

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137824.11 - phase: 0
(316 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
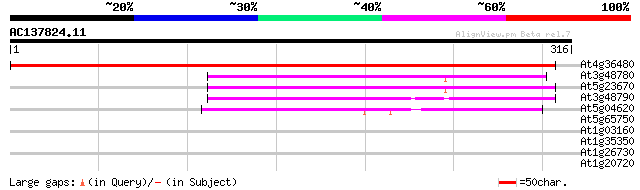
Score E
Sequences producing significant alignments: (bits) Value
At4g36480 serine palmitoyltransferase (AtLCB1) 488 e-138
At3g48780 serine palmitoyltransferase-like protein 105 2e-23
At5g23670 serine palmitoyltransferase 103 1e-22
At3g48790 serine palmitoyltransferase-like protein 90 1e-18
At5g04620 8-amino-7-oxononanoate synthase - like protein 72 3e-13
At5g65750 2-oxoglutarate dehydrogenase, E1 component 31 1.0
At1g03160 hypothetical protein 30 1.7
At1g35350 hypothetical protein 28 5.0
At1g26730 unknown protein 28 5.0
At1g20720 hypothetical protein 28 8.5
>At4g36480 serine palmitoyltransferase (AtLCB1)
Length = 482
Score = 488 bits (1256), Expect = e-138
Identities = 234/307 (76%), Positives = 269/307 (87%)
Query: 1 MASSFINFVNTTIDLVTYALHAPSARAVVFGFNIGGHLFIEVLLLVVILFLLSQKSYKPP 60
MAS+ + N ++ VT L +PSAR V+FG I GH F+E LL VVI+ LL++KSYKPP
Sbjct: 1 MASNLVEMFNAALNWVTMILESPSARVVLFGVPIRGHFFVEGLLGVVIIILLTRKSYKPP 60
Query: 61 KRPLSNKEIDELCDEWVPQPLIPSLNDEMPYEPPVLESAAGPHTIVNGKEVVNFASANYL 120
KRPL+ +EIDELCDEWVP+PLIP + ++M +EPPVLESAAGPHT VNGK+VVNFASANYL
Sbjct: 61 KRPLTEQEIDELCDEWVPEPLIPPITEDMKHEPPVLESAAGPHTTVNGKDVVNFASANYL 120
Query: 121 GLIGHQKLLDSCSSALEKYGVGSCGPRGFYGTIDVHLDCEGRISKFLGTPDSILYSYGLS 180
GLIGH+KLL+SC+SALEKYGVGSCGPRGFYGTIDVHLDCE RISKFLGTPDSILYSYGLS
Sbjct: 121 GLIGHEKLLESCTSALEKYGVGSCGPRGFYGTIDVHLDCETRISKFLGTPDSILYSYGLS 180
Query: 181 TMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSRSTVVYFKHNNMDSLRETLENITSIYK 240
TMFS IP F KKGD+IVADEGVHWGIQNGL LSRST+VYFKHN+M+SLR TLE I + YK
Sbjct: 181 TMFSTIPCFCKKGDVIVADEGVHWGIQNGLQLSRSTIVYFKHNDMESLRITLEKIMTKYK 240
Query: 241 RTKNLRRYIVIEALYQNSGQIAPLDEIIKLKEKYCFRILLDESNSFGVLGSSGRGLTEHY 300
R+KNLRRYIV EA+YQNSGQIAPLDEI+KLKEKY FR++LDESNSFGVLG SGRGL EH+
Sbjct: 241 RSKNLRRYIVAEAVYQNSGQIAPLDEIVKLKEKYRFRVILDESNSFGVLGRSGRGLAEHH 300
Query: 301 GVPVCRV 307
VP+ ++
Sbjct: 301 SVPIEKI 307
>At3g48780 serine palmitoyltransferase-like protein
Length = 489
Score = 105 bits (263), Expect = 2e-23
Identities = 63/195 (32%), Positives = 101/195 (51%), Gaps = 4/195 (2%)
Query: 112 VNFASANYLGLIGHQKLLDS-CSSALEKYGVGSCGPRGFYGTIDVHLDCEGRISKFLGTP 170
+N S NYLG + +L+K+ +C R GT VH + E ++K++G P
Sbjct: 103 LNLGSYNYLGFGSFDEYCTPRVIESLKKFSASTCSSRVDAGTTSVHAELEDCVAKYVGQP 162
Query: 171 DSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSRSTVVYFKHNNMDSLRE 230
++++ G +T + IP KG +I++D H I NG S +T+ F+HN L +
Sbjct: 163 AAVIFGMGYATNSAIIPVLIGKGGLIISDSLNHTSIVNGARGSGATIRVFQHNTPGHLEK 222
Query: 231 TL-ENITSIYKRTKN--LRRYIVIEALYQNSGQIAPLDEIIKLKEKYCFRILLDESNSFG 287
L E I RT + +V+E +Y G+I L EI+ + +KY + LDE++S G
Sbjct: 223 VLKEQIAEGQPRTHRPWKKIIVVVEGIYSMEGEICHLPEIVSICKKYKAYVYLDEAHSIG 282
Query: 288 VLGSSGRGLTEHYGV 302
+G +GRG+ E GV
Sbjct: 283 AIGKTGRGVCELLGV 297
>At5g23670 serine palmitoyltransferase
Length = 489
Score = 103 bits (257), Expect = 1e-22
Identities = 62/200 (31%), Positives = 101/200 (50%), Gaps = 4/200 (2%)
Query: 112 VNFASANYLGLIGHQKLLDS-CSSALEKYGVGSCGPRGFYGTIDVHLDCEGRISKFLGTP 170
+N S NYLG + +L+K+ +C R GT VH + E +++F+G P
Sbjct: 103 LNLGSYNYLGFGSFDEYCTPRVIESLKKFSASTCSSRVDAGTTSVHAELEECVTRFVGKP 162
Query: 171 DSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSRSTVVYFKHNNMDSLRE 230
++++ G +T + IP KG +I++D H I NG S +T+ F+HN L
Sbjct: 163 AAVVFGMGYATNSAIIPVLIGKGGLIISDSLNHSSIVNGARGSGATIRVFQHNTPSHLER 222
Query: 231 TL-ENITSIYKRTKN--LRRYIVIEALYQNSGQIAPLDEIIKLKEKYCFRILLDESNSFG 287
L E I RT + +V+E +Y G+I L E++ + +KY + LDE++S G
Sbjct: 223 VLREQIAEGQPRTHRPWKKIIVVVEGIYSMEGEICHLPEVVAICKKYKAYVYLDEAHSIG 282
Query: 288 VLGSSGRGLTEHYGVPVCRV 307
+G +G+G+ E GV V
Sbjct: 283 AIGKTGKGICELLGVDTADV 302
>At3g48790 serine palmitoyltransferase-like protein
Length = 386
Score = 90.1 bits (222), Expect = 1e-18
Identities = 57/197 (28%), Positives = 97/197 (48%), Gaps = 5/197 (2%)
Query: 112 VNFASANYLGLIGHQKLLDS-CSSALEKYGVGSCGPRGFYGTIDVHLDCEGRISKFLGTP 170
++ S NYLG + +L+K+ +C GT VH + E ++KF+G P
Sbjct: 3 LSLGSYNYLGFCPFDEYCTPRVIESLKKFSASTCSSCVDAGTTAVHGELEECVAKFVGKP 62
Query: 171 DSILYSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSRSTVVYFKHNNMDSLRE 230
++++ G T + I KG +I++D H I NG S +T+ F+HN +
Sbjct: 63 AAVVFGMGYLTNSAIISVLIGKGGLIISDSLNHTSIINGARGSGATIRVFQHNILK--EH 120
Query: 231 TLENITSIYKRTKNLRRYIVIEALYQNSGQIAPLDEIIKLKEKYCFRILLDESNSFGVLG 290
+E ++ K + +V+E +Y G+I L EI+ + +Y + LDE++S G +G
Sbjct: 121 IIEGQPRTHRPWKKI--IVVVEGIYSMEGEICDLPEIVSVCSEYKAYVYLDEAHSIGAIG 178
Query: 291 SSGRGLTEHYGVPVCRV 307
+GRG+ E GV V
Sbjct: 179 KTGRGVCELLGVDTTEV 195
>At5g04620 8-amino-7-oxononanoate synthase - like protein
Length = 476
Score = 72.4 bits (176), Expect = 3e-13
Identities = 47/208 (22%), Positives = 101/208 (47%), Gaps = 21/208 (10%)
Query: 109 KEVVNFASANYLGLIGHQKLLDSCSSALEKYGVGSCGPRGFYGTIDVHLDCEGRISKFLG 168
K+++ F+ +YLGL H + ++ ++A+++YG+G G G H E +++
Sbjct: 102 KKLLLFSGNDYLGLSSHPTISNAAANAVKEYGMGPKGSALICGYTTYHRLLESSLAQLKK 161
Query: 169 TPDSILYSYGLSTMFSAIPAFSKKGDIIVA--------------DEGVHWGIQNGLYLS- 213
D ++ G + +A+ A ++ A D H I +G+ L+
Sbjct: 162 KEDCLVCPTGFAANMAAMVAIGSVASLLAASGKPLKNEKVAIFSDALNHASIIDGVRLAE 221
Query: 214 -RSTVVYFKHNNMDSLRETLENITSIYKRTKNLRRYIVIEALYQNSGQIAPLDEIIKLKE 272
+ V F + + D + ++ S+ K R+ +V ++L+ G AP++E+ +L++
Sbjct: 222 RQGNVEVFVYRHCD-----MYHLNSLLSNCKMKRKVVVTDSLFSMDGDFAPMEELSQLRK 276
Query: 273 KYCFRILLDESNSFGVLGSSGRGLTEHY 300
KY F +++D+++ V G +G G+ E +
Sbjct: 277 KYGFLLVIDDAHGTFVCGENGGGVAEEF 304
>At5g65750 2-oxoglutarate dehydrogenase, E1 component
Length = 1025
Score = 30.8 bits (68), Expect = 1.0
Identities = 23/104 (22%), Positives = 42/104 (40%), Gaps = 15/104 (14%)
Query: 189 FSKKGDIIVADEGVHWGIQNGLYLSRSTVV--YFKHNNMDSLRETLENITSIYKRTKNLR 246
+ ++ +I + EG+ WG+ L + V + + + D R T + S+ +
Sbjct: 626 YEQRAQMIESGEGIDWGLGEALAFATLVVEGNHVRLSGQDVERGTFSHRHSVLHDQETGE 685
Query: 247 RYIVIEALYQNSGQIAPLDEIIKLKEKYCFRILLDESNSFGVLG 290
Y PLD +IK ++ F + + FGVLG
Sbjct: 686 EY-------------CPLDHLIKNQDPEMFTVSNSSLSEFGVLG 716
>At1g03160 hypothetical protein
Length = 642
Score = 30.0 bits (66), Expect = 1.7
Identities = 26/120 (21%), Positives = 51/120 (41%), Gaps = 6/120 (5%)
Query: 165 KFLGTPDSILYSYGLSTMFSA---IPAFSKKGDIIVADEGVHWGIQNGLYLSRSTVVYFK 221
K L T + ILY + A + + D+ +AD G +W +Q+ L + +
Sbjct: 508 KLLNTENVILYPVSARSALEAKLSTASLVGRDDLEIADPGSNWRVQSFNELEKFLYSFLD 567
Query: 222 HN---NMDSLRETLENITSIYKRTKNLRRYIVIEALYQNSGQIAPLDEIIKLKEKYCFRI 278
+ M+ +R LE +I +R + +V + +A D+II ++Y ++
Sbjct: 568 SSTATGMERIRLKLETPMAIAERLLSSVEALVRQDCLAAREDLASADKIISRTKEYALKM 627
>At1g35350 hypothetical protein
Length = 747
Score = 28.5 bits (62), Expect = 5.0
Identities = 14/50 (28%), Positives = 26/50 (52%), Gaps = 1/50 (2%)
Query: 206 IQNGLYLSRSTVVYFKHNNMDSLRETLENI-TSIYKRTKNLRRYIVIEAL 254
I+N L LS + F N+ + E L+N+ Y++ ++L+ Y + L
Sbjct: 226 IRNVLKLSNKEDIKFTKENLKKIEERLKNVFIEFYRKLRHLKNYSFLNTL 275
>At1g26730 unknown protein
Length = 750
Score = 28.5 bits (62), Expect = 5.0
Identities = 14/50 (28%), Positives = 26/50 (52%), Gaps = 1/50 (2%)
Query: 206 IQNGLYLSRSTVVYFKHNNMDSLRETLENI-TSIYKRTKNLRRYIVIEAL 254
I+N L LS + F N+ + E L+N+ Y++ ++L+ Y + L
Sbjct: 226 IKNVLKLSNQEELKFTRENLKKIEERLKNVFIEFYRKLRHLKNYSFLNTL 275
>At1g20720 hypothetical protein
Length = 986
Score = 27.7 bits (60), Expect = 8.5
Identities = 17/59 (28%), Positives = 28/59 (46%)
Query: 175 YSYGLSTMFSAIPAFSKKGDIIVADEGVHWGIQNGLYLSRSTVVYFKHNNMDSLRETLE 233
YSY ++ + A KG II+ DE + + L + ST + H+NM+ + E
Sbjct: 256 YSYIVNPVIRAGVEVDLKGAIIIFDEAHFFSMLLMLMIRPSTKSFTVHSNMEDIAREAE 314
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.141 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,323,378
Number of Sequences: 26719
Number of extensions: 329675
Number of successful extensions: 743
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 731
Number of HSP's gapped (non-prelim): 11
length of query: 316
length of database: 11,318,596
effective HSP length: 99
effective length of query: 217
effective length of database: 8,673,415
effective search space: 1882131055
effective search space used: 1882131055
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC137824.11


