

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135161.1 - phase: 0
(820 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
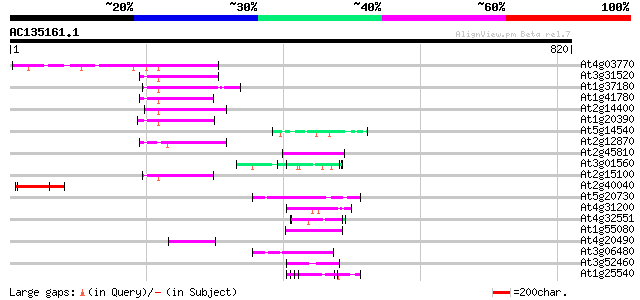
Score E
Sequences producing significant alignments: (bits) Value
At4g03770 hypothetical protein 70 6e-12
At3g31520 hypothetical protein 64 3e-10
At1g37180 hypothetical protein 62 1e-09
At1g41780 hypothetical protein 55 2e-07
At2g14400 putative retroelement pol polyprotein 53 8e-07
At1g20390 hypothetical protein 53 8e-07
At5g14540 unknown protein 51 2e-06
At2g12870 putative retroelement pol polyprotein 51 2e-06
At2g45810 putative ATP-dependent RNA helicase 50 4e-06
At3g01560 unknown protein 49 9e-06
At2g15100 putative retroelement pol polyprotein 49 9e-06
At2g40040 unknown protein 49 1e-05
At5g20730 auxin response factor 7 (ARF7) 48 3e-05
At4g31200 predicted protein 48 3e-05
At4g32551 Leunig protein 46 1e-04
At1g55080 unknown protein 46 1e-04
At4g20490 putative protein 45 1e-04
At3g06480 putative RNA helicase 45 2e-04
At3g52460 putative protein 45 2e-04
At1g25540 hypothetical protein 45 2e-04
>At4g03770 hypothetical protein
Length = 464
Score = 69.7 bits (169), Expect = 6e-12
Identities = 83/328 (25%), Positives = 133/328 (40%), Gaps = 41/328 (12%)
Query: 4 MAELIKTMAETQTQAQAQIQAQ---AQALAQAQTQAQAVTEAQAQSQAPPPPPPIRTQAE 60
+AEL MA+ Q + Q QA AQ L A +Q Q T R A+
Sbjct: 34 LAELRSMMAQLQQKVNDQEQANRSLAQQLEAATSQGQIRTTRFGARHLQDR----RAAAD 89
Query: 61 ASSSWTLCADTPRQSAPQRSAPWFPPFTAGEIFRPITCEAQMP------THQYTAQVPLP 114
+ + + + P R TA EI R T A + T + Q+P P
Sbjct: 90 LNPTRFVFHTPGNTTRPVRR-------TAPEIGRDRTEPAILGNRETKRTERNKPQLPPP 142
Query: 115 AMRVTPATMTYSAPVIHTIPQTEEPIFHSGNVEAYEEVSDLREKYDELRRDMKALREKGK 174
VT A + EE I + +E+S ++ + +MK +R +
Sbjct: 143 RAEVTEADHIG----VSDDKDLEENIRWAEEYATEQEISAIKLSLAKAENEMKLVRSQMH 198
Query: 175 FGKTA---YDLCLVPSVQVPHKFKIPD----------FEKYKGSSCPEEHLKMYV----R 217
++ D L S P I + E + GSS P+ HLK+++ R
Sbjct: 199 NAVSSAPNIDRILEESHNTPFTHMISNAIISDPGKLRIEYFNGSSDPKGHLKLFIISVAR 258
Query: 218 RMPAYAQDDQILIYYFQESLTGPASKWYTNLDKTRVQTFRDLCEAFVEQYSYNVDMTPDR 277
+ D L + F E L GPA W+T L V +F++L F++QYS +D +
Sbjct: 259 AKFRPEERDAGLCHLFVEHLKGPALNWFTRLKGNSVDSFQELSTLFLKQYSVLIDPSTSD 318
Query: 278 SDLQAMTQGDKETFKEYAQRWRDTAAQV 305
+DL +++Q E +++ ++R T A+V
Sbjct: 319 ADLWSLSQQPNEPLRDFLAKFRSTLAKV 346
>At3g31520 hypothetical protein
Length = 528
Score = 63.9 bits (154), Expect = 3e-10
Identities = 37/119 (31%), Positives = 64/119 (53%), Gaps = 7/119 (5%)
Query: 191 PHKFKIPDFEKYKGSSCPEEHLKMYV----RRMPAYAQDDQILIYYFQESLTGPASKWYT 246
P K +I E + GSS P+ HLK ++ R + D L + F E L GPA W++
Sbjct: 239 PGKLRI---EYFNGSSDPKRHLKSFIISVARTKFRPEERDAGLCHLFVEHLKGPALCWFS 295
Query: 247 NLDKTRVQTFRDLCEAFVEQYSYNVDMTPDRSDLQAMTQGDKETFKEYAQRWRDTAAQV 305
L+ + +F++L F++QYS +D+ +DL +++Q E +++ + R T A+V
Sbjct: 296 RLEGNSMDSFQELSTLFLKQYSVLIDLGTSDADLWSLSQQPNEPLRDFLAKLRSTLAKV 354
>At1g37180 hypothetical protein
Length = 661
Score = 62.0 bits (149), Expect = 1e-09
Identities = 42/148 (28%), Positives = 72/148 (48%), Gaps = 9/148 (6%)
Query: 194 FKIPDFEKYKGSSCPEEHLKMY---VRRMPAYA-QDDQILIYYFQESLTGPASKWYTNLD 249
FK+P Y G P EHL + VRR+P ++D L F E+L+GPA W+T L+
Sbjct: 314 FKLP---VYNGKGDPNEHLTSFQVIVRRVPLEPYEEDAGLCKLFSENLSGPALTWFTQLE 370
Query: 250 KTRVQTFRDLCEAFVEQYSYNVDMTPDRSDLQAMTQGDKETFKEYAQRWRDTAAQVSPRI 309
+ + F+ L AF++QY Y + + L ++Q + + Y +++ Q+ P +
Sbjct: 371 EGSIDNFKQLSTAFIKQYGYFIKSDITEAHLWNLSQSADDPLRTYIIVFKEIMVQI-PSL 429
Query: 310 EEKEMTKLFLKTLNHFYYKKMVGSTPKS 337
+ + LK K+ S+PK+
Sbjct: 430 SD-STAQFALKNGTFSVEKESSKSSPKA 456
>At1g41780 hypothetical protein
Length = 246
Score = 54.7 bits (130), Expect = 2e-07
Identities = 33/112 (29%), Positives = 55/112 (48%), Gaps = 7/112 (6%)
Query: 191 PHKFKIPDFEKYKGSSCPEEHLKMY----VRRMPAYAQDDQILIYYFQESLTGPASKWYT 246
P K +I E + GSS P+ HLK + R + D L + F E L GP W++
Sbjct: 138 PEKLRI---EYFSGSSDPKGHLKSFKISVARAKFKPEESDAGLCHLFVEHLKGPVLDWFS 194
Query: 247 NLDKTRVQTFRDLCEAFVEQYSYNVDMTPDRSDLQAMTQGDKETFKEYAQRW 298
L+ V +F++L F++QYS +D DL +++Q E ++ ++
Sbjct: 195 RLEGNSVDSFQELSTLFLKQYSVLIDPGTSDVDLWSLSQQPNEPLSDFLTKF 246
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 52.8 bits (125), Expect = 8e-07
Identities = 31/124 (25%), Positives = 61/124 (49%), Gaps = 4/124 (3%)
Query: 198 DFEKYKGSSCPEEHLKMYV----RRMPAYAQDDQILIYYFQESLTGPASKWYTNLDKTRV 253
+ E Y G P+ +L ++ R A +D F E+L G A W+T L+ +
Sbjct: 161 NLESYNGLEDPKGYLAAFLIAAGRVDLNEADEDVRYCKLFSENLCGQALMWFTQLEPGSI 220
Query: 254 QTFRDLCEAFVEQYSYNVDMTPDRSDLQAMTQGDKETFKEYAQRWRDTAAQVSPRIEEKE 313
F +L F++QYS +D + +DL ++QG ET + + +++ +++S ++
Sbjct: 221 SNFNELSVVFLKQYSILMDKSISDTDLWNLSQGPNETLRAFITKFKYVLSKLSRISQQSA 280
Query: 314 MTKL 317
++ L
Sbjct: 281 LSAL 284
>At1g20390 hypothetical protein
Length = 1791
Score = 52.8 bits (125), Expect = 8e-07
Identities = 31/117 (26%), Positives = 58/117 (49%), Gaps = 7/117 (5%)
Query: 187 SVQVPHKFKIPDFEKYKGSSCPEEHLKMY---VRRMPAYAQD-DQILIYYFQESLTGPAS 242
S++ K K+ E Y G + P+E L + + R + D F E LTGPA
Sbjct: 165 SIRGAQKIKL---ESYNGRNDPKEFLTSFNVAINRAELTIDNFDAGRCQIFIEHLTGPAH 221
Query: 243 KWYTNLDKTRVQTFRDLCEAFVEQYSYNVDMTPDRSDLQAMTQGDKETFKEYAQRWR 299
W++ L + +F L +F++ Y+ ++ +DL +++QG KE+ + + R++
Sbjct: 222 NWFSRLKPNSIDSFHQLTSSFLKHYAPLIENQTSNADLWSISQGAKESLRSFVDRFK 278
>At5g14540 unknown protein
Length = 530
Score = 51.2 bits (121), Expect = 2e-06
Identities = 53/163 (32%), Positives = 65/163 (39%), Gaps = 45/163 (27%)
Query: 384 MVTHGGPQQTY---PAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNV 440
+ G P Q + PA QH ++P S PQ+P Q P FP PS Q+
Sbjct: 239 LTQQGLPPQQFIQPPASQH--GLSPPSLQL------PQLPNQFSPQQEPYFP--PSGQSQ 288
Query: 441 QPQNVQQ--------QNFQQQPYQQYPYQ-QYPQQ-----------NFQQQPYQQRPQQP 480
P +Q Q+ Q PYQ P Q QYPQQ N ++ PY Q+ P
Sbjct: 289 PPPTIQPPYQPPPPTQSLHQPPYQPPPQQPQYPQQPPPQLQHPSGYNPEEPPYPQQSYPP 348
Query: 481 RPPRMPINPIPVTYAELLPGLLKKNLVQTRTAPPIPEKLPSWY 523
PPR P + P PG Q APP P PS Y
Sbjct: 349 NPPRQPPSHPP-------PGSAPSQ--QYYNAPPTP---PSMY 379
>At2g12870 putative retroelement pol polyprotein
Length = 411
Score = 51.2 bits (121), Expect = 2e-06
Identities = 38/135 (28%), Positives = 63/135 (46%), Gaps = 15/135 (11%)
Query: 191 PHKFKIPDFEKYKGSSCPEEHLKMYVRRMPAYAQDDQI--------LIYYFQESLTGPAS 242
P K +I + + GSS P+ HLK ++ YA D+ L + F E L GP
Sbjct: 74 PGKLRI---DYFNGSSYPKGHLKSFI----IYAARDKFKPEERDAGLCHLFVEHLKGPVL 126
Query: 243 KWYTNLDKTRVQTFRDLCEAFVEQYSYNVDMTPDRSDLQAMTQGDKETFKEYAQRWRDTA 302
+ L+ V +F +L F++Q+S +D SDL +++Q E + ++R T
Sbjct: 127 VLFLRLEGNFVDSFEELSTLFLKQFSVLIDPGTLDSDLWSLSQQPNEPLMDLLTKFRSTL 186
Query: 303 AQVSPRIEEKEMTKL 317
A V I+ ++ L
Sbjct: 187 ASVEGIIDVAALSAL 201
>At2g45810 putative ATP-dependent RNA helicase
Length = 528
Score = 50.4 bits (119), Expect = 4e-06
Identities = 39/93 (41%), Positives = 46/93 (48%), Gaps = 3/93 (3%)
Query: 400 IAAITPTSHP-FQQTN-NHPQIPQYPQI-PQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQ 456
I A P P FQ N N PQ QY Q +PQ PQ PQ +Q Q+ QQ Q+ Q
Sbjct: 15 IGAAGPGPDPNFQSRNPNPPQPQQYLQSRTPFPQQPQPQPPQYLQSQSDAQQYVQRGYPQ 74
Query: 457 QYPYQQYPQQNFQQQPYQQRPQQPRPPRMPINP 489
Q QQ QQ QQQ QQ Q R ++P +P
Sbjct: 75 QIQQQQQLQQQQQQQQQQQEQQWSRRAQLPGDP 107
>At3g01560 unknown protein
Length = 511
Score = 49.3 bits (116), Expect = 9e-06
Identities = 48/178 (26%), Positives = 71/178 (38%), Gaps = 30/178 (16%)
Query: 332 GSTPKSFAEMVGMGVQLEEGVR---------EGRLVKNTTPASGTKKTGNHFPRKKEQEV 382
GST ++ + V+++ GV+ E +L K+ K H +
Sbjct: 182 GSTDGKMRQLKNILVEVQSGVQLLKDKQEILEAQLSKHQVSNQHAKTHSLHVDPTAQSPA 241
Query: 383 GMVTHGGPQQTYPAYQHIAAITPTSHPFQQTNNH--PQIPQ--------YPQIPQYPQFP 432
+ P Q +P + T+ P Q ++ PQ+P Y P +PQ P
Sbjct: 242 PV-----PMQQFPLTSFPQPPSSTAAPSQPPSSQLPPQLPTQFSSQQEPYCPPPSHPQPP 296
Query: 433 -QNPSP-QNVQPQNVQQQNFQQQPYQ-QYPYQQYPQQNF---QQQPYQQRPQQPRPPR 484
NP P Q Q Q Q ++Q P Q QYP Q P + +Q PYQ + P PPR
Sbjct: 297 PSNPPPYQAPQTQTPHQPSYQSPPQQPQYPQQPPPSSGYNPEEQPPYQMQSYPPNPPR 354
Score = 43.9 bits (102), Expect = 4e-04
Identities = 28/89 (31%), Positives = 36/89 (39%), Gaps = 13/89 (14%)
Query: 405 PTSHPFQQTNNHPQI----------PQYPQIPQYPQFPQNPSPQN-VQPQNVQQQNFQQQ 453
P SHP +N P P Y PQ PQ+PQ P P + P+ +Q +Q Q
Sbjct: 289 PPSHPQPPPSNPPPYQAPQTQTPHQPSYQSPPQQPQYPQQPPPSSGYNPE--EQPPYQMQ 346
Query: 454 PYQQYPYQQYPQQNFQQQPYQQRPQQPRP 482
Y P +Q P P QP+P
Sbjct: 347 SYPPNPPRQQPPAGSTPSQQFYNPPQPQP 375
Score = 43.9 bits (102), Expect = 4e-04
Identities = 31/107 (28%), Positives = 42/107 (38%), Gaps = 16/107 (14%)
Query: 392 QTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQ 451
+T+ + A +P P QQ + +PQ P P P + PQ Q + Q
Sbjct: 227 KTHSLHVDPTAQSPAPVPMQQF----PLTSFPQPPSSTAAPSQPPSSQLPPQLPTQFSSQ 282
Query: 452 QQPY------------QQYPYQQYPQQNFQQQPYQQRPQQPRPPRMP 486
Q+PY PYQ Q Q YQ PQQP+ P+ P
Sbjct: 283 QEPYCPPPSHPQPPPSNPPPYQAPQTQTPHQPSYQSPPQQPQYPQQP 329
>At2g15100 putative retroelement pol polyprotein
Length = 1329
Score = 49.3 bits (116), Expect = 9e-06
Identities = 32/108 (29%), Positives = 51/108 (46%), Gaps = 7/108 (6%)
Query: 194 FKIPDFEKYKGSSCPEEHLKMY---VRRMPAYA-QDDQILIYYFQESLTGPASKWYTNLD 249
FK+P Y G +EHL + R+P ++D L F E+L G A W+T L+
Sbjct: 173 FKLP---VYNGKGDLKEHLTSFQVIAGRVPLEPHEEDAGLCKLFSENLFGLALTWFTQLE 229
Query: 250 KTRVQTFRDLCEAFVEQYSYNVDMTPDRSDLQAMTQGDKETFKEYAQR 297
+ + F+ L AF++QY Y ++ + L +Q E + Y R
Sbjct: 230 EGSIDNFKQLSTAFIKQYEYFINSDITEAHLWNFSQSADEPLRTYIYR 277
>At2g40040 unknown protein
Length = 839
Score = 48.5 bits (114), Expect = 1e-05
Identities = 23/69 (33%), Positives = 48/69 (69%), Gaps = 5/69 (7%)
Query: 12 AETQTQAQAQIQAQAQALAQAQTQAQAVTEAQAQSQAPPPPPPIRTQAEASSSWTLCADT 71
++TQ+Q+Q+Q Q+Q+Q+ +Q+Q+Q+Q+ +++Q+QSQ+P +TQ ++ S A +
Sbjct: 774 SQTQSQSQSQSQSQSQSQSQSQSQSQSQSQSQSQSQSPS-----QTQTQSPSQTQAQAQS 828
Query: 72 PRQSAPQRS 80
P +P ++
Sbjct: 829 PSSQSPSQT 837
Score = 45.4 bits (106), Expect = 1e-04
Identities = 19/50 (38%), Positives = 38/50 (76%)
Query: 9 KTMAETQTQAQAQIQAQAQALAQAQTQAQAVTEAQAQSQAPPPPPPIRTQ 58
++ +++Q+Q+Q+Q Q+Q+Q+ + +QTQ Q+ ++ QAQ+Q+P P +TQ
Sbjct: 789 QSQSQSQSQSQSQSQSQSQSQSPSQTQTQSPSQTQAQAQSPSSQSPSQTQ 838
Score = 42.4 bits (98), Expect = 0.001
Identities = 19/49 (38%), Positives = 36/49 (72%)
Query: 9 KTMAETQTQAQAQIQAQAQALAQAQTQAQAVTEAQAQSQAPPPPPPIRT 57
++ +++Q+Q+Q+Q Q+Q+Q+ +Q QTQ+ + T+AQAQS + P +T
Sbjct: 791 QSQSQSQSQSQSQSQSQSQSPSQTQTQSPSQTQAQAQSPSSQSPSQTQT 839
Score = 36.6 bits (83), Expect = 0.058
Identities = 28/96 (29%), Positives = 49/96 (50%), Gaps = 8/96 (8%)
Query: 15 QTQAQAQIQAQAQALAQAQTQAQAVTEAQAQSQAPPPPPPIRTQAEASSSWTLCADTPRQ 74
QTQ Q+Q +Q +A + +Q QAQ+ ++ Q+QSQ+ ++Q+++ S + + Q
Sbjct: 749 QTQTQSQSPSQTRAQSPSQAQAQSPSQTQSQSQS-------QSQSQSQSQSQSQSQSQSQ 801
Query: 75 SAPQRSAPWFPPFTAGEIFRPITCEAQMPTHQYTAQ 110
S Q S P T + +AQ P+ Q +Q
Sbjct: 802 SQSQ-SQSQSPSQTQTQSPSQTQAQAQSPSSQSPSQ 836
>At5g20730 auxin response factor 7 (ARF7)
Length = 1164
Score = 47.8 bits (112), Expect = 3e-05
Identities = 43/160 (26%), Positives = 66/160 (40%), Gaps = 13/160 (8%)
Query: 356 RLVKNTTPASGTKKTGNHFPRKKEQEVGMVTHGGPQQTYPAYQHIAAITPTS--HPFQQT 413
+L+ TP G + F K+ Q+ M P T Q + + +S H QQ+
Sbjct: 462 KLLSFQTPHGGISSSNLQF-NKQNQQAPMSQLPQPPTTLSQQQQLQQLLHSSLNHQQQQS 520
Query: 414 NNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPY 473
+ Q Q + Q Q N Q Q+ QQQ QQ QQ QQ+ QQP
Sbjct: 521 QSQQQQQQQQLLQQQQQLQSQQHSNNNQSQSQQQQQLLQQQQQQQLQQQH------QQPL 574
Query: 474 QQRPQQPRPPRMPINPIPVTYAELLPGLLKKNLVQTRTAP 513
QQ+ QQ + P+ +++ P L+++ +Q P
Sbjct: 575 QQQTQQQQLRTQPLQ----SHSHPQPQQLQQHKLQQLQVP 610
Score = 31.6 bits (70), Expect = 1.9
Identities = 33/160 (20%), Positives = 54/160 (33%), Gaps = 37/160 (23%)
Query: 391 QQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQN----------- 439
+Q A+ H+ A T S + + + + P PQ QF + Q
Sbjct: 667 KQLQQAHHHLGASTSQSSVIETSKSSSNLMSAP--PQETQFSRQVEQQQPPGLNGQNQQT 724
Query: 440 --------VQPQNVQQQNFQQQPYQQY----------------PYQQYPQQNFQQQPYQQ 475
Q Q + QQ+ +QP+ Q+ P Q P Q Q QQ
Sbjct: 725 LLQQKAHQAQAQQIFQQSLLEQPHIQFQLLQRLQQQQQQQFLSPQSQLPHHQLQSQQLQQ 784
Query: 476 RPQQPRPPRMPINPIPVTYAELLPGLLKKNLVQTRTAPPI 515
P + + P + + L P + + Q + PP+
Sbjct: 785 LPTLSQGHQFPSSCTNNGLSTLQPPQMLVSRPQEKQNPPV 824
Score = 30.8 bits (68), Expect = 3.2
Identities = 27/98 (27%), Positives = 39/98 (39%), Gaps = 5/98 (5%)
Query: 377 KKEQEVGMVTHGGPQQTYPAYQHIAAITPTSHPFQQTNNHP--QIPQYPQIPQYP-QFPQ 433
+++Q++ H Q+ Q QQ + P Q Q Q+ P Q
Sbjct: 533 QQQQQLQSQQHSNNNQSQSQQQQQLLQQQQQQQLQQQHQQPLQQQTQQQQLRTQPLQSHS 592
Query: 434 NPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQ 471
+P PQ +Q +QQ Q Q Y QQ QQ+ QQ
Sbjct: 593 HPQPQQLQQHKLQQLQVPQN--QLYNGQQAAQQHQSQQ 628
Score = 30.0 bits (66), Expect = 5.5
Identities = 16/42 (38%), Positives = 21/42 (49%)
Query: 15 QTQAQAQIQAQAQALAQAQTQAQAVTEAQAQSQAPPPPPPIR 56
Q Q Q Q+Q Q Q Q QTQ Q + QS + P P ++
Sbjct: 559 QQQQQQQLQQQHQQPLQQQTQQQQLRTQPLQSHSHPQPQQLQ 600
>At4g31200 predicted protein
Length = 650
Score = 47.8 bits (112), Expect = 3e-05
Identities = 37/105 (35%), Positives = 44/105 (41%), Gaps = 12/105 (11%)
Query: 405 PTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQ---PQNVQQQNF-------QQQP 454
P + QQ + Q Q PQ +PQ PQ PSP N P + Q F QQQ
Sbjct: 15 PYAQQQQQQGPNFQQQQQPQFGFHPQHPQYPSPMNASGFIPPHPSMQQFPYQHPMHQQQQ 74
Query: 455 YQQYPYQQYPQQNFQQQPYQQRPQQPRPPRMPINPIPVTYAELLP 499
Q P+ +PQ QQQP P P P +P P P Y P
Sbjct: 75 PQHLPHPPHPQMFGQQQPQAFLPHLP-PHHLP-PPFPGPYDSAPP 117
Score = 30.0 bits (66), Expect = 5.5
Identities = 34/136 (25%), Positives = 49/136 (36%), Gaps = 16/136 (11%)
Query: 376 RKKEQEVGMVTHGGPQQTYPAYQHIAAITPTSHPFQQTNNHPQIP--QYPQIPQYPQFPQ 433
R+K+ GG Y Y+H ++ P PF P P P I P
Sbjct: 151 RQKDNPDYAFLFGGEGHGYYRYKHFLSMHPPGGPFD-----PPFPSSSMPMIHHPPNPMM 205
Query: 434 NPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRP-------QQPRPPRMP 486
+PS NV P + +Q P+ + + + Q QP+ P Q R
Sbjct: 206 SPSMNNV-PGALAVPPIRQPPFPPF-HDHHQLQQHLPQPHPFAPHARPDFDQSTHAFRGL 263
Query: 487 INPIPVTYAELLPGLL 502
P+P A L G+L
Sbjct: 264 SGPLPADVAMELNGVL 279
>At4g32551 Leunig protein
Length = 931
Score = 45.8 bits (107), Expect = 1e-04
Identities = 30/88 (34%), Positives = 35/88 (39%), Gaps = 8/88 (9%)
Query: 412 QTNNHPQIPQYPQIPQYPQFPQNP--------SPQNVQPQNVQQQNFQQQPYQQYPYQQY 463
Q + HPQ+ Q Q Q Q Q Q Q+ Q QQQ QQ QQ
Sbjct: 92 QQSQHPQVSQQQQQQQQQQIQMQQLLLQRAQQQQQQQQQQHHHHQQQQQQQQQQQQQQQQ 151
Query: 464 PQQNFQQQPYQQRPQQPRPPRMPINPIP 491
QQ Q QP Q+ QQ P+ P P
Sbjct: 152 QQQQHQNQPPSQQQQQQSTPQHQQQPTP 179
Score = 43.5 bits (101), Expect = 5e-04
Identities = 30/76 (39%), Positives = 34/76 (44%), Gaps = 1/76 (1%)
Query: 411 QQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQ 470
QQ + Q Q Q Q Q+ Q Q Q QQQ QQQ QQ+ Q P Q QQ
Sbjct: 109 QQIQMQQLLLQRAQQQQQQQQQQHHHHQQQQQQQQQQQQQQQQQQQQH-QNQPPSQQQQQ 167
Query: 471 QPYQQRPQQPRPPRMP 486
Q Q QQP P + P
Sbjct: 168 QSTPQHQQQPTPQQQP 183
Score = 37.7 bits (86), Expect = 0.026
Identities = 24/62 (38%), Positives = 28/62 (44%)
Query: 418 QIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRP 477
Q Q Q + Q Q Q Q Q QQQ Q QP Q QQ Q+ QQ QQ+P
Sbjct: 124 QQQQQQQQHHHHQQQQQQQQQQQQQQQQQQQQHQNQPPSQQQQQQSTPQHQQQPTPQQQP 183
Query: 478 QQ 479
Q+
Sbjct: 184 QR 185
Score = 37.0 bits (84), Expect = 0.045
Identities = 32/106 (30%), Positives = 40/106 (37%), Gaps = 4/106 (3%)
Query: 376 RKKEQEVGMVTHGGPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNP 435
+ +EQ++ H PQ + Q Q Q Q Q + Q Q
Sbjct: 85 KAREQQLQQSQH--PQVSQQQQQQQQQQIQMQQLLLQRAQQQQQQQQQQHHHHQQQQQQQ 142
Query: 436 SPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQPR 481
Q Q Q QQQ+ Q P QQ Q PQ QQQP Q+ Q R
Sbjct: 143 QQQQQQQQQQQQQHQNQPPSQQQQQQSTPQH--QQQPTPQQQPQRR 186
Score = 31.6 bits (70), Expect = 1.9
Identities = 25/77 (32%), Positives = 34/77 (43%), Gaps = 3/77 (3%)
Query: 3 EMAELIKTMAETQTQAQAQIQAQAQALAQAQTQAQAVTEAQAQSQAPPPPPPIRTQAEAS 62
+M +L+ A+ Q Q Q Q Q Q Q Q Q + Q Q Q PP + Q + S
Sbjct: 112 QMQQLLLQRAQQQQQQQQQQHHHHQQQQQQQQQQQQ--QQQQQQQQHQNQPPSQQQQQQS 169
Query: 63 SSWTLCADTPRQSAPQR 79
+ TP+Q PQR
Sbjct: 170 TPQHQQQPTPQQQ-PQR 185
>At1g55080 unknown protein
Length = 244
Score = 45.8 bits (107), Expect = 1e-04
Identities = 34/89 (38%), Positives = 41/89 (45%), Gaps = 6/89 (6%)
Query: 404 TPTSHPFQQTNNHPQIPQYPQIPQYPQF--PQNPSPQNVQPQNVQQQNFQQQPYQQYPYQ 461
TPT+H Q Q+ Q Q Q QF Q Q Q Q QQ + Q YQQ+ Q
Sbjct: 25 TPTNHDQQLFLQQQQLQQQQQQQQQQQFHLQQQQQTQQQQQQFQPQQQQEMQQYQQFQQQ 84
Query: 462 QY--PQQNFQQQP--YQQRPQQPRPPRMP 486
Q+ QQ FQQQ Q P QP+ + P
Sbjct: 85 QHFIQQQQFQQQQRLLQSPPLQPQSLQSP 113
Score = 29.3 bits (64), Expect = 9.3
Identities = 30/104 (28%), Positives = 41/104 (38%), Gaps = 19/104 (18%)
Query: 367 TKKTGNHFPRKKEQEVGMVTHGGPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIP 426
T++ F +++QE+ QQ + Q FQQ Q P P
Sbjct: 60 TQQQQQQFQPQQQQEMQQYQQFQQQQHFIQQQQ----------FQQQQRLLQSP-----P 104
Query: 427 QYPQFPQNPSPQNVQ---PQNVQQQNFQQQPYQQYPYQQYPQQN 467
PQ Q+P PQ PQ++ QQQ Q P Q PQQ+
Sbjct: 105 LQPQSLQSPPPQQTMVHTPQSMMHTPQQQQQLVQTPVQT-PQQH 147
>At4g20490 putative protein
Length = 853
Score = 45.4 bits (106), Expect = 1e-04
Identities = 23/68 (33%), Positives = 36/68 (52%)
Query: 233 FQESLTGPASKWYTNLDKTRVQTFRDLCEAFVEQYSYNVDMTPDRSDLQAMTQGDKETFK 292
F E LTG A ++ L+ + F L AF++QY + SDL +MTQ + ET
Sbjct: 225 FVEGLTGNALTCFSRLEANSIDNFTQLSTAFLKQYRVFIQPGASSSDLWSMTQENGETLN 284
Query: 293 EYAQRWRD 300
+Y R+++
Sbjct: 285 DYLGRFKE 292
>At3g06480 putative RNA helicase
Length = 1088
Score = 45.1 bits (105), Expect = 2e-04
Identities = 39/124 (31%), Positives = 52/124 (41%), Gaps = 10/124 (8%)
Query: 356 RLVKNTTPASGTKKTGNHFPRKKEQEVGMVT--HGGPQQT--YPAYQHIAAITPTSHPFQ 411
+L + P+SG H ++ + VG V+ HG QQ +P+ QH+ HP Q
Sbjct: 64 KLAQIPVPSSGQGHQAQH---EQAKPVGHVSQQHGFQQQPQQFPS-QHVRPQMMQQHPAQ 119
Query: 412 QT--NNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQ 469
Q + Q PQ P +PS Q QP QQQ Q + Q P Q Q
Sbjct: 120 QMPQQSGQQFPQQQSQSMVPHPHGHPSVQTYQPTTQQQQQGMQNQHSQMPQQLSHQYAHS 179
Query: 470 QQPY 473
QQ Y
Sbjct: 180 QQHY 183
Score = 40.4 bits (93), Expect = 0.004
Identities = 38/121 (31%), Positives = 50/121 (40%), Gaps = 27/121 (22%)
Query: 383 GMVTHGGPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQP 442
G++ + P+ Y+ +A P S P+ QIP P Q Q+ Q
Sbjct: 35 GILYYWNPETNVTQYERPSAPPPHS---------ATTPKLAQIP-VPSSGQGHQAQHEQA 84
Query: 443 QNV----QQQNFQQQPYQ------------QYPYQQYPQQNFQQQPYQQ-RPQQPRPPRM 485
+ V QQ FQQQP Q Q+P QQ PQQ+ QQ P QQ + P P
Sbjct: 85 KPVGHVSQQHGFQQQPQQFPSQHVRPQMMQQHPAQQMPQQSGQQFPQQQSQSMVPHPHGH 144
Query: 486 P 486
P
Sbjct: 145 P 145
Score = 32.0 bits (71), Expect = 1.4
Identities = 29/87 (33%), Positives = 31/87 (35%), Gaps = 13/87 (14%)
Query: 405 PTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYP----- 459
P H QQ Q Q+P PQ Q P PQ QQ QQQ P
Sbjct: 86 PVGHVSQQHGFQQQPQQFPSQHVRPQMMQQ-HPAQQMPQQSGQQFPQQQSQSMVPHPHGH 144
Query: 460 -----YQQYPQQNFQ--QQPYQQRPQQ 479
YQ QQ Q Q + Q PQQ
Sbjct: 145 PSVQTYQPTTQQQQQGMQNQHSQMPQQ 171
Score = 29.6 bits (65), Expect = 7.1
Identities = 24/73 (32%), Positives = 30/73 (40%), Gaps = 9/73 (12%)
Query: 425 IPQYPQFPQNPSPQNVQPQNVQQ-------QNFQQQPYQQYPYQQYPQQN-FQQQPYQQR 476
+ QY + P P P + + Q Q Q Q Q P QQ+ FQQQP Q
Sbjct: 46 VTQYER-PSAPPPHSATTPKLAQIPVPSSGQGHQAQHEQAKPVGHVSQQHGFQQQPQQFP 104
Query: 477 PQQPRPPRMPINP 489
Q RP M +P
Sbjct: 105 SQHVRPQMMQQHP 117
>At3g52460 putative protein
Length = 300
Score = 44.7 bits (104), Expect = 2e-04
Identities = 29/82 (35%), Positives = 34/82 (41%), Gaps = 13/82 (15%)
Query: 405 PTSHPFQQTNNHPQIPQ--YPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQ 462
P P Q PQ Q YP + YP + Q P P N+ PYQQYPY Q
Sbjct: 28 PPPPPPQSQPPPPQTQQQTYPPVMGYPGYHQPPPPY---------PNYPNAPYQQYPYAQ 78
Query: 463 YPQQNF--QQQPYQQRPQQPRP 482
P ++ P QQ P RP
Sbjct: 79 APPASYYGSSYPAQQNPVYQRP 100
Score = 33.1 bits (74), Expect = 0.65
Identities = 22/55 (40%), Positives = 24/55 (43%), Gaps = 4/55 (7%)
Query: 432 PQNPSPQNVQPQNVQQQNFQQQPYQQYP-YQQYPQQ--NFQQQPYQQRPQQPRPP 483
P P PQ+ QP Q Q P YP Y Q P N+ PYQQ P PP
Sbjct: 28 PPPPPPQS-QPPPPQTQQQTYPPVMGYPGYHQPPPPYPNYPNAPYQQYPYAQAPP 81
>At1g25540 hypothetical protein
Length = 809
Score = 44.7 bits (104), Expect = 2e-04
Identities = 28/75 (37%), Positives = 32/75 (42%)
Query: 405 PTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYP 464
P QQ H Q Q QI Q Q Q+ Q + QQQ QQQ QQ+ Q
Sbjct: 652 PNQQQQQQQQLHQQQQQQQQIQQQQQQQQHLQQQQMPQLQQQQQQHQQQQQQQHQLSQLQ 711
Query: 465 QQNFQQQPYQQRPQQ 479
QQQ QQ+ QQ
Sbjct: 712 HHQQQQQQQQQQQQQ 726
Score = 43.1 bits (100), Expect = 6e-04
Identities = 24/58 (41%), Positives = 27/58 (46%)
Query: 417 PQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQ 474
PQIP Q Q Q Q +Q Q QQQ+ QQQ Q QQ Q QQQ +Q
Sbjct: 649 PQIPNQQQQQQQQLHQQQQQQQQIQQQQQQQQHLQQQQMPQLQQQQQQHQQQQQQQHQ 706
Score = 43.1 bits (100), Expect = 6e-04
Identities = 36/108 (33%), Positives = 46/108 (42%), Gaps = 12/108 (11%)
Query: 411 QQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQ 470
QQ QI Q Q Q+ Q Q P Q Q Q+ QQQ Q Q Q +QQ QQ QQ
Sbjct: 664 QQQQQQQQIQQQQQQQQHLQQQQMPQLQQQQQQHQQQQQQQHQLSQLQHHQQQQQQQQQQ 723
Query: 471 QPYQQRPQ-----QPRPPRMPINPIPVTYAELLPGLLKKNLVQTRTAP 513
Q Q Q Q + P+N + + L N +Q +T+P
Sbjct: 724 QQQHQLTQLQHHHQQQQQASPLNQMQQQTSPL-------NQMQQQTSP 764
Score = 42.7 bits (99), Expect = 8e-04
Identities = 28/58 (48%), Positives = 31/58 (53%), Gaps = 4/58 (6%)
Query: 423 PQIPQYPQFPQNP-SPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQ 479
PQIP Q Q Q Q Q +QQQ QQQ QQ QQ PQ QQQ +QQ+ QQ
Sbjct: 649 PQIPNQQQQQQQQLHQQQQQQQQIQQQQQQQQHLQQ---QQMPQLQQQQQQHQQQQQQ 703
Score = 40.8 bits (94), Expect = 0.003
Identities = 28/93 (30%), Positives = 33/93 (35%)
Query: 390 PQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQN 449
P Q Q + QQ Q Q Q+PQ Q Q Q Q + Q
Sbjct: 652 PNQQQQQQQQLHQQQQQQQQIQQQQQQQQHLQQQQMPQLQQQQQQHQQQQQQQHQLSQLQ 711
Query: 450 FQQQPYQQYPYQQYPQQNFQQQPYQQRPQQPRP 482
QQ QQ QQ Q Q Q + Q+ QQ P
Sbjct: 712 HHQQQQQQQQQQQQQHQLTQLQHHHQQQQQASP 744
Score = 33.1 bits (74), Expect = 0.65
Identities = 29/115 (25%), Positives = 41/115 (35%), Gaps = 14/115 (12%)
Query: 377 KKEQEVGMVTHGGPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPS 436
+++Q+ + QQ + Q + + QQ Q Q Q+ + Q Q
Sbjct: 664 QQQQQQQQIQQQQQQQQHLQQQQMPQLQQQQQQHQQQQQ--QQHQLSQLQHHQQQQQQQQ 721
Query: 437 PQNVQPQNVQQQNFQQQPYQQYPYQQYPQQN------------FQQQPYQQRPQQ 479
Q Q Q Q Q+ QQ Q P Q QQ Q QQ+PQQ
Sbjct: 722 QQQQQHQLTQLQHHHQQQQQASPLNQMQQQTSPLNQMQQQTSPLNQMQQQQQPQQ 776
Score = 32.3 bits (72), Expect = 1.1
Identities = 20/40 (50%), Positives = 24/40 (60%), Gaps = 3/40 (7%)
Query: 446 QQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQPRPPRM 485
QQQ QQQ +QQ QQ QQ QQQ QQ QQ + P++
Sbjct: 654 QQQQQQQQLHQQ---QQQQQQIQQQQQQQQHLQQQQMPQL 690
Score = 31.2 bits (69), Expect = 2.5
Identities = 22/51 (43%), Positives = 25/51 (48%), Gaps = 1/51 (1%)
Query: 431 FPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQPR 481
FP + Q N QQQ QQ QQ QQ QQ QQQ QQ+ Q P+
Sbjct: 640 FPGDMVVFKPQIPNQQQQQQQQLHQQQQQQQQIQQQQQQQQHLQQQ-QMPQ 689
Score = 30.4 bits (67), Expect = 4.2
Identities = 20/51 (39%), Positives = 22/51 (42%), Gaps = 11/51 (21%)
Query: 432 PQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQPRP 482
PQ P+ Q Q Q + QQ QQQ QQ QQQ Q QQ P
Sbjct: 649 PQIPNQQQQQQQQLHQQQQQQQQIQQ-----------QQQQQQHLQQQQMP 688
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.133 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,444,414
Number of Sequences: 26719
Number of extensions: 939454
Number of successful extensions: 6400
Number of sequences better than 10.0: 216
Number of HSP's better than 10.0 without gapping: 95
Number of HSP's successfully gapped in prelim test: 126
Number of HSP's that attempted gapping in prelim test: 4585
Number of HSP's gapped (non-prelim): 950
length of query: 820
length of database: 11,318,596
effective HSP length: 108
effective length of query: 712
effective length of database: 8,432,944
effective search space: 6004256128
effective search space used: 6004256128
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 64 (29.3 bits)
Medicago: description of AC135161.1


