5/38 (13%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGG G+ G G GGGG
Sbjct: 71 GGGGGGGGGGGGGGGGGGGGGGWGW-----GGGGGGGG 103
Score = 31.2 bits (69), Expect = 0.087
Identities = 19/40 (47%), Positives = 20/40 (49%), Gaps = 3/40 (7%)
Query: 79 GTGWQTTGRKGGGSG---KGGKGGGGGYRIPIPGVGKGGG 115
G G G GGG G GG GGGG Y+ G GKG G
Sbjct: 78 GGGGGGGGGGGGGGGWGWGGGGGGGGWYKWGCGGGGKGKG 117
Score = 28.5 bits (62), Expect = 0.56
Identities = 12/22 (54%), Positives = 13/22 (58%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGG 100
G GW G GGG GKG +G G
Sbjct: 101 GGGWYKWGCGGGGKGKGREGRG 122
Score = 25.0 bits (53), Expect = 6.2
Identities = 14/33 (42%), Positives = 15/33 (45%)
Query: 84 TTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
TTG + GG G YR G G GGGG
Sbjct: 46 TTGVVVRDKKRSYNGGSGSYRWGWGGGGGGGGG 78
>At4g39260 glycine-rich protein (clone AtGRP8)
Length = 169
Score = 39.3 bits (90), Expect = 3e-04
Identities = 31/88 (35%), Positives = 35/88 (39%), Gaps = 16/88 (18%)
Query: 30 DRNTNECGGAILKGNEGKSN-DGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRK 88
+ N E G ++ NE +S G G G GY G G G G
Sbjct: 66 EMNGKELDGRVITVNEAQSRGSGGGGGGRGGSGGGYRSG-----------GGG----GYS 110
Query: 89 GGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
GGG G GGGGGY G G GGGG
Sbjct: 111 GGGGGGYSGGGGGGYERRSGGYGSGGGG 138
Score = 35.0 bits (79), Expect = 0.006
Identities = 30/97 (30%), Positives = 38/97 (38%), Gaps = 11/97 (11%)
Query: 21 GKGTLLQEGDRNTNECGGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGT 80
G+ + E + GG G+ G G G G GY G G
Sbjct: 74 GRVITVNEAQSRGSGGGGGGRGGSGGGYRSGGGGGYSGGGGGGYSGGG----------GG 123
Query: 81 GWQT-TGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G++ +G G G G GG+G GGG R G G G GG
Sbjct: 124 GYERRSGGYGSGGGGGGRGYGGGGRREGGGYGGGDGG 160
Score = 26.2 bits (56), Expect = 2.8
Identities = 12/25 (48%), Positives = 14/25 (56%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGY 103
G G + G GGG G GGGGG+
Sbjct: 145 GGGRREGGGYGGGDGGSYGGGGGGW 169
>At2g30560 putative glycine-rich protein
Length = 171
Score = 38.9 bits (89), Expect = 4e-04
Identities = 19/31 (61%), Positives = 21/31 (67%), Gaps = 4/31 (12%)
Query: 86 GRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G+ G GSG GGKGGGGG G G+GGGG
Sbjct: 3 GKGGSGSGGGGKGGGGGG----SGGGRGGGG 29
Score = 37.4 bits (85), Expect = 0.001
Identities = 21/39 (53%), Positives = 23/39 (58%), Gaps = 1/39 (2%)
Query: 79 GTGWQTTGRKGGGSGKG-GKGGGGGYRIPIPGVGKGGGG 116
G+G G GGGSG G G GGGGG + G GK GGG
Sbjct: 8 GSGGGGKGGGGGGSGGGRGGGGGGGAKGGCGGGGKSGGG 46
Score = 36.6 bits (83), Expect = 0.002
Identities = 18/36 (50%), Positives = 21/36 (58%)
Query: 81 GWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G ++ G GGG GG GGGGG + G G GGGG
Sbjct: 103 GGKSGGGGGGGKNGGGCGGGGGGKGGKSGGGSGGGG 138
Score = 36.2 bits (82), Expect = 0.003
Identities = 31/91 (34%), Positives = 36/91 (39%), Gaps = 18/91 (19%)
Query: 43 GNEGKSNDGCGDGIEIHC----RKGYVDGTIWDKDISITKGTGWQTTGRKGG-------- 90
G GKS G G G + R Y+ ++ D G G + G GG
Sbjct: 38 GGGGKSGGGGGGGGYMVAPGSNRSSYISRDNFESDPKGGSGGGGKGGGGGGGISGGGAGG 97
Query: 91 ----GSGKGGKGGGGGYRIPIPGVGKGGGGK 117
G GK G GGGGG G G GGGGK
Sbjct: 98 KSGCGGGKSGGGGGGGKN--GGGCGGGGGGK 126
Score = 34.3 bits (77), Expect = 0.010
Identities = 21/55 (38%), Positives = 22/55 (39%), Gaps = 16/55 (29%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRI----------------PIPGVGKGGGGK 117
G G G GGG GG GGGGGY + P G GGGGK
Sbjct: 28 GGGGGAKGGCGGGGKSGGGGGGGGYMVAPGSNRSSYISRDNFESDPKGGSGGGGK 82
Score = 34.3 bits (77), Expect = 0.010
Identities = 20/39 (51%), Positives = 23/39 (58%), Gaps = 4/39 (10%)
Query: 79 GTGWQTTGRKGGGSGKGGKGG-GGGYRIPIPGVGKGGGG 116
G G + G +GGG G G KGG GGG + G G GGGG
Sbjct: 16 GGGGGSGGGRGGGGGGGAKGGCGGGGK---SGGGGGGGG 51
>At2g36120 unknown protein
Length = 255
Score = 38.5 bits (88), Expect = 5e-04
Identities = 31/79 (39%), Positives = 33/79 (41%), Gaps = 3/79 (3%)
Query: 37 GGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGG 96
GG GN G G G+G H GY G + G G G GGG G GG
Sbjct: 168 GGGGGGGNGGGGGGGSGEG-GAH-GGGYGAGGGAGEGYGGGAGAGGHGGGG-GGGGGSGG 224
Query: 97 KGGGGGYRIPIPGVGKGGG 115
GGGGG G G GGG
Sbjct: 225 GGGGGGGYAAASGYGHGGG 243
Score = 35.8 bits (81), Expect = 0.004
Identities = 17/28 (60%), Positives = 18/28 (63%)
Query: 89 GGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
GGG+G GG GGGGG G G GGGG
Sbjct: 204 GGGAGAGGHGGGGGGGGGSGGGGGGGGG 231
Score = 34.3 bits (77), Expect = 0.010
Identities = 23/67 (34%), Positives = 27/67 (39%)
Query: 49 NDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGGKGGGGGYRIPIP 108
N G G+ + G G+ + G G G GGG G G GG GG P
Sbjct: 41 NSGLSAGLGVGIGGGPGGGSGYGGGSGEGGGAGGHGEGHIGGGGGGGHGGGAGGGGGGGP 100
Query: 109 GVGKGGG 115
G G GGG
Sbjct: 101 GGGYGGG 107
Score = 32.3 bits (72), Expect = 0.039
Identities = 19/37 (51%), Positives = 19/37 (51%), Gaps = 1/37 (2%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
G G G GGG G GG GGGGG G G GGG
Sbjct: 115 GYGGGEAGGHGGGGG-GGAGGGGGGGGGAHGGGYGGG 150
Score = 32.3 bits (72), Expect = 0.039
Identities = 18/37 (48%), Positives = 19/37 (50%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
G G G GGGSG+GG G GG G G GGG
Sbjct: 95 GGGGGPGGGYGGGSGEGGGAGYGGGEAGGHGGGGGGG 131
Score = 32.0 bits (71), Expect = 0.051
Identities = 15/28 (53%), Positives = 15/28 (53%)
Query: 89 GGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
GGG G GG GGGG G G G GG
Sbjct: 170 GGGGGNGGGGGGGSGEGGAHGGGYGAGG 197
Score = 31.6 bits (70), Expect = 0.066
Identities = 15/27 (55%), Positives = 15/27 (55%)
Query: 89 GGGSGKGGKGGGGGYRIPIPGVGKGGG 115
GGG G GG GGGG P G G G G
Sbjct: 83 GGGGGHGGGAGGGGGGGPGGGYGGGSG 109
Score = 31.6 bits (70), Expect = 0.066
Identities = 20/39 (51%), Positives = 21/39 (53%), Gaps = 2/39 (5%)
Query: 79 GTGWQTTGRKGGGSGKG-GKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G G G+GGG GY G G GGGG
Sbjct: 91 GAGGGGGGGPGGGYGGGSGEGGGAGYGGGEAG-GHGGGG 128
Score = 31.2 bits (69), Expect = 0.087
Identities = 16/35 (45%), Positives = 17/35 (47%)
Query: 81 GWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
G + G GGG G G GGGGG G G G G
Sbjct: 118 GGEAGGHGGGGGGGAGGGGGGGGGAHGGGYGGGQG 152
Score = 30.8 bits (68), Expect = 0.11
Identities = 18/38 (47%), Positives = 18/38 (47%), Gaps = 6/38 (15%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GGGGG G G GGGG
Sbjct: 150 GQGAGAGGGYGGGGAGGHGGGGGG------GNGGGGGG 181
Score = 30.8 bits (68), Expect = 0.11
Identities = 18/43 (41%), Positives = 24/43 (54%), Gaps = 10/43 (23%)
Query: 79 GTGWQTTGRKGGGSGKGGKG-----GGGGYRIPIPGVGKGGGG 116
G+G+ +GGG+G G+G GGGG+ G G GGGG
Sbjct: 59 GSGYGGGSGEGGGAGGHGEGHIGGGGGGGH-----GGGAGGGG 96
Score = 30.4 bits (67), Expect = 0.15
Identities = 27/80 (33%), Positives = 28/80 (34%), Gaps = 5/80 (6%)
Query: 37 GGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGG 96
GG G G G G G GY G G G G GGG G
Sbjct: 89 GGGAGGGGGGGPGGGYGGGSGEGGGAGYGGGEAGGHG-----GGGGGGAGGGGGGGGGAH 143
Query: 97 KGGGGGYRIPIPGVGKGGGG 116
GG GG + G G GGGG
Sbjct: 144 GGGYGGGQGAGAGGGYGGGG 163
Score = 30.4 bits (67), Expect = 0.15
Identities = 25/79 (31%), Positives = 30/79 (37%), Gaps = 17/79 (21%)
Query: 37 GGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGG 96
GG+ G G+ G G +G++ G G G G GGG G G
Sbjct: 58 GGSGYGGGSGEGGGAGGHG------EGHIGG-----------GGGGGHGGGAGGGGGGGP 100
Query: 97 KGGGGGYRIPIPGVGKGGG 115
GG GG G G GGG
Sbjct: 101 GGGYGGGSGEGGGAGYGGG 119
Score = 30.4 bits (67), Expect = 0.15
Identities = 27/72 (37%), Positives = 30/72 (41%), Gaps = 11/72 (15%)
Query: 43 GNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGGKGGGGG 102
G G + G G G H GY G + G G G GGG G GG GGGGG
Sbjct: 127 GGGGGAGGGGGGGGGAH-GGGYGGG----QGAGAGGGYGGGGAGGHGGGGG-GGNGGGGG 180
Query: 103 YRIPIPGVGKGG 114
G G+GG
Sbjct: 181 -----GGSGEGG 187
Score = 30.0 bits (66), Expect = 0.19
Identities = 26/80 (32%), Positives = 29/80 (35%), Gaps = 9/80 (11%)
Query: 37 GGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGG 96
GG G+ G G G G G G + G G+ G G G G GG
Sbjct: 117 GGGEAGGHGGGGGGGAGGG---GGGGGGAHGGGYGGGQGAGAGGGYGGGGAGGHGGGGGG 173
Query: 97 KGGGGGYRIPIPGVGKGGGG 116
GGGG G G G GG
Sbjct: 174 GNGGGG------GGGSGEGG 187
Score = 28.9 bits (63), Expect = 0.43
Identities = 19/47 (40%), Positives = 20/47 (42%), Gaps = 9/47 (19%)
Query: 79 GTGWQTTGRKGGGSGKG---------GKGGGGGYRIPIPGVGKGGGG 116
G G G GGGSG+G G G G GY G GGGG
Sbjct: 170 GGGGGNGGGGGGGSGEGGAHGGGYGAGGGAGEGYGGGAGAGGHGGGG 216
Score = 28.5 bits (62), Expect = 0.56
Identities = 17/38 (44%), Positives = 19/38 (49%), Gaps = 7/38 (18%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G+ G G G G GG G GG+ G G GGGG
Sbjct: 190 GGGYGAGG--GAGEGYGGGAGAGGH-----GGGGGGGG 220
Score = 26.2 bits (56), Expect = 2.8
Identities = 16/38 (42%), Positives = 16/38 (42%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G G G G G GGG G G GGGG
Sbjct: 184 GEGGAHGGGYGAGGGAGEGYGGGAGAGGHGGGGGGGGG 221
Score = 25.8 bits (55), Expect = 3.6
Identities = 11/25 (44%), Positives = 12/25 (48%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGY 103
G G+ G G G GG G GGY
Sbjct: 229 GGGYAAASGYGHGGGAGGGEGSGGY 253
>At3g24410 hypothetical protein
Length = 240
Score = 38.1 bits (87), Expect = 7e-04
Identities = 28/80 (35%), Positives = 34/80 (42%), Gaps = 1/80 (1%)
Query: 37 GGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGG-SGKG 95
GG +KG EG +G G + G +G + G G G GGG +G
Sbjct: 156 GGEGVKGGEGVVGEGVKGGEGVVGDGGRGEGVKGGGGEGVEGGIGGTVVGGGGGGRAGGR 215
Query: 96 GKGGGGGYRIPIPGVGKGGG 115
GGGGG R VG GGG
Sbjct: 216 AVGGGGGGRAGGRAVGGGGG 235
Score = 32.0 bits (71), Expect = 0.051
Identities = 26/71 (36%), Positives = 32/71 (44%), Gaps = 8/71 (11%)
Query: 48 SNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGGKGGGGGYRIPI 107
S + C G E G VDG + D G G +GGG G G GG G +
Sbjct: 44 SLEDCAGGEE----PGVVDGVLGDLPPEGLAGGGGLRLPTRGGGVGVEGGGGDLG---GV 96
Query: 108 PGV-GKGGGGK 117
GV G+GGGG+
Sbjct: 97 EGVAGEGGGGE 107
Score = 27.3 bits (59), Expect = 1.3
Identities = 22/67 (32%), Positives = 29/67 (42%), Gaps = 6/67 (8%)
Query: 53 GDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSG--KGGKGGGGGYRIPIPGV 110
G G+ + R G V D+ +G G GGG KG GGG G + +
Sbjct: 72 GGGLRLPTRGGGVGVEGGGGDLGGVEGVA----GEGGGGEEGLKGEDGGGEGVKGGEGML 127
Query: 111 GKGGGGK 117
G GGGG+
Sbjct: 128 GDGGGGE 134
Score = 25.0 bits (53), Expect = 6.2
Identities = 16/36 (44%), Positives = 19/36 (52%), Gaps = 4/36 (11%)
Query: 86 GRKGGGSG-KGGK---GGGGGYRIPIPGVGKGGGGK 117
G GGG G KGG+ G GGG + G+G G K
Sbjct: 112 GEDGGGEGVKGGEGMLGDGGGGEGVVGDGGRGDGVK 147
>At1g05135 unknown protein
Length = 384
Score = 37.7 bits (86), Expect = 0.001
Identities = 28/79 (35%), Positives = 32/79 (40%), Gaps = 10/79 (12%)
Query: 37 GGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGG 96
GG + G G G G G + G+ G G G G GGG+G GG
Sbjct: 240 GGGVGGGAGGGGGGGGGGGGGANGGSGHGSGF----------GAGGGVGGGVGGGAGGGG 289
Query: 97 KGGGGGYRIPIPGVGKGGG 115
GGGGG G G GGG
Sbjct: 290 GGGGGGGGGVGGGSGHGGG 308
Score = 35.0 bits (79), Expect = 0.006
Identities = 29/96 (30%), Positives = 32/96 (33%), Gaps = 8/96 (8%)
Query: 21 GKGTLLQEGDRNTNECGGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGT 80
G G+ G GG + G G G G G G+ G
Sbjct: 188 GHGSGFGAGGGIGGGAGGGVGGGGGGGGGGGGGGGAN--------GGSGHGSGFGAGGGV 239
Query: 81 GWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G GGG G GG GGGG G G G GG
Sbjct: 240 GGGVGGGAGGGGGGGGGGGGGANGGSGHGSGFGAGG 275
Score = 34.7 bits (78), Expect = 0.008
Identities = 29/81 (35%), Positives = 33/81 (39%), Gaps = 10/81 (12%)
Query: 37 GGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGG 96
GG + G G G G G + G+ G G G G GGG G GG
Sbjct: 278 GGGVGGGAGGGGGGGGGGGGGVGGGSGHGGG------FGAGGGLGGGAGGGLGGGGGAGG 331
Query: 97 KGGGG-GYRIPIPGVGKGGGG 116
GGGG G+ GVG G GG
Sbjct: 332 GGGGGLGHG---GGVGGGHGG 349
Score = 33.5 bits (75), Expect = 0.017
Identities = 28/79 (35%), Positives = 31/79 (38%), Gaps = 14/79 (17%)
Query: 37 GGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGG 96
GG G G +N G G G G G + G G G GGG G GG
Sbjct: 250 GGGGGGGGGGGANGGSGHGSGFGAGGGVGGG--------VGGGAGG---GGGGGGGGGGG 298
Query: 97 KGGGGGYRIPIPGVGKGGG 115
GGG G+ G G GGG
Sbjct: 299 VGGGSGHG---GGFGAGGG 314
Score = 32.3 bits (72), Expect = 0.039
Identities = 18/37 (48%), Positives = 18/37 (48%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
G G G GGG G GG GGGG G G GGG
Sbjct: 81 GAGGGVGGGAGGGLGGGGGAGGGGGGGIGGGSGHGGG 117
Score = 32.0 bits (71), Expect = 0.051
Identities = 27/79 (34%), Positives = 30/79 (37%), Gaps = 14/79 (17%)
Query: 37 GGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGG 96
GG G G +N G G G G G + G G G GGG G GG
Sbjct: 212 GGGGGGGGGGGANGGSGHGSGFGAGGGVGGG--------VGGGAGG---GGGGGGGGGGG 260
Query: 97 KGGGGGYRIPIPGVGKGGG 115
GG G+ G G GGG
Sbjct: 261 ANGGSGHG---SGFGAGGG 276
Score = 32.0 bits (71), Expect = 0.051
Identities = 28/80 (35%), Positives = 29/80 (36%), Gaps = 12/80 (15%)
Query: 37 GGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGG 96
GG G G G G G I G+ G G G G GGG G GG
Sbjct: 87 GGGAGGGLGGGGGAGGGGGGGIGGGSGHGGG------FGAGGGVGGGAGGGIGGGGGAGG 140
Query: 97 KGGGGGYRIPIPGVGKGGGG 116
GGGG G G G GG
Sbjct: 141 GGGGG------VGGGSGHGG 154
Score = 31.6 bits (70), Expect = 0.066
Identities = 19/39 (48%), Positives = 19/39 (48%), Gaps = 2/39 (5%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGG--GGYRIPIPGVGKGGG 115
G G G GGG G GG GGG GG G G GGG
Sbjct: 85 GVGGGAGGGLGGGGGAGGGGGGGIGGGSGHGGGFGAGGG 123
Score = 31.6 bits (70), Expect = 0.066
Identities = 27/79 (34%), Positives = 32/79 (40%), Gaps = 9/79 (11%)
Query: 37 GGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGG 96
GG I G G + G G G+ G G + + G G G GG G GG
Sbjct: 129 GGGI--GGGGGAGGGGGGGVG----GGSGHGGGFGAGGGVGGGAGGIGGGGGAGGGGGGG 182
Query: 97 KGGGGGYRIPIPGVGKGGG 115
GGG G+ G G GGG
Sbjct: 183 VGGGSGHG---SGFGAGGG 198
Score = 31.6 bits (70), Expect = 0.066
Identities = 18/38 (47%), Positives = 20/38 (52%), Gaps = 6/38 (15%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G + G GGG G+GG G GG GVG G GG
Sbjct: 61 GGGRGSGGGIGGGGGQGGGFGAGG------GVGGGAGG 92
Score = 31.2 bits (69), Expect = 0.087
Identities = 20/45 (44%), Positives = 21/45 (46%), Gaps = 7/45 (15%)
Query: 79 GTGWQTTGRKGGGSGKG-------GKGGGGGYRIPIPGVGKGGGG 116
G G G GGGSG G G GGG G + G G GGGG
Sbjct: 174 GAGGGGGGGVGGGSGHGSGFGAGGGIGGGAGGGVGGGGGGGGGGG 218
Score = 29.6 bits (65), Expect = 0.25
Identities = 21/42 (50%), Positives = 21/42 (50%), Gaps = 4/42 (9%)
Query: 79 GTGWQTTGRKGGGSGKG---GKGGG-GGYRIPIPGVGKGGGG 116
G G G GGGSG G G GGG GG I G G GGG
Sbjct: 137 GAGGGGGGGVGGGSGHGGGFGAGGGVGGGAGGIGGGGGAGGG 178
Score = 29.6 bits (65), Expect = 0.25
Identities = 24/68 (35%), Positives = 25/68 (36%)
Query: 37 GGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGG 96
GG G G G G G + G G I I G G G G GSG G
Sbjct: 316 GGGAGGGLGGGGGAGGGGGGGLGHGGGVGGGHGGGVGIGIGIGIGVGVGGGSGQGSGSGS 375
Query: 97 KGGGGGYR 104
GGGG R
Sbjct: 376 GSGGGGGR 383
Score = 29.3 bits (64), Expect = 0.33
Identities = 17/35 (48%), Positives = 20/35 (56%), Gaps = 2/35 (5%)
Query: 81 GWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
G + GR G G+GG GG GG R G+G GGG
Sbjct: 43 GRRCAGR--GRFGRGGGGGFGGGRGSGGGIGGGGG 75
Score = 28.9 bits (63), Expect = 0.43
Identities = 18/39 (46%), Positives = 20/39 (51%), Gaps = 1/39 (2%)
Query: 79 GTGWQTTGRKGGGSGKGGK-GGGGGYRIPIPGVGKGGGG 116
G G G +GGG G GG GGG G + G GGGG
Sbjct: 67 GGGIGGGGGQGGGFGAGGGVGGGAGGGLGGGGGAGGGGG 105
Score = 28.9 bits (63), Expect = 0.43
Identities = 23/64 (35%), Positives = 27/64 (41%), Gaps = 4/64 (6%)
Query: 53 GDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGGK-GGGGGYRIPIPGVG 111
GDG + + D W + + G G GGG G GG GGGGG G G
Sbjct: 25 GDGSVVGPARFRDDDCRWGRRCAGRGRFGRGGGGGFGGGRGSGGGIGGGGGQG---GGFG 81
Query: 112 KGGG 115
GGG
Sbjct: 82 AGGG 85
Score = 28.5 bits (62), Expect = 0.56
Identities = 19/41 (46%), Positives = 19/41 (46%), Gaps = 3/41 (7%)
Query: 79 GTGWQTTGRKGGGSG---KGGKGGGGGYRIPIPGVGKGGGG 116
G G G G GSG GG GGG G G G GGGG
Sbjct: 256 GGGGGANGGSGHGSGFGAGGGVGGGVGGGAGGGGGGGGGGG 296
Score = 28.5 bits (62), Expect = 0.56
Identities = 15/28 (53%), Positives = 15/28 (53%)
Query: 89 GGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
GGG G GG GGG I G GGGG
Sbjct: 153 GGGFGAGGGVGGGAGGIGGGGGAGGGGG 180
Score = 27.7 bits (60), Expect = 0.96
Identities = 17/39 (43%), Positives = 19/39 (48%), Gaps = 4/39 (10%)
Query: 78 KGTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
+G+G G G G G G GG GG G G GGGG
Sbjct: 64 RGSGGGIGGGGGQGGGFGAGGGVGGG----AGGGLGGGG 98
Score = 27.7 bits (60), Expect = 0.96
Identities = 15/28 (53%), Positives = 15/28 (53%), Gaps = 5/28 (17%)
Query: 89 GGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
GGG G G G GGG G G GGGG
Sbjct: 159 GGGVGGGAGGIGGG-----GGAGGGGGG 181
Score = 26.9 bits (58), Expect = 1.6
Identities = 17/31 (54%), Positives = 17/31 (54%), Gaps = 1/31 (3%)
Query: 86 GRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
GR G G G GG GGG G I G G GGG
Sbjct: 50 GRFGRGGG-GGFGGGRGSGGGIGGGGGQGGG 79
Score = 26.2 bits (56), Expect = 2.8
Identities = 17/38 (44%), Positives = 17/38 (44%), Gaps = 5/38 (13%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G G G G GG GG G G G GGGG
Sbjct: 147 GGGSGHGGGFGAGGGVGGGAGGIG-----GGGGAGGGG 179
>At5g35660 unknown protein
Length = 343
Score = 37.4 bits (85), Expect = 0.001
Identities = 22/38 (57%), Positives = 24/38 (62%), Gaps = 8/38 (21%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G GWQ G +GGG KGG GGGGG+R G GGGG
Sbjct: 304 GGGWQGGGGRGGG-WKGG-GGGGGWR------GGGGGG 333
Score = 37.0 bits (84), Expect = 0.002
Identities = 28/77 (36%), Positives = 38/77 (48%), Gaps = 7/77 (9%)
Query: 42 KGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGG--KGG 99
KG G+ G G G E+ + ++G ++I G G G +GGG G GG +GG
Sbjct: 225 KGGGGRGGGG-GGGAELENEE--INGA--SEEIQWQGGGGGHGGGWQGGGGGHGGGWQGG 279
Query: 100 GGGYRIPIPGVGKGGGG 116
GGG+ G G GGG
Sbjct: 280 GGGHGGGWQGGGGRGGG 296
Score = 36.6 bits (83), Expect = 0.002
Identities = 22/43 (51%), Positives = 25/43 (57%), Gaps = 7/43 (16%)
Query: 78 KGTGWQTTGRKGGG----SGKGGKGGGGGYRIPIPGVGKGGGG 116
KG G Q G KGGG KGG G GGG++ G G+GGGG
Sbjct: 195 KGGGGQGGGWKGGGGQGGGWKGGGGQGGGWK---GGGGRGGGG 234
Score = 36.2 bits (82), Expect = 0.003
Identities = 28/74 (37%), Positives = 34/74 (45%), Gaps = 5/74 (6%)
Query: 44 NEGKSNDGCGD-GIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGGKGGGGG 102
N+ K + GD G+E K + D+ KG G G KGGG GG GGGG
Sbjct: 49 NDAKDSLNKGDVGVEATVEKEEENNYEIDQHGGGWKGGGGHGGGWKGGGGHGGGWKGGGG 108
Query: 103 YRIPIPGVGKGGGG 116
+ G KGGGG
Sbjct: 109 H----GGGWKGGGG 118
Score = 35.8 bits (81), Expect = 0.004
Identities = 31/84 (36%), Positives = 38/84 (44%), Gaps = 15/84 (17%)
Query: 42 KGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGT---GW-----QTTGRKGGGSG 93
KG G G G G E+ + +D D+D+ +G GW Q G KGGG
Sbjct: 124 KGGGGGRGGGGGGGAELETVE--IDSAS-DEDVQNYRGGHGGGWKEGGGQGGGWKGGGGQ 180
Query: 94 KGGKGGGGGYRIPIPGVGKGGGGK 117
GG GGGG G KGGGG+
Sbjct: 181 GGGWKGGGGQ----GGGWKGGGGQ 200
Score = 35.4 bits (80), Expect = 0.005
Identities = 21/40 (52%), Positives = 22/40 (54%), Gaps = 4/40 (10%)
Query: 78 KGTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGGK 117
KG G Q G KGGG GG GGGG G KGGGG+
Sbjct: 185 KGGGGQGGGWKGGGGQGGGWKGGGGQ----GGGWKGGGGQ 220
Score = 35.4 bits (80), Expect = 0.005
Identities = 21/40 (52%), Positives = 22/40 (54%), Gaps = 4/40 (10%)
Query: 78 KGTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGGK 117
KG G Q G KGGG GG GGGG G KGGGG+
Sbjct: 175 KGGGGQGGGWKGGGGQGGGWKGGGGQ----GGGWKGGGGQ 210
Score = 35.0 bits (79), Expect = 0.006
Identities = 18/37 (48%), Positives = 21/37 (56%), Gaps = 3/37 (8%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
G GWQ G GG +GG G GGG++ G G GGG
Sbjct: 273 GGGWQGGGGGHGGGWQGGGGRGGGWK---GGGGHGGG 306
Score = 34.7 bits (78), Expect = 0.008
Identities = 20/45 (44%), Positives = 24/45 (52%), Gaps = 12/45 (26%)
Query: 78 KGTGWQTTG------RKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
+G GW+ G + GGG G G KGGGGG G +GGGG
Sbjct: 293 RGGGWKGGGGHGGGWQGGGGRGGGWKGGGGG------GGWRGGGG 331
Score = 34.7 bits (78), Expect = 0.008
Identities = 21/43 (48%), Positives = 23/43 (52%), Gaps = 4/43 (9%)
Query: 78 KGTGWQTTGRKGGG----SGKGGKGGGGGYRIPIPGVGKGGGG 116
KG G G KGGG KGG G GGG++ G G GGGG
Sbjct: 94 KGGGGHGGGWKGGGGHGGGWKGGGGHGGGWKGGGGGRGGGGGG 136
Score = 34.3 bits (77), Expect = 0.010
Identities = 15/24 (62%), Positives = 16/24 (66%)
Query: 78 KGTGWQTTGRKGGGSGKGGKGGGG 101
KG G G KGGG G+GG GGGG
Sbjct: 114 KGGGGHGGGWKGGGGGRGGGGGGG 137
Score = 32.0 bits (71), Expect = 0.051
Identities = 20/42 (47%), Positives = 20/42 (47%), Gaps = 5/42 (11%)
Query: 79 GTGWQTTGR-----KGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
G GWQ G KGGG GG GGGG G G GGG
Sbjct: 284 GGGWQGGGGRGGGWKGGGGHGGGWQGGGGRGGGWKGGGGGGG 325
Score = 31.2 bits (69), Expect = 0.087
Identities = 14/29 (48%), Positives = 18/29 (61%), Gaps = 4/29 (13%)
Query: 78 KGTGWQTTGR----KGGGSGKGGKGGGGG 102
+G GW+ G +GGG G G +GGGGG
Sbjct: 313 RGGGWKGGGGGGGWRGGGGGGGWRGGGGG 341
Score = 29.6 bits (65), Expect = 0.25
Identities = 20/46 (43%), Positives = 20/46 (43%), Gaps = 8/46 (17%)
Query: 78 KGTGWQTTGRKGGGSGKGGKGGG--------GGYRIPIPGVGKGGG 115
KG G Q G KGGG GG GGG G I G GGG
Sbjct: 215 KGGGGQGGGWKGGGGRGGGGGGGAELENEEINGASEEIQWQGGGGG 260
Score = 28.9 bits (63), Expect = 0.43
Identities = 13/23 (56%), Positives = 15/23 (64%), Gaps = 2/23 (8%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGG 101
G GW+ G GGG +GG GGGG
Sbjct: 323 GGGWRGGG--GGGGWRGGGGGGG 343
>At4g36230 putative glycine-rich cell wall protein
Length = 221
Score = 37.4 bits (85), Expect = 0.001
Identities = 19/31 (61%), Positives = 19/31 (61%), Gaps = 6/31 (19%)
Query: 86 GRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
GR GGG G GG GGGGG G G GGGG
Sbjct: 151 GRGGGGGGGGGGGGGGG------GGGGGGGG 175
Score = 36.6 bits (83), Expect = 0.002
Identities = 20/38 (52%), Positives = 21/38 (54%), Gaps = 6/38 (15%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G+G G GGG G GG GGGGG G G GGGG
Sbjct: 147 GSGRGRGGGGGGGGGGGGGGGGGG------GGGGGGGG 178
Score = 36.2 bits (82), Expect = 0.003
Identities = 35/115 (30%), Positives = 44/115 (37%), Gaps = 13/115 (11%)
Query: 2 VKQKKSKINIGVHEYANWLGKGTLLQEGDRNTNECGGAILKGNEGKSNDGCGDGIEIHCR 61
V KS NI E+ N L + T +C G G G G G +
Sbjct: 55 VLDSKSTANISTVEFENALD-----DQNVNKTEKCESRAGGGGGGGGGGGGGGGGQGSGG 109
Query: 62 KGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G KD S + +++G GGG G GG G G G G G+GGGG
Sbjct: 110 GGGEGNGGNGKDNSHKRN---KSSGGGGGGGGGGGGGSGNG-----SGRGRGGGG 156
Score = 35.4 bits (80), Expect = 0.005
Identities = 19/38 (50%), Positives = 22/38 (57%), Gaps = 6/38 (15%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G+G + +GGG G GG GGGGG G G GGGG
Sbjct: 143 GSGNGSGRGRGGGGGGGGGGGGGG------GGGGGGGG 174
Score = 34.7 bits (78), Expect = 0.008
Identities = 19/39 (48%), Positives = 20/39 (50%), Gaps = 8/39 (20%)
Query: 78 KGTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
+G G G GGG G GG GGGGG G GGGG
Sbjct: 150 RGRGGGGGGGGGGGGGGGGGGGGGG--------GGGGGG 180
Score = 33.9 bits (76), Expect = 0.013
Identities = 17/32 (53%), Positives = 18/32 (56%), Gaps = 5/32 (15%)
Query: 86 GRKGGGSGKGGKGGGGGYRIPIPGVGKGGGGK 117
G GGG G GG+G GGG G G GG GK
Sbjct: 94 GGGGGGGGGGGQGSGGG-----GGEGNGGNGK 120
Score = 33.1 bits (74), Expect = 0.023
Identities = 17/36 (47%), Positives = 18/36 (49%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGG 114
G G G GGG G GKGGG G+ G G GG
Sbjct: 167 GGGGGGGGGGGGGGGSDGKGGGWGFGFGWGGDGPGG 202
Score = 32.3 bits (72), Expect = 0.039
Identities = 19/37 (51%), Positives = 19/37 (51%), Gaps = 4/37 (10%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
G G G GGG G GG GGGGG GKGGG
Sbjct: 156 GGGGGGGGGGGGGGGGGGGGGGGGG----GSDGKGGG 188
Score = 31.6 bits (70), Expect = 0.066
Identities = 18/37 (48%), Positives = 18/37 (48%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
G G G GGG G GG GGGGG G G G G
Sbjct: 154 GGGGGGGGGGGGGGGGGGGGGGGGGGGGSDGKGGGWG 190
Score = 29.6 bits (65), Expect = 0.25
Identities = 16/38 (42%), Positives = 17/38 (44%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G G G GGG+ G G GG
Sbjct: 165 GGGGGGGGGGGGGGGGGSDGKGGGWGFGFGWGGDGPGG 202
>At5g59630 glycine-rich protein - like
Length = 253
Score = 37.0 bits (84), Expect = 0.002
Identities = 21/39 (53%), Positives = 21/39 (53%), Gaps = 5/39 (12%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGGK 117
G G G GGG G GG GGGGG G G GGGGK
Sbjct: 88 GGGGGGGGGGGGGGGGGGGGGGGG-----GGGGGGGGGK 121
Score = 36.6 bits (83), Expect = 0.002
Identities = 19/40 (47%), Positives = 22/40 (54%), Gaps = 6/40 (15%)
Query: 77 TKGTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
+ +G + G GGG G GG GGGGG G G GGGG
Sbjct: 64 SSNSGGSSGGGGGGGGGGGGGGGGGG------GGGGGGGG 97
Score = 35.8 bits (81), Expect = 0.004
Identities = 20/46 (43%), Positives = 22/46 (47%), Gaps = 6/46 (13%)
Query: 71 DKDISITKGTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
D + + G G GGG G GG GGGGG G G GGGG
Sbjct: 59 DNGLDSSNSGGSSGGGGGGGGGGGGGGGGGGG------GGGGGGGG 98
Score = 35.4 bits (80), Expect = 0.005
Identities = 20/38 (52%), Positives = 20/38 (52%), Gaps = 6/38 (15%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 81 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 112
Score = 35.4 bits (80), Expect = 0.005
Identities = 20/38 (52%), Positives = 20/38 (52%), Gaps = 6/38 (15%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 77 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 108
Score = 35.4 bits (80), Expect = 0.005
Identities = 20/38 (52%), Positives = 20/38 (52%), Gaps = 6/38 (15%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 82 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 113
Score = 35.4 bits (80), Expect = 0.005
Identities = 20/38 (52%), Positives = 20/38 (52%), Gaps = 6/38 (15%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GGG G GG GGGGG G G GGGG
Sbjct: 76 GGGGGGGGGGGGGGGGGGGGGGGG------GGGGGGGG 107
Score = 32.3 bits (72), Expect = 0.039
Identities = 15/32 (46%), Positives = 19/32 (58%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGV 110
G G G GGG G GG GGGGG ++ + G+
Sbjct: 96 GGGGGGGGGGGGGGGGGGGGGGGGGKLQLLGL 127
>At5g46730 unknown protein
Length = 268
Score = 37.0 bits (84), Expect = 0.002
Identities = 26/80 (32%), Positives = 30/80 (37%)
Query: 37 GGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGG 96
GG G G+S G G G GY + G G GG + G
Sbjct: 46 GGVSSGGYGGESGGGYGGGSGEGAGGGYGGAEGYASGGGSGHGGGGGGAASSGGYASGAG 105
Query: 97 KGGGGGYRIPIPGVGKGGGG 116
+GGGGGY G GGGG
Sbjct: 106 EGGGGGYGGAAGGHAGGGGG 125
Score = 35.8 bits (81), Expect = 0.004
Identities = 33/102 (32%), Positives = 36/102 (34%), Gaps = 6/102 (5%)
Query: 16 YANWLGKGTLLQEGDRNTNECGGAILKGNEGKSNDGCGDGIE-IHCRKGYVDGTIWDKDI 74
Y N G+G G + GG G G G G H GY G
Sbjct: 145 YGNGAGEG-----GGAGASGYGGGAYGGGGGHGGGGGGGSAGGAHGGSGYGGGEGGGAGG 199
Query: 75 SITKGTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
+ G G GGGSG GG GGGG G G G GG
Sbjct: 200 GGSHGGAGGYGGGGGGGSGGGGAYGGGGAHGGGYGSGGGEGG 241
Score = 34.7 bits (78), Expect = 0.008
Identities = 28/83 (33%), Positives = 31/83 (36%), Gaps = 8/83 (9%)
Query: 37 GGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGG---GSG 93
GG G + +G G G GY G G G G GG G G
Sbjct: 138 GGEHASGYGNGAGEGGGAGAS-----GYGGGAYGGGGGHGGGGGGGSAGGAHGGSGYGGG 192
Query: 94 KGGKGGGGGYRIPIPGVGKGGGG 116
+GG GGGG G G GGGG
Sbjct: 193 EGGGAGGGGSHGGAGGYGGGGGG 215
Score = 32.7 bits (73), Expect = 0.030
Identities = 19/39 (48%), Positives = 23/39 (58%), Gaps = 4/39 (10%)
Query: 79 GTGWQTTGRKGGGSGKGGKGG-GGGYRIPIPGVGKGGGG 116
G G+ + G +GGG G G GG GGG+ G G GGGG
Sbjct: 230 GGGYGSGGGEGGGYGGGAAGGYGGGHG---GGGGHGGGG 265
Score = 30.8 bits (68), Expect = 0.11
Identities = 14/25 (56%), Positives = 14/25 (56%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGY 103
G G G GGG G GG GGGGY
Sbjct: 242 GYGGGAAGGYGGGHGGGGGHGGGGY 266
Score = 30.0 bits (66), Expect = 0.19
Identities = 20/48 (41%), Positives = 24/48 (49%), Gaps = 10/48 (20%)
Query: 79 GTGWQTTGRKGGGSGKG-----GKGGGGGY-----RIPIPGVGKGGGG 116
G+G ++G GG SG G G+G GGGY G G GGGG
Sbjct: 44 GSGGVSSGGYGGESGGGYGGGSGEGAGGGYGGAEGYASGGGSGHGGGG 91
Score = 30.0 bits (66), Expect = 0.19
Identities = 15/38 (39%), Positives = 18/38 (46%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G + G G+G+GG G GY G G G GG
Sbjct: 136 GAGGEHASGYGNGAGEGGGAGASGYGGGAYGGGGGHGG 173
Score = 29.3 bits (64), Expect = 0.33
Identities = 30/90 (33%), Positives = 33/90 (36%), Gaps = 20/90 (22%)
Query: 37 GGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGG--GSGK 94
GGA + G S G G G GY G +G G G GG G G
Sbjct: 74 GGAEGYASGGGSGHGGGGG-GAASSGGYASGA--------GEGGGGGYGGAAGGHAGGGG 124
Query: 95 GGKGGGGGYRIPI---------PGVGKGGG 115
GG GGGGG G G+GGG
Sbjct: 125 GGSGGGGGSAYGAGGEHASGYGNGAGEGGG 154
Score = 28.9 bits (63), Expect = 0.43
Identities = 18/34 (52%), Positives = 18/34 (52%), Gaps = 4/34 (11%)
Query: 86 GRKGGGSGKGGKGGGGGYRIPIP---GVGKGGGG 116
G GGG G GG G GGGY G G GGGG
Sbjct: 227 GAHGGGYGSGG-GEGGGYGGGAAGGYGGGHGGGG 259
Score = 28.9 bits (63), Expect = 0.43
Identities = 14/30 (46%), Positives = 15/30 (49%)
Query: 86 GRKGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
G+ GG G GG GG GG G GGG
Sbjct: 31 GQASGGGGHGGGGGSGGVSSGGYGGESGGG 60
Score = 28.1 bits (61), Expect = 0.73
Identities = 18/40 (45%), Positives = 19/40 (47%), Gaps = 5/40 (12%)
Query: 79 GTGWQTTGRKGGGSGKGGKGG--GGGYRIPIPGVGKGGGG 116
G G G GG GG GG GGGY G G+G GG
Sbjct: 35 GGGGHGGGGGSGGVSSGGYGGESGGGYG---GGSGEGAGG 71
Score = 26.6 bits (57), Expect = 2.1
Identities = 32/107 (29%), Positives = 38/107 (34%), Gaps = 18/107 (16%)
Query: 16 YANWLGKGTLLQEGDRNTNECGGAILKGNEGKSNDGCGD--GIEIHCRKGYVDGTIWDKD 73
YA+ G+G G G G G S G G G GY +G
Sbjct: 100 YASGAGEG-----GGGGYGGAAGGHAGGGGGGSGGGGGSAYGAGGEHASGYGNGA----- 149
Query: 74 ISITKGTGWQTTGRKGGGSGKG---GKGGGGGYRIPIPGVGKGGGGK 117
+G G +G GG G G G GGGGG G GGG+
Sbjct: 150 ---GEGGGAGASGYGGGAYGGGGGHGGGGGGGSAGGAHGGSGYGGGE 193
>At2g42520 putative ATP-dependent RNA helicase
Length = 633
Score = 36.6 bits (83), Expect = 0.002
Identities = 19/32 (59%), Positives = 20/32 (62%), Gaps = 3/32 (9%)
Query: 85 TGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
+GR G G G GG GGGGGY G G GGGG
Sbjct: 582 SGRGGYGGGGGGYGGGGGYG---GGGGYGGGG 610
Score = 33.1 bits (74), Expect = 0.023
Identities = 18/38 (47%), Positives = 18/38 (47%)
Query: 79 GTGWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGGG 116
G G G GG G GG GGGGGY G GG G
Sbjct: 588 GGGGGGYGGGGGYGGGGGYGGGGGYGGGYGGASSGGYG 625
Score = 33.1 bits (74), Expect = 0.023
Identities = 19/42 (45%), Positives = 22/42 (52%), Gaps = 3/42 (7%)
Query: 79 GTGWQTTGRKGGGSGKG-GKGGGGGYRIPI--PGVGKGGGGK 117
G G +G + G G G GGGGGYR PG G G GG+
Sbjct: 55 GYGGPPSGSRWAPGGSGVGVGGGGGYRADAGRPGSGSGYGGR 96
Score = 32.3 bits (72), Expect = 0.039
Identities = 21/58 (36%), Positives = 28/58 (48%), Gaps = 11/58 (18%)
Query: 48 SNDGCGDGIEIHCRKGYVDGTIW---DKDISITKGTGWQT-TGRKGGGSGKGGKGGGG 101
+ND G G G G+ W + + G G++ GR G GSG GG+GGGG
Sbjct: 50 ANDRVGYG-------GPPSGSRWAPGGSGVGVGGGGGYRADAGRPGSGSGYGGRGGGG 100
Score = 31.6 bits (70), Expect = 0.066
Identities = 20/46 (43%), Positives = 23/46 (49%), Gaps = 15/46 (32%)
Query: 79 GTGWQTTGRKGG---------GSGKGGKGGGGGYRIPIPGVGKGGG 115
G ++ GR GG GSG+GG GGGGG G G GGG
Sbjct: 560 GKNRRSGGRFGGRDFRREGSFGSGRGGYGGGGG------GYGGGGG 599
Score = 31.6 bits (70), Expect = 0.066
Identities = 20/38 (52%), Positives = 21/38 (54%), Gaps = 4/38 (10%)
Query: 79 GTGWQTTGRKGGG-SGKGGKGGGGGYRIPIPGVGKGGG 115
G+G G GGG G GG GGGGGY G G GGG
Sbjct: 581 GSGRGGYGGGGGGYGGGGGYGGGGGYG---GGGGYGGG 615
>At1g15840 hypothetical protein
Length = 126
Score = 36.6 bits (83), Expect = 0.002
Identities = 26/73 (35%), Positives = 29/73 (39%), Gaps = 27/73 (36%)
Query: 43 GNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGTGWQTTGRKGGGSGKGGKGGGGG 102
G EGK +G G+G G G T+G GGG G G KGGG G
Sbjct: 31 GGEGKKKNGGGEG-----------------------GGGEGTSGEGGGGGGDGTKGGGDG 67
Query: 103 YRIPIPGVGKGGG 115
I G G G G
Sbjct: 68 ----ISGGGHGDG 76
Score = 33.9 bits (76), Expect = 0.013
Identities = 31/95 (32%), Positives = 40/95 (41%), Gaps = 11/95 (11%)
Query: 21 GKGTLLQEGDRNTNECGGAILKGNEGKSNDGCGDGIEIHCRKGYVDGTIWDKDISITKGT 80
G T+ G + GG G EG S +G G G + KG DG G
Sbjct: 23 GGNTITGGGGEGKKKNGGGEGGGGEGTSGEGGGGGGD--GTKGGGDGISGGGH-----GD 75
Query: 81 GWQTTGRKGGGSGKGGKGGGGGYRIPIPGVGKGGG 115
G +G G G+ GG+ G G R G+G+GGG
Sbjct: 76 GLGCSGGGGDGTKGGGRRGDGLGR----GLGRGGG 106
Score = 33.5 bits (75), Expect = 0.017
Identities = 23/44 (52%), Positives = 24/44 (54%), Gaps = 9/44 (20%)
Query: 77 TKGTGWQTTGRKGGGSGK----GGKGGGGGYRIPIPGVGKGGGG 116
TKG G TG GGG GK GG+GGGG G G GGGG
Sbjct: 20 TKGGGNTITG--GGGEGKKKNGGGEGGGGE---GTSGEGGGGGG 58
Score = 29.3 bits (64), Expect = 0.33
Identities = 16/34 (47%), Positives = 19/34 (55%), Gaps = 3/34 (8%)
Query: 86 GRKGGG---SGKGGKGGGGGYRIPIPGVGKGGGG 116
G GGG SG+GG GGG G + G+ GG G
Sbjct: 41 GEGGGGEGTSGEGGGGGGDGTKGGGDGISGGGHG 74
Score = 28.5 bits (62), Expect = 0.56
Identities = 16/33 (48%), Positives = 18/33 (54%), Gaps = 4/33 (12%)
Query: 89 GGGSGKGGKG----GGGGYRIPIPGVGKGGGGK 117
GGG G G G GGGG G G+GGGG+
Sbjct: 15 GGGDGTKGGGNTITGGGGEGKKKNGGGEGGGGE 47
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.311 0.143 0.454
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,704,490
Number of Sequences: 26719
Number of extensions: 211161
Number of successful extensions: 5026
Number of sequences better than 10.0: 340
Number of HSP's better than 10.0 without gapping: 284
Number of HSP's successfully gapped in prelim test: 57
Number of HSP's that attempted gapping in prelim test: 693
Number of HSP's gapped (non-prelim): 1666
length of query: 117
length of database: 11,318,596
effective HSP length: 93
effective length of query: 24
effective length of database: 8,833,729
effective search space: 212009496
effective search space used: 212009496
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 52 (24.6 bits)
Medicago: description of AC138451.8


