

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136450.2 + phase: 0 /pseudo
(198 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
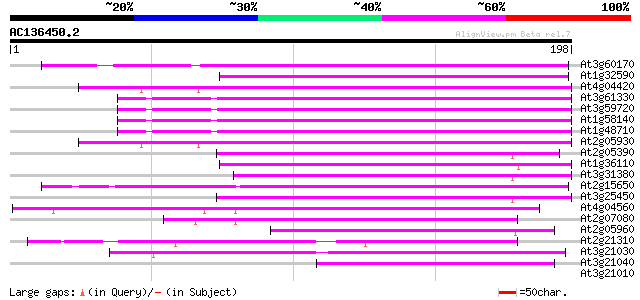
Score E
Sequences producing significant alignments: (bits) Value
At3g60170 putative protein 89 2e-18
At1g32590 hypothetical protein, 5' partial 64 7e-11
At4g04420 putative transposon protein 63 1e-10
At3g61330 copia-type polyprotein 63 1e-10
At3g59720 copia-type reverse transcriptase-like protein 63 1e-10
At1g58140 hypothetical protein 63 1e-10
At1g48710 hypothetical protein 63 1e-10
At2g05930 copia-like retroelement pol polyprotein 61 3e-10
At2g05390 putative retroelement pol polyprotein 61 4e-10
At1g36110 hypothetical protein 60 6e-10
At3g31380 hypothetical protein 59 2e-09
At2g15650 putative retroelement pol polyprotein 59 2e-09
At3g25450 hypothetical protein 57 6e-09
At4g04560 putative transposon protein 56 1e-08
At2g07080 putative gag-protease polyprotein 46 1e-05
At2g05960 putative retroelement pol polyprotein 45 2e-05
At2g21310 putative retroelement pol polyprotein 44 6e-05
At3g21030 hypothetical protein 42 2e-04
At3g21040 hypothetical protein 40 6e-04
At3g21010 unknown protein 39 0.002
>At3g60170 putative protein
Length = 1339
Score = 88.6 bits (218), Expect = 2e-18
Identities = 61/186 (32%), Positives = 100/186 (52%), Gaps = 8/186 (4%)
Query: 12 VPKFNGDLEEFS*WKTNFYNHVMSLDEELWDILEGGIGDLVVDEEGADVDRRKHTPAQKK 71
+P+F+G + +S NF ELW ++E GI +VV +R A ++
Sbjct: 12 IPRFDGYYDFWSMTMENFLR-----SRELWRLVEEGIPAIVVGTTPVSEAQRS---AVEE 63
Query: 72 LYKKHHTIRGALVKAIPKAEYMKMSDKSTAKSMFASLCANYEGSKKVREAKALMLVHQYE 131
K ++ L +AI + + DKST+K+++ S+ Y+GS KV+ A+ L ++E
Sbjct: 64 AKLKDLKVKNFLFQAIDREILETILDKSTSKAIWESMKKKYQGSTKVKRAQLQALRKEFE 123
Query: 132 LFKMKDGESIEQMYSRFHTLVSGLQILKKSCVASDHVSKILRSLPARWRPKVTAIEETKD 191
L MK+GE I+ R T+V+ ++ + S VSKILRSL ++ V +IEE+ D
Sbjct: 124 LLAMKEGEKIDTFLGRTLTVVNKMKTNGEVMEQSTIVSKILRSLTPKFNYVVCSIEESND 183
Query: 192 LNTLSV 197
L+TLS+
Sbjct: 184 LSTLSI 189
>At1g32590 hypothetical protein, 5' partial
Length = 1263
Score = 63.5 bits (153), Expect = 7e-11
Identities = 37/123 (30%), Positives = 67/123 (54%)
Query: 75 KHHTIRGALVKAIPKAEYMKMSDKSTAKSMFASLCANYEGSKKVREAKALMLVHQYELFK 134
K H ++ L +I K + K T+K ++ S+ Y+G+ +V+ A+ L +E+ +
Sbjct: 24 KDHKVKNYLFASIDKTILKTILQKETSKDLWESMKRKYQGNDRVQSAQLQRLRRSFEVLE 83
Query: 135 MKDGESIEQMYSRFHTLVSGLQILKKSCVASDHVSKILRSLPARWRPKVTAIEETKDLNT 194
MK GE+I +SR + + ++ L + S V KILR+L ++ V AIEE+ ++
Sbjct: 84 MKIGETITGYFSRVMEITNDMRNLGEDMPDSKVVEKILRTLVEKFTYVVCAIEESNNIKE 143
Query: 195 LSV 197
L+V
Sbjct: 144 LTV 146
>At4g04420 putative transposon protein
Length = 1008
Score = 62.8 bits (151), Expect = 1e-10
Identities = 42/177 (23%), Positives = 81/177 (45%), Gaps = 3/177 (1%)
Query: 25 WKTNFYNHVMSLDEELWDILE-GGIGDLVVDEEGADVDRRKH--TPAQKKLYKKHHTIRG 81
WK + L +E W G ++ E+G DV + K A++ K +
Sbjct: 24 WKVKMRALIRGLGKEAWIATSIGWKAPVIKGEDGEDVLKTKDQWNDAEEAKAKANSRALS 83
Query: 82 ALVKAIPKAEYMKMSDKSTAKSMFASLCANYEGSKKVREAKALMLVHQYELFKMKDGESI 141
+ + + ++ ++ + +AK + L YEG+ V+ ++ ML Q+E M++ E+I
Sbjct: 84 LIFNFVNQNQFKRIQNCESAKEAWDKLAKAYEGTSSVKRSRIDMLASQFENLSMEETENI 143
Query: 142 EQMYSRFHTLVSGLQILKKSCVASDHVSKILRSLPARWRPKVTAIEETKDLNTLSVQ 198
E+ + + S L K V K+LR LP+R+ K TA+ + D +++ +
Sbjct: 144 EEFSGKISAIASEAHNLGKKYKDKKLVKKLLRCLPSRFESKRTAMGTSLDTDSIDFE 200
>At3g61330 copia-type polyprotein
Length = 1352
Score = 62.8 bits (151), Expect = 1e-10
Identities = 39/160 (24%), Positives = 84/160 (52%), Gaps = 4/160 (2%)
Query: 39 ELWDILEGGIGDLVVDEEGADVDRRKHTPAQKKLYKKHHTIRGALVKAIPKAEYMKMSDK 98
++W+I+E G + + EG+ +K + K+ + + + + + K+ +
Sbjct: 33 DVWEIVEKGF--IEPENEGSLSQTQKDGLRDSR--KRDKKALCLIYQGLDEDTFEKVVEA 88
Query: 99 STAKSMFASLCANYEGSKKVREAKALMLVHQYELFKMKDGESIEQMYSRFHTLVSGLQIL 158
++AK + L +Y+G+ +V++ + L ++E +MK+GE + +SR T+ + L+
Sbjct: 89 TSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDYFSRVLTVTNNLKRN 148
Query: 159 KKSCVASDHVSKILRSLPARWRPKVTAIEETKDLNTLSVQ 198
+ + K+LRSL ++ VT IEETKDL ++++
Sbjct: 149 GEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIE 188
>At3g59720 copia-type reverse transcriptase-like protein
Length = 1272
Score = 62.8 bits (151), Expect = 1e-10
Identities = 39/160 (24%), Positives = 84/160 (52%), Gaps = 4/160 (2%)
Query: 39 ELWDILEGGIGDLVVDEEGADVDRRKHTPAQKKLYKKHHTIRGALVKAIPKAEYMKMSDK 98
++W+I+E G + + EG+ +K + K+ + + + + + K+ +
Sbjct: 33 DVWEIVEKGF--IEPENEGSLSQTQKDGLRDSR--KRDKKALCLIYQGLDEDTFEKVVEA 88
Query: 99 STAKSMFASLCANYEGSKKVREAKALMLVHQYELFKMKDGESIEQMYSRFHTLVSGLQIL 158
++AK + L +Y+G+ +V++ + L ++E +MK+GE + +SR T+ + L+
Sbjct: 89 TSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDYFSRVLTVTNNLKRN 148
Query: 159 KKSCVASDHVSKILRSLPARWRPKVTAIEETKDLNTLSVQ 198
+ + K+LRSL ++ VT IEETKDL ++++
Sbjct: 149 GEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIE 188
>At1g58140 hypothetical protein
Length = 1320
Score = 62.8 bits (151), Expect = 1e-10
Identities = 39/160 (24%), Positives = 84/160 (52%), Gaps = 4/160 (2%)
Query: 39 ELWDILEGGIGDLVVDEEGADVDRRKHTPAQKKLYKKHHTIRGALVKAIPKAEYMKMSDK 98
++W+I+E G + + EG+ +K + K+ + + + + + K+ +
Sbjct: 33 DVWEIVEKGF--IEPENEGSLSQTQKDGLRDSR--KRDKKALCLIYQGLDEDTFEKVVEA 88
Query: 99 STAKSMFASLCANYEGSKKVREAKALMLVHQYELFKMKDGESIEQMYSRFHTLVSGLQIL 158
++AK + L +Y+G+ +V++ + L ++E +MK+GE + +SR T+ + L+
Sbjct: 89 TSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDYFSRVLTVTNNLKRN 148
Query: 159 KKSCVASDHVSKILRSLPARWRPKVTAIEETKDLNTLSVQ 198
+ + K+LRSL ++ VT IEETKDL ++++
Sbjct: 149 GEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIE 188
>At1g48710 hypothetical protein
Length = 1352
Score = 62.8 bits (151), Expect = 1e-10
Identities = 39/160 (24%), Positives = 84/160 (52%), Gaps = 4/160 (2%)
Query: 39 ELWDILEGGIGDLVVDEEGADVDRRKHTPAQKKLYKKHHTIRGALVKAIPKAEYMKMSDK 98
++W+I+E G + + EG+ +K + K+ + + + + + K+ +
Sbjct: 33 DVWEIVEKGF--IEPENEGSLSQTQKDGLRDSR--KRDKKALCLIYQGLDEDTFEKVVEA 88
Query: 99 STAKSMFASLCANYEGSKKVREAKALMLVHQYELFKMKDGESIEQMYSRFHTLVSGLQIL 158
++AK + L +Y+G+ +V++ + L ++E +MK+GE + +SR T+ + L+
Sbjct: 89 TSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDYFSRVLTVTNNLKRN 148
Query: 159 KKSCVASDHVSKILRSLPARWRPKVTAIEETKDLNTLSVQ 198
+ + K+LRSL ++ VT IEETKDL ++++
Sbjct: 149 GEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIE 188
>At2g05930 copia-like retroelement pol polyprotein
Length = 916
Score = 61.2 bits (147), Expect = 3e-10
Identities = 41/177 (23%), Positives = 81/177 (45%), Gaps = 3/177 (1%)
Query: 25 WKTNFYNHVMSLDEELWDILE-GGIGDLVVDEEGADVDRRKH--TPAQKKLYKKHHTIRG 81
WK + L +E W G ++ E+G DV + + A++ +
Sbjct: 36 WKVKMRALIRGLGKEAWIATSIGWKAPVIKGEDGEDVLKTEDQWNDAEEAKATANSRALS 95
Query: 82 ALVKAIPKAEYMKMSDKSTAKSMFASLCANYEGSKKVREAKALMLVHQYELFKMKDGESI 141
+ ++ + ++ ++ + +AK + L YEG+ V+ ++ ML Q+E M++ E+I
Sbjct: 96 LIFNSVNQNQFKQIQNCESAKEAWDKLAKAYEGTSSVKRSRIDMLASQFENLTMEETENI 155
Query: 142 EQMYSRFHTLVSGLQILKKSCVASDHVSKILRSLPARWRPKVTAIEETKDLNTLSVQ 198
E+ + + S L K V K+LR LP+R+ K TA+ + D N++ +
Sbjct: 156 EEFSGKISAIASEAHNLGKKYKDKKLVKKLLRCLPSRFESKRTAMGTSLDTNSIDFE 212
>At2g05390 putative retroelement pol polyprotein
Length = 1307
Score = 60.8 bits (146), Expect = 4e-10
Identities = 33/122 (27%), Positives = 69/122 (56%), Gaps = 1/122 (0%)
Query: 74 KKHHTIRGALVKAIPKAEYMKMSDKSTAKSMFASLCANYEGSKKVREAKALMLVHQYELF 133
+K+ R L +++P++ +++ T+K+M+ ++ G+++V+EAK L+ +++
Sbjct: 22 EKNDMARALLFQSVPESTILQVGKHKTSKAMWEAIKTRNLGAERVKEAKLQTLMAEFDRL 81
Query: 134 KMKDGESIEQMYSRFHTLVSGLQILKKSCVASDHVSKILRSLP-ARWRPKVTAIEETKDL 192
MKD E+I++ R + + + L + S V K L+SLP ++ + A+E+ DL
Sbjct: 82 NMKDNETIDEFVGRISEISTKSESLGEEIEESKIVKKFLKSLPRKKYIHIIAALEQILDL 141
Query: 193 NT 194
NT
Sbjct: 142 NT 143
>At1g36110 hypothetical protein
Length = 745
Score = 60.5 bits (145), Expect = 6e-10
Identities = 36/125 (28%), Positives = 67/125 (52%), Gaps = 1/125 (0%)
Query: 75 KHHTIRGALVKAIPKAEYMKMSDKSTAKSMFASLCANYEGSKKVREAKALMLVHQYELFK 134
K+ RG + ++IP++ +++ +TAK ++ S+ Y G+ +V+EA+ L+ ++E K
Sbjct: 56 KNTMARGLIFQSIPESLTLQVGTLATAKLVWDSIKTRYVGADRVKEARLQTLMAEFEKMK 115
Query: 135 MKDGESIEQMYSRFHTLVSGLQILKKSCVASDHVSKILRSLPAR-WRPKVTAIEETKDLN 193
MK+ E I+ R L + L + S V K L +LP R + + ++E+ DLN
Sbjct: 116 MKESEKIDVFAGRLAELATRSDALGSNIETSKLVKKFLNALPLRKYIHIIASLEQVLDLN 175
Query: 194 TLSVQ 198
S +
Sbjct: 176 NTSFE 180
>At3g31380 hypothetical protein
Length = 262
Score = 58.5 bits (140), Expect = 2e-09
Identities = 35/120 (29%), Positives = 65/120 (54%), Gaps = 1/120 (0%)
Query: 80 RGALVKAIPKAEYMKMSDKSTAKSMFASLCANYEGSKKVREAKALMLVHQYELFKMKDGE 139
RG LV++IP+A +++ + TAK ++ S+ + G+++V+EA+ L+ +E KMK+ E
Sbjct: 3 RGLLVQSIPEAFTLQVGNLQTAKEVWDSIKTRHVGAERVKEARVQTLMADFEKMKMKEAE 62
Query: 140 SIEQMYSRFHTLVSGLQILKKSCVASDHVSKILRSLP-ARWRPKVTAIEETKDLNTLSVQ 198
I+ R L + L + V K L SLP ++ + ++E+ DLN + +
Sbjct: 63 KIDDFAGRLSELSTKSAPLGVNIEVPKLVKKFLNSLPRKKYIHIIASLEQVLDLNNTTFE 122
>At2g15650 putative retroelement pol polyprotein
Length = 1347
Score = 58.5 bits (140), Expect = 2e-09
Identities = 46/186 (24%), Positives = 87/186 (46%), Gaps = 5/186 (2%)
Query: 12 VPKFNGDLEEFS*WKTNFYNHVMSLDEELWDILEGGIGDLVVDEEGADVDRRKHTPAQKK 71
+P F+G+ +F W + +LW ++E G+ V E R T ++
Sbjct: 9 IPIFDGEKYDF--WSIKMATIFRT--RKLWSVVEEGVPVEPVQAEETPETARAKTLREEA 64
Query: 72 LYKKHHTIRGALVKAIPKAEYMKMSDKSTAKSMFASLCANYEGSKKVREAKALMLVHQYE 131
+ ++ L A+ + +++ S++K + L Y+GS +VR K L +YE
Sbjct: 65 VTNDTMALQ-ILQTAVTDQIFSRIAAASSSKEAWDVLKDEYQGSPQVRLVKLQSLRREYE 123
Query: 132 LFKMKDGESIEQMYSRFHTLVSGLQILKKSCVASDHVSKILRSLPARWRPKVTAIEETKD 191
KM D ++I+ + L L + + + KIL SLPA++ V+ +E+T+D
Sbjct: 124 NLKMYDNDNIKTFTDKLIVLEIQLTYHGEKKTNTQLIQKILISLPAKFDSIVSVLEQTRD 183
Query: 192 LNTLSV 197
L+ L++
Sbjct: 184 LDALTM 189
>At3g25450 hypothetical protein
Length = 1343
Score = 57.0 bits (136), Expect = 6e-09
Identities = 32/126 (25%), Positives = 70/126 (55%), Gaps = 1/126 (0%)
Query: 74 KKHHTIRGALVKAIPKAEYMKMSDKSTAKSMFASLCANYEGSKKVREAKALMLVHQYELF 133
+K+ R L ++IP++ +++ + T+ +++ ++ + G+++V+EA+ L+ +++
Sbjct: 58 EKNDMARALLFQSIPESLILQVGKQKTSSAVWEAIKSRNLGAERVKEARLQTLMAEFDKL 117
Query: 134 KMKDGESIEQMYSRFHTLVSGLQILKKSCVASDHVSKILRSLP-ARWRPKVTAIEETKDL 192
KMKD E+I+ R + + L + S V K L+SLP ++ V A+E+ DL
Sbjct: 118 KMKDSETIDDYVGRISEITTKAAALGEDIEESKIVKKFLKSLPRKKYIHIVAALEQVLDL 177
Query: 193 NTLSVQ 198
T + +
Sbjct: 178 KTTTFE 183
>At4g04560 putative transposon protein
Length = 590
Score = 56.2 bits (134), Expect = 1e-08
Identities = 41/190 (21%), Positives = 84/190 (43%), Gaps = 4/190 (2%)
Query: 2 GNNKKSDFGKVPK-FNGDLEEFS*WKTNFYNHVMSLDEELWDILEGGIGDLVVDEEGADV 60
G K F +PK D E + WK + S+D + W +E G + D+
Sbjct: 332 GKEKAQRFIAIPKPLKLDAEHYGYWKVLIKRSIQSIDMDAWFAVEDGWMPPTTKDAKRDI 391
Query: 61 DRRKHTP--AQKKLYKKHHT-IRGALVKAIPKAEYMKMSDKSTAKSMFASLCANYEGSKK 117
+ T A +K H++ + ++ + ++ ++ +AK ++ L ++E +
Sbjct: 392 VSKSRTEWIADEKTAANHNSQALSVIFGSLLRNKFTQVQGCLSAKEVWEILQVSFECTNN 451
Query: 118 VREAKALMLVHQYELFKMKDGESIEQMYSRFHTLVSGLQILKKSCVASDHVSKILRSLPA 177
V+ + ML ++E M+ ES++ + ++ +L K+ V K LRSLP
Sbjct: 452 VKRTRLDMLASEFENLTMEAEESVDDFNGKLSSITQEAVVLGKTYKDKKMVKKFLRSLPD 511
Query: 178 RWRPKVTAIE 187
+++ +AI+
Sbjct: 512 KFQSHKSAID 521
Score = 37.0 bits (84), Expect = 0.007
Identities = 22/68 (32%), Positives = 38/68 (55%)
Query: 129 QYELFKMKDGESIEQMYSRFHTLVSGLQILKKSCVASDHVSKILRSLPARWRPKVTAIEE 188
+YE KMK+ ++I ++ + + L++ + V KIL SLP R+ V +++
Sbjct: 97 EYENLKMKESDNINTFMTKLIEMGNQLRVHGEEKSDYQIVQKILISLPKRFDIIVAMMKQ 156
Query: 189 TKDLNTLS 196
TKDL +LS
Sbjct: 157 TKDLTSLS 164
>At2g07080 putative gag-protease polyprotein
Length = 627
Score = 45.8 bits (107), Expect = 1e-05
Identities = 30/127 (23%), Positives = 63/127 (48%), Gaps = 2/127 (1%)
Query: 55 EEGADVDRRK-HTPAQKKLYKKHHT-IRGALVKAIPKAEYMKMSDKSTAKSMFASLCANY 112
E+G + + K + A++KL K + A+ + + E+ + +AK + +L ++
Sbjct: 36 EDGFKITKPKANWTAEEKLQSKFNARAMKAIFNGVDEDEFKLIQGCKSAKQAWDTLQKSH 95
Query: 113 EGSKKVREAKALMLVHQYELFKMKDGESIEQMYSRFHTLVSGLQILKKSCVASDHVSKIL 172
EG+ V+ + + Q+E KM+ E I + S+ L + +++ K+ V K+L
Sbjct: 96 EGTSSVKRTRLDHIATQFEYLKMEPDEKIVKFSSKISALANEAEVMGKTYKDQKLVKKLL 155
Query: 173 RSLPARW 179
R LP ++
Sbjct: 156 RCLPPKF 162
>At2g05960 putative retroelement pol polyprotein
Length = 1200
Score = 45.4 bits (106), Expect = 2e-05
Identities = 27/101 (26%), Positives = 56/101 (54%), Gaps = 1/101 (0%)
Query: 93 MKMSDKSTAKSMFASLCANYEGSKKVREAKALMLVHQYELFKMKDGESIEQMYSRFHTLV 152
+++ T+K+++ + + G+++V+EAK L+ +++ KMKD E+I++ R +
Sbjct: 35 LQVGKLKTSKAVWDKIQSRNLGAERVKEAKLKTLMAEFDKLKMKDNETIDECAGRLSEIS 94
Query: 153 SGLQILKKSCVASDHVSKILRSLPA-RWRPKVTAIEETKDL 192
+ L + + V K L+SLP ++ V A+E+ DL
Sbjct: 95 TKSTSLGEDIEETKVVKKFLKSLPTKKYIHIVAALEQVLDL 135
>At2g21310 putative retroelement pol polyprotein
Length = 838
Score = 43.9 bits (102), Expect = 6e-05
Identities = 47/179 (26%), Positives = 77/179 (42%), Gaps = 19/179 (10%)
Query: 7 SDFGKVPKFNGDLEEFS*WKTNFYNHVMSLDEELWDILEGGIGDLVVDEEG--ADVDRRK 64
S +V K +G+ ++ WK H+ EL +LEG D ++EE A+ D
Sbjct: 3 SGHSEVEKLDGE-GDYVLWKEKLLAHI-----ELLGLLEGLEEDEAIEEEESTAETDSLL 56
Query: 65 HTPAQKKLYKKHHTIRGALVKAIPKAEYMKMSDKSTAKSMFASLCANYEGSKKVREAKAL 124
K L +K R ++ ++ K+ + TA M L K+ AK+L
Sbjct: 57 TKTEDKVLKEKRGKARSTVILSLGNHVLRKVIKEKTAAGMIRVL-------DKLFMAKSL 109
Query: 125 ----MLVHQYELFKMKDGESIEQMYSRFHTLVSGLQILKKSCVASDHVSKILRSLPARW 179
L + +KM D +IE+ + F L+S L+ +K S D +L SLP ++
Sbjct: 110 PNRIYLKQRLYGYKMSDSMTIEENVNDFFKLISDLENVKVSVPDEDQAIVLLMSLPKQF 168
>At3g21030 hypothetical protein
Length = 411
Score = 42.0 bits (97), Expect = 2e-04
Identities = 33/162 (20%), Positives = 70/162 (42%), Gaps = 5/162 (3%)
Query: 36 LDEELWDILEGGIG-DLVVDEEGADVDRRKHTPAQKKLYKKHHTIRGALVKAIPKAEYMK 94
++ +WD+++ G+ + + E A + + L K + + ++P + + K
Sbjct: 27 VENGVWDVVQNGVSPNPTTNPELAATIKAVDLAQWRNLVVKDNKALKIMQSSLPDSVFRK 86
Query: 95 MSDKSTAKSMFASLCANYEGSKKVREAKALMLVHQYELFKMKDGESIEQMYSRFHTLVSG 154
++AK ++ L + +EAK L Q+E M +GE ++ R ++
Sbjct: 87 TISIASAKELWDLL----KKGNDTKEAKLRRLEKQFENLMMYEGERMKLYLKRLEKIIEE 142
Query: 155 LQILKKSCVASDHVSKILRSLPARWRPKVTAIEETKDLNTLS 196
L +L ++K+L SLP + V ++E L L+
Sbjct: 143 LFVLGNPISDDKVIAKLLTSLPRSYDDSVPVLKEFMTLPELT 184
>At3g21040 hypothetical protein
Length = 534
Score = 40.4 bits (93), Expect = 6e-04
Identities = 22/84 (26%), Positives = 39/84 (46%)
Query: 109 CANYEGSKKVREAKALMLVHQYELFKMKDGESIEQMYSRFHTLVSGLQILKKSCVASDHV 168
C Y+ + +++AK L L Q+E M DGES++ R + + D +
Sbjct: 224 CRGYDKNNPIKDAKLLRLEKQFEKLMMYDGESMDSYLERVLDTIEQFRASGNPLSDDDVI 283
Query: 169 SKILRSLPARWRPKVTAIEETKDL 192
+K+L SL + + +EE +L
Sbjct: 284 AKLLTSLSWPYDDAIPVLEELMNL 307
Score = 35.4 bits (80), Expect = 0.020
Identities = 29/163 (17%), Positives = 69/163 (41%), Gaps = 6/163 (3%)
Query: 36 LDEELWDILEGGIGD--LVVDEEGADVDRRKHTPAQKKLYKKHHTIRGALVKAIPKAEYM 93
++ +WD+++ G+ E A + T + ++ + + ++ + A+P + +
Sbjct: 27 VENGVWDVVQNGVSPNPTTDPEVAATIKVEDLTQWRNRMIEDNKALK-IMQSALPDSVFR 85
Query: 94 KMSDKSTAKSMFASLCANYEGSKKVREAKALMLVHQYELFKMKDGESIEQMYSRFHTLVS 153
K +++K ++ L +G+ K L Q E M +GE ++ R ++
Sbjct: 86 KTISVASSKELWDLL---KKGNDNEEAKKLRRLEKQLENLTMYEGERMKSYLKRVEKIIE 142
Query: 154 GLQILKKSCVASDHVSKILRSLPARWRPKVTAIEETKDLNTLS 196
+ ++K+L SLP + +T ++E L L+
Sbjct: 143 EFFVWGNPISDEKVIAKLLTSLPRPYDDSITVLKEFMTLPDLT 185
>At3g21010 unknown protein
Length = 438
Score = 38.5 bits (88), Expect = 0.002
Identities = 37/164 (22%), Positives = 72/164 (43%), Gaps = 8/164 (4%)
Query: 37 DEELWDILEGGI--GDLVVDEEGADVDRRKHTPAQKKLYKKHHTIRGALVKAIPKAEYMK 94
++ LWD++E G+ V E A + + + + L +K L ++ + + K
Sbjct: 30 EKGLWDVVENGVPPDPSKVPELAATIQAEEFSK-WRDLAEKDMKALQILQSSLTDSAFRK 88
Query: 95 MSDKSTAKSMFASLCANYEGSKKVREAKALMLVHQYELFKMKDGESIEQMYSRFHTLVSG 154
S+AK ++ SL +G+ + +AK L Q+E M +GE I R +V
Sbjct: 89 TLSASSAKDLWDSL---EKGNNE--QAKLRRLEKQFEELSMNEGEPINPYIDRVIEIVEQ 143
Query: 155 LQILKKSCVASDHVSKILRSLPARWRPKVTAIEETKDLNTLSVQ 198
L+ K + + K+L +L + + + DL +S++
Sbjct: 144 LRRFKIAKSDYQVIEKVLATLSGSYDDVAPVLGDLVDLKNMSLK 187
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.134 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,459,099
Number of Sequences: 26719
Number of extensions: 183758
Number of successful extensions: 516
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 30
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 476
Number of HSP's gapped (non-prelim): 50
length of query: 198
length of database: 11,318,596
effective HSP length: 94
effective length of query: 104
effective length of database: 8,807,010
effective search space: 915929040
effective search space used: 915929040
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC136450.2


