

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134823.6 + phase: 0
(662 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
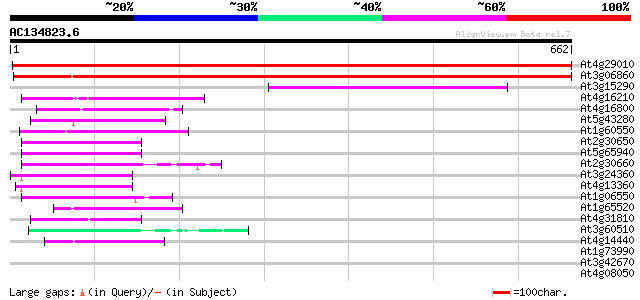
Score E
Sequences producing significant alignments: (bits) Value
At4g29010 AIM1 protein 992 0.0
At3g06860 fatty acid multifunctional protein (AtMFP2) 773 0.0
At3g15290 putative 3-hydroxybutyryl-CoA dehydrogenase 132 8e-31
At4g16210 enoyl-CoA hydratase like protein 105 6e-23
At4g16800 enoyl-CoA hydratase 103 4e-22
At5g43280 enoyl CoA hydratase-like protein 87 4e-17
At1g60550 enoyl-CoA hydratase/isomerase like protein 84 3e-16
At2g30650 3-hydroxyisobutyryl-coenzyme A hydrolase 59 1e-08
At5g65940 3-hydroxyisobutyryl-coenzyme A hydrolase 55 1e-07
At2g30660 3-hydroxyisobutyryl-coenzyme A hydrolase 54 2e-07
At3g24360 3-hydroxyisobutyryl-coenzyme A hydrolase like protein 53 5e-07
At4g13360 3-hydroxyisobutyryl-coenzyme A hydrolase - like protein 49 7e-06
At1g06550 3-hydroxyisobutyryl-coenzyme A hydrolase, putative 49 9e-06
At1g65520 hypothetical protein 47 3e-05
At4g31810 enoyl-CoA hydratase like protein 47 4e-05
At3g60510 enoyl-CoA-hydratase - like protein 45 1e-04
At4g14440 carnitine racemase like protein 44 2e-04
At1g73990 putative protease SppA (SppA) 31 1.9
At3g42670 putative protein 30 3.3
At4g08050 30 4.3
>At4g29010 AIM1 protein
Length = 721
Score = 992 bits (2564), Expect = 0.0
Identities = 488/661 (73%), Positives = 581/661 (87%), Gaps = 2/661 (0%)
Query: 4 VKVDFEVGNDGVAVITMCNPPVNALAIPIIRGLKNKFEEAARRNDVKAIVLTGKGGRFSG 63
+ V EVGNDGVAVIT+ NPPVN+LA PII GLK KF +A +RNDVKAIVL G GRFSG
Sbjct: 5 IGVTMEVGNDGVAVITISNPPVNSLASPIISGLKEKFRDANQRNDVKAIVLIGNNGRFSG 64
Query: 64 GFDISVMQKVHQTGDITLVPDVSVELVVNSIEDSKKPVVAAVEGLALGGGLELAMGCHAR 123
GFDI+V Q+VH+TGD++L+P+VSVELV N +EDS+KPVVAAVEGLALGGGLELAM CHAR
Sbjct: 65 GFDINVFQQVHKTGDLSLMPEVSVELVCNLMEDSRKPVVAAVEGLALGGGLELAMACHAR 124
Query: 124 VAAPKAQLGLPELTLGIIPGFGGTQRLPRLVGTSKAVEMMLTSKPITAEEGQKLGLIDAI 183
VAAPKAQLGLPELTLG+IPGFGGTQRLPRLVG +KA +M+L SK I++EEG KLGLIDA+
Sbjct: 125 VAAPKAQLGLPELTLGVIPGFGGTQRLPRLVGLAKATDMILLSKSISSEEGHKLGLIDAL 184
Query: 184 VSPAELLKLSRQWALEIAEQRRPWIRSLHITDKLGS--DAREVLRTARQHVKKTAPHLPQ 241
V P ++L SR+WAL+IAE R+P+++SLH TDK+GS +AR +L+ +RQ KK AP++PQ
Sbjct: 185 VPPGDVLSTSRKWALDIAEGRKPFLQSLHRTDKIGSLSEARAILKNSRQLAKKIAPNMPQ 244
Query: 242 QQACIDVIEHGILHGGYSGVLREAEVFKKLVLSETAKGLIHVFFAQRTISKIPGVTDIGL 301
ACI+VIE GI+HGGYSGVL+EAEVFK+LVLS+TAKGL+HVFFAQR SK+P VTD+GL
Sbjct: 245 HHACIEVIEEGIIHGGYSGVLKEAEVFKQLVLSDTAKGLVHVFFAQRATSKVPNVTDVGL 304
Query: 302 KPRNVRKAAVIGGGLMGSGIATALILGNIRVILKEVNSEYLQKGIKTIEANVRGLVTRKK 361
KPR ++K AVIGGGLMGSGIATAL+L NIRV+LKE+NSE+L KGIK++EAN++ LV+R K
Sbjct: 305 KPRPIKKVAVIGGGLMGSGIATALLLSNIRVVLKEINSEFLMKGIKSVEANMKSLVSRGK 364
Query: 362 LTQQKAEGALSLLKGVLDYAEFKDVDMVIEAVIEKVSLKQDIFSDLEKICPPHCILASNT 421
LTQ KA ALSL KGVLDY EF DVDMVIEAVIE + LKQ+IF ++EK+C PHCILASNT
Sbjct: 365 LTQDKAGKALSLFKGVLDYTEFNDVDMVIEAVIENIQLKQNIFKEIEKVCSPHCILASNT 424
Query: 422 STIDLNIIGEKISSQDRVIGAHFFSPAHIMPLLEIVRTNKTSAQVILDLVTVGKIIKKSP 481
STIDL++IGEK +S+DR++GAHFFSPAH+MPLLEIVR+ TSAQVILDL+ VGK IKK P
Sbjct: 425 STIDLDVIGEKTNSKDRIVGAHFFSPAHLMPLLEIVRSKNTSAQVILDLMAVGKAIKKVP 484
Query: 482 VVVGNCTGFAVNRTFFPYAQGAHFLANLGVDVFRIDRLISNFGLPMGPFQLQDLSGYGVA 541
VVVGNC GFAVNRTFFPY+Q AH LANLGVD+FRID +I++FGLP+GPFQL DL+G+G+
Sbjct: 485 VVVGNCIGFAVNRTFFPYSQAAHMLANLGVDLFRIDSVITSFGLPLGPFQLGDLAGHGIG 544
Query: 542 VAVGKEFAGSFAGRTFPTPLLDLLIKSGRNGKNNGKGYYIYEKGSKPKPDPSVLPIVEES 601
+AVG +A + R F +P+ +LL+KSGRNGK NG+GYYIYEKGSKPKPDPSVL IVE+S
Sbjct: 545 LAVGPIYAKVYGDRMFRSPMTELLLKSGRNGKINGRGYYIYEKGSKPKPDPSVLSIVEKS 604
Query: 602 RRLSNIMPNGKPISITDQEIVEMILFPVVNEACRVLEEGIVIRASDLDIASVLGMSFPSY 661
R+L+NIMP GKPIS+TD+EIVEMILFPVVNEACRVL+EG+VIRASDLDIASVLGMSFPSY
Sbjct: 605 RKLTNIMPGGKPISVTDKEIVEMILFPVVNEACRVLDEGVVIRASDLDIASVLGMSFPSY 664
Query: 662 R 662
R
Sbjct: 665 R 665
>At3g06860 fatty acid multifunctional protein (AtMFP2)
Length = 725
Score = 773 bits (1995), Expect = 0.0
Identities = 387/662 (58%), Positives = 503/662 (75%), Gaps = 5/662 (0%)
Query: 5 KVDFEVGNDGVAVITMCNPPVNALAIPIIRGLKNKFEEAARRNDVKAIVLTGKGGRFSGG 64
K EVG DGVAVIT+ NPPVN+L+ ++ LK+ +EEA RNDVKAIV+TG GRFSGG
Sbjct: 8 KTVMEVGGDGVAVITLINPPVNSLSFDVLYNLKSNYEEALSRNDVKAIVITGAKGRFSGG 67
Query: 65 FDISVMQKVHQTGDIT--LVPDVSVELVVNSIEDSKKPVVAAVEGLALGGGLELAMGCHA 122
FDIS ++ Q G++ +S++++ + +E ++KP VAA++GLALGGGLELAM CHA
Sbjct: 68 FDISGFGEM-QKGNVKEPKAGYISIDIITDLLEAARKPSVAAIDGLALGGGLELAMACHA 126
Query: 123 RVAAPKAQLGLPELTLGIIPGFGGTQRLPRLVGTSKAVEMMLTSKPITAEEGQKLGLIDA 182
R++AP AQLGLPEL LG+IPGFGGTQRLPRLVG +KA+EM+LTSKP+ AEEG LGLIDA
Sbjct: 127 RISAPAAQLGLPELQLGVIPGFGGTQRLPRLVGLTKALEMILTSKPVKAEEGHSLGLIDA 186
Query: 183 IVSPAELLKLSRQWALEIAEQRRPWIRSLHITDKLG--SDAREVLRTARQHVKKTAPHLP 240
+V PAEL+ +R+WAL+I +R+PW+ S+ TDKL +ARE+L A+ K AP++
Sbjct: 187 VVPPAELVTTARRWALDIVGRRKPWVSSVSKTDKLPPLGEAREILTFAKAQTLKRAPNMK 246
Query: 241 QQQACIDVIEHGILHGGYSGVLREAEVFKKLVLSETAKGLIHVFFAQRTISKIPGVTDIG 300
C+D IE GI+ G +G+ +EAEV ++V +T KGLIHVFF+QR +K+PGVTD G
Sbjct: 247 HPLMCLDAIEVGIVSGPRAGLEKEAEVASQVVKLDTTKGLIHVFFSQRGTAKVPGVTDRG 306
Query: 301 LKPRNVRKAAVIGGGLMGSGIATALILGNIRVILKEVNSEYLQKGIKTIEANVRGLVTRK 360
L PR ++K A+IGGGLMGSGIATALIL N VILKEVN ++L+ GI ++AN++ V +
Sbjct: 307 LVPRKIKKVAIIGGGLMGSGIATALILSNYPVILKEVNEKFLEAGIGRVKANLQSRVRKG 366
Query: 361 KLTQQKAEGALSLLKGVLDYAEFKDVDMVIEAVIEKVSLKQDIFSDLEKICPPHCILASN 420
++Q+K E +SLLKG LDY F+DVDMVIEAVIE +SLKQ IF+DLEK CP HCILASN
Sbjct: 367 SMSQEKFEKTMSLLKGSLDYESFRDVDMVIEAVIENISLKQQIFADLEKYCPQHCILASN 426
Query: 421 TSTIDLNIIGEKISSQDRVIGAHFFSPAHIMPLLEIVRTNKTSAQVILDLVTVGKIIKKS 480
TSTIDLN IGE+ SQDR++GAHFFSPAHIMPLLEIVRTN TSAQVI+DL+ VGK IKK+
Sbjct: 427 TSTIDLNKIGERTKSQDRIVGAHFFSPAHIMPLLEIVRTNHTSAQVIVDLLDVGKKIKKT 486
Query: 481 PVVVGNCTGFAVNRTFFPYAQGAHFLANLGVDVFRIDRLISNFGLPMGPFQLQDLSGYGV 540
PVVVGNCTGFAVNR FFPY Q A FL G D + IDR IS FG+PMGPF+L DL G+GV
Sbjct: 487 PVVVGNCTGFAVNRMFFPYTQAAMFLVECGADPYLIDRAISKFGMPMGPFRLCDLVGFGV 546
Query: 541 AVAVGKEFAGSFAGRTFPTPLLDLLIKSGRNGKNNGKGYYIYEKGSKPKPDPSVLPIVEE 600
A+A +F +F+ RT+ + ++ L+ + R G+ KG+Y+Y+ K KPDP + +E+
Sbjct: 547 AIATATQFIENFSERTYKSMIIPLMQEDKRAGEATRKGFYLYDDKRKAKPDPELKKYIEK 606
Query: 601 SRRLSNIMPNGKPISITDQEIVEMILFPVVNEACRVLEEGIVIRASDLDIASVLGMSFPS 660
+R +S + + K ++++++I+EM FPVVNEACRV EGI ++A+DLDIA ++GM FP
Sbjct: 607 ARSISGVKLDPKLANLSEKDIIEMTFFPVVNEACRVFAEGIAVKAADLDIAGIMGMGFPP 666
Query: 661 YR 662
YR
Sbjct: 667 YR 668
>At3g15290 putative 3-hydroxybutyryl-CoA dehydrogenase
Length = 294
Score = 132 bits (331), Expect = 8e-31
Identities = 85/285 (29%), Positives = 139/285 (47%), Gaps = 3/285 (1%)
Query: 306 VRKAAVIGGGLMGSGIATALILGNIRVILKEVNSEYLQKGIKTIEANVRGLVTRKKLTQQ 365
++ V+G G MGSGIA + V L + + + L + I ++V+ V++ ++++
Sbjct: 4 MKSVGVVGAGQMGSGIAQLAATSGLDVWLMDADRDALSRATAAISSSVKRFVSKGLISKE 63
Query: 366 KAEGALSLLKGVLDYAEFKDVDMVIEAVIEKVSLKQDIFSDLEKICPPHCILASNTSTID 425
+ A+ L+ + + D+++EA++E +K+ +F DL+ I ILASNTS+I
Sbjct: 64 VGDDAMHRLRLTSNLEDLCSADIIVEAIVESEDIKKKLFKDLDGIAKSSAILASNTSSIS 123
Query: 426 LNIIGEKISSQDRVIGAHFFSPAHIMPLLEIVRTNKTSAQVILDLVTVGKIIKKSPVVVG 485
+ + +VIG HF +P IM L+EI+R TS + L + + K+ V
Sbjct: 124 ITRLASATRRPSQVIGMHFMNPPPIMKLVEIIRGADTSEETFLATKVLAERFGKTTVCSQ 183
Query: 486 NCTGFAVNRTFFPYAQGAHFLANLGVDVFR-IDR-LISNFGLPMGPFQLQDLSGYGVAVA 543
+ GF VNR P A GV ID + PMGP +L DL G V ++
Sbjct: 184 DYAGFVVNRILMPMINEAFHTLYTGVATKEDIDSGMKHGTNHPMGPLELADLIGLDVCLS 243
Query: 544 VGKEF-AGSFAGRTFPTPLLDLLIKSGRNGKNNGKGYYIYEKGSK 587
V K G + P PLL + +GR G+ G G Y Y + ++
Sbjct: 244 VMKVLHEGLGDSKYAPCPLLVQYVDAGRLGRKRGVGVYDYREATQ 288
>At4g16210 enoyl-CoA hydratase like protein
Length = 265
Score = 105 bits (263), Expect = 6e-23
Identities = 70/220 (31%), Positives = 115/220 (51%), Gaps = 8/220 (3%)
Query: 14 GVAVITMCNPP-VNALAIPIIRGLKNKFEEAARRNDVKAIVLTGKGGRFSGGFDISVMQK 72
G+AVIT+ P +N+L ++ L F++ V+ ++ TG G F G D++ +
Sbjct: 18 GIAVITINRPKSLNSLTRAMMVDLAKAFKDMDSDESVQVVIFTGSGRSFCSGVDLTAAES 77
Query: 73 VHQTGDITLVPDVSVELVVNSIEDSKKPVVAAVEGLALGGGLELAMGCHARVAAPKAQLG 132
V + GD V D + VV +E +KP++ A+ G A+ G ELA+ C VA+ A+
Sbjct: 78 VFK-GD---VKDPETDPVVQ-MERLRKPIIGAINGFAITAGFELALACDILVASRGAKFM 132
Query: 133 LPELTLGIIPGFGGTQRLPRLVGTSKAVEMMLTSKPITAEEGQKLGLIDAIVSPAELLKL 192
GI P +G +Q+L R++G +KA E+ LTS P+TA+ KLG ++ +V E LK
Sbjct: 133 DTHARFGIFPSWGLSQKLSRIIGANKAREVSLTSMPLTADVAGKLGFVNHVVEEGEALKK 192
Query: 193 SRQWALEIAEQRRPWIRSLH--ITDKLGSDAREVLRTARQ 230
+R+ A I + + + + I D L D L ++
Sbjct: 193 AREIAEAIIKNEQGMVLRIKSVINDGLKLDLGHALTLEKE 232
>At4g16800 enoyl-CoA hydratase
Length = 229
Score = 103 bits (256), Expect = 4e-22
Identities = 63/173 (36%), Positives = 96/173 (55%), Gaps = 7/173 (4%)
Query: 32 IIRGLKNKFEEAARRNDVKAIVLTGK-GGRFSGGFDISVMQKVHQTGDITLVPDVSVELV 90
+++ L+N FE + N + +++ G F G D+ + + + T V S+ +
Sbjct: 1 MLKSLQNAFESIHQDNSARVVMIRSLVPGVFCAGADLKERRTMSPSEVHTYVN--SLRYM 58
Query: 91 VNSIEDSKKPVVAAVEGLALGGGLELAMGCHARVAAPKAQLGLPELTLGIIPGFGGTQRL 150
+ IE P +AA+EG ALGGGLE+A+ C R+ A GLPE L IIPG GGTQRL
Sbjct: 59 FSFIEALSIPTIAAIEGAALGGGLEMALACDLRICGENAVFGLPETGLAIIPGAGGTQRL 118
Query: 151 PRLVGTSKAVEMMLTSKPITAEEGQKLGLIDAIVSPAELLKLSRQWALEIAEQ 203
RLVG S + E++ T + I A E GL++ V+ E + + A+E+A+Q
Sbjct: 119 SRLVGRSVSKELIFTGRKIDAIEAANKGLVNICVTAGE----AHEKAIEMAQQ 167
>At5g43280 enoyl CoA hydratase-like protein
Length = 278
Score = 86.7 bits (213), Expect = 4e-17
Identities = 46/169 (27%), Positives = 83/169 (48%), Gaps = 10/169 (5%)
Query: 25 VNALAIPIIRGLKNKFEEAARRNDVKAIVLTGKGGRFSGGFDISVMQKVH---------- 74
+NAL++ + DV I+L+G G F G D++ + +
Sbjct: 31 LNALSLDFFIEFPKALSSLDQNPDVSVIILSGAGKHFCSGIDLNSLSSISTQSSSGNDRG 90
Query: 75 QTGDITLVPDVSVELVVNSIEDSKKPVVAAVEGLALGGGLELAMGCHARVAAPKAQLGLP 134
++ + S++ + +IE +KPV+AA+ G +GGG++L C R + A +
Sbjct: 91 RSSEQLRRKIKSMQAAITAIEQCRKPVIAAIHGACIGGGVDLITACDIRYCSEDAFFSIK 150
Query: 135 ELTLGIIPGFGGTQRLPRLVGTSKAVEMMLTSKPITAEEGQKLGLIDAI 183
E+ L I+ G QRLP +VG + A+E+ LT++ + E + LGL+ +
Sbjct: 151 EVDLAIVADLGTLQRLPSIVGYANAMELALTARRFSGSEAKDLGLVSKV 199
>At1g60550 enoyl-CoA hydratase/isomerase like protein
Length = 337
Score = 83.6 bits (205), Expect = 3e-16
Identities = 67/202 (33%), Positives = 97/202 (47%), Gaps = 3/202 (1%)
Query: 12 NDGVAVITMCNPPV-NALAIPIIRGLKNKFEEAARRNDVKAIVLTGKGGR-FSGGFDISV 69
++G+A IT+ P NA ++ L F +A + V I+LTGKG + F G D
Sbjct: 84 DEGIAKITINRPERRNAFRPQTVKELMRAFNDARDDSSVGVIILTGKGTKAFCSGGD-QA 142
Query: 70 MQKVHQTGDITLVPDVSVELVVNSIEDSKKPVVAAVEGLALGGGLELAMGCHARVAAPKA 129
++ D V ++V + I KPV+A V G A+GGG L M C +AA A
Sbjct: 143 LRTQDGYADPNDVGRLNVLDLQVQIRRLPKPVIAMVAGYAVGGGHILHMVCDLTIAADNA 202
Query: 130 QLGLPELTLGIIPGFGGTQRLPRLVGTSKAVEMMLTSKPITAEEGQKLGLIDAIVSPAEL 189
G +G G+ + RLVG KA EM ++ TA E +K+GLI+ +V +L
Sbjct: 203 IFGQTGPKVGSFDAGYGSSIMSRLVGPKKAREMWFMTRFYTASEAEKMGLINTVVPLEDL 262
Query: 190 LKLSRQWALEIAEQRRPWIRSL 211
K + +W EI IR L
Sbjct: 263 EKETVKWCREILRNSPTAIRVL 284
>At2g30650 3-hydroxyisobutyryl-coenzyme A hydrolase
Length = 410
Score = 58.5 bits (140), Expect = 1e-08
Identities = 43/144 (29%), Positives = 68/144 (46%), Gaps = 3/144 (2%)
Query: 15 VAVITMCNPP-VNALAIPIIRGLKNKFEEAARRNDVKAIVLTGKGGRFSGGFDIS-VMQK 72
V ++T P +NAL+ ++ L F VK +VL G+G FS G DI +++
Sbjct: 59 VRILTFNRPKQLNALSFHMVSRLLQLFLAYEEDPSVKLVVLKGQGRAFSAGGDIPPIVRD 118
Query: 73 VHQTGDITLVPDVSVELVVNSIEDS-KKPVVAAVEGLALGGGLELAMGCHARVAAPKAQL 131
+ Q I V +N + + +KP V+ + G+ +GGG L+ R+A
Sbjct: 119 ILQGKLIRGAHYFKVGYTLNYVLSTYRKPQVSILNGIVMGGGAGLSTNGRFRIATENTVF 178
Query: 132 GLPELTLGIIPGFGGTQRLPRLVG 155
+PE LG+ P G + L RL G
Sbjct: 179 AMPETALGLFPDVGASYFLSRLPG 202
>At5g65940 3-hydroxyisobutyryl-coenzyme A hydrolase
Length = 378
Score = 55.1 bits (131), Expect = 1e-07
Identities = 40/144 (27%), Positives = 69/144 (47%), Gaps = 3/144 (2%)
Query: 15 VAVITMCNPP-VNALAIPIIRGLKNKFEEAARRNDVKAIVLTGKGGRFSGGFDISVMQKV 73
V ++T+ P +NAL+ +I L F VK ++L G G F G D++ + +
Sbjct: 19 VRILTLNRPKQLNALSFHMISRLLQLFLAFEEDPSVKLVILKGHGRAFCAGGDVAAVVRD 78
Query: 74 HQTGDITLVPDV-SVELVVNSIEDS-KKPVVAAVEGLALGGGLELAMGCHARVAAPKAQL 131
G+ L + S E ++N + + K V+ + G+ +GGG +++ R+A
Sbjct: 79 INQGNWRLGANYFSSEYMLNYVMATYSKAQVSILNGIVMGGGAGVSVHGRFRIATENTVF 138
Query: 132 GLPELTLGIIPGFGGTQRLPRLVG 155
+PE LG+ P G + L RL G
Sbjct: 139 AMPETALGLFPDVGASYFLSRLPG 162
>At2g30660 3-hydroxyisobutyryl-coenzyme A hydrolase
Length = 376
Score = 54.3 bits (129), Expect = 2e-07
Identities = 67/248 (27%), Positives = 104/248 (41%), Gaps = 39/248 (15%)
Query: 15 VAVITMCNPP-VNALAIPIIRGLKNKFEEAARRNDVKAIVLTGKGGRFSGGFDISVMQKV 73
V ++T+ P +NAL +I L F VK ++L G+G F G D+ + +
Sbjct: 15 VRILTLNRPKQLNALCFNMISRLLQLFRAYEEDPSVKLVILKGQGRAFCAGGDVPPVVQN 74
Query: 74 HQTGDITLVPDV-SVELVVNSIEDS-KKPVVAAVEGLALGGGLELAMGCHARVAAPKAQL 131
G L D + +N + + KP V+ + G+ +G G +++ R+A
Sbjct: 75 MVQGKWRLGADFFRDQYTLNYVMATYSKPQVSILNGIVMGAGAGVSIHGRFRIATENTVF 134
Query: 132 GLPELTLGIIPGFGGTQRLPRLVGTSKAVEMMLTSKPITAEEGQKLGLIDAIVSPAELLK 191
+PE +LG+ P G + L RL G G+ +GL A + AELL
Sbjct: 135 AMPETSLGLFPDVGASYFLSRLPGFF----------------GEYVGLTGARLDGAELL- 177
Query: 192 LSRQWALEIAEQRRPWIRSLHI-TD--KLGSD----AREVLRTARQHVKKTAPHLPQQQA 244
A +A P R + TD K+GS +L QH PHL Q+ A
Sbjct: 178 -----ACGLATHFVPSTRLTALETDLCKVGSSDPSFVSTILDAYTQH-----PHLKQKSA 227
Query: 245 C--IDVIE 250
+DVI+
Sbjct: 228 YHRLDVID 235
>At3g24360 3-hydroxyisobutyryl-coenzyme A hydrolase like protein
Length = 418
Score = 53.1 bits (126), Expect = 5e-07
Identities = 45/153 (29%), Positives = 76/153 (49%), Gaps = 8/153 (5%)
Query: 1 MASVKVDFEVGN---DGVAVITMCNPP-VNALAIPIIRGLKNKFEEAARRNDVKAIVLTG 56
MAS +F GN +GVA+IT+ P +NA+ + + K+ +E VK +V+ G
Sbjct: 39 MASGSDEFVKGNVYPNGVALITLDRPKALNAMNLEMDLKYKSLLDEWEYDPGVKCVVVEG 98
Query: 57 KGGR-FSGGFDIS-VMQKVHQTGDITLVPDVSVE--LVVNSIEDSKKPVVAAVEGLALGG 112
R F G DI V+ ++ + +LV V ++ I +KP ++ ++G+ +G
Sbjct: 99 STSRAFCAGMDIKGVVAEILMDKNTSLVQKVFTAEYSLICKIAGYRKPYISLMDGITMGF 158
Query: 113 GLELAMGCHARVAAPKAQLGLPELTLGIIPGFG 145
GL L+ RV + L +PE +G+ P G
Sbjct: 159 GLGLSGHGRYRVITERTVLAMPENGIGLFPDVG 191
>At4g13360 3-hydroxyisobutyryl-coenzyme A hydrolase - like protein
Length = 381
Score = 49.3 bits (116), Expect = 7e-06
Identities = 42/147 (28%), Positives = 72/147 (48%), Gaps = 8/147 (5%)
Query: 7 DFEVGN---DGVAVITMCNPP-VNALAIPIIRGLKNKFEEAARRNDVKAIVLTGKGGR-F 61
DF GN +GVA+IT+ +NA+ + + K+ +E VK +++ G R F
Sbjct: 8 DFVKGNVFPNGVALITLDRTKALNAMNLDMDIKYKSFLDEWESDPRVKCVIVEGSTSRAF 67
Query: 62 SGGFDIS-VMQKVHQTGDITLVPDVSVE--LVVNSIEDSKKPVVAAVEGLALGGGLELAM 118
G DI V ++ + + LV V ++ +I KKP ++ ++G+ +G GL L+
Sbjct: 68 CAGMDIKGVAAEIQKDKNTPLVQKVFTAEYTLICAIAAYKKPYISLMDGITMGFGLGLSG 127
Query: 119 GCHARVAAPKAQLGLPELTLGIIPGFG 145
RV + L +PE +G+ P G
Sbjct: 128 HGRYRVITERTVLAMPENGIGLFPDVG 154
>At1g06550 3-hydroxyisobutyryl-coenzyme A hydrolase, putative
Length = 374
Score = 48.9 bits (115), Expect = 9e-06
Identities = 46/184 (25%), Positives = 75/184 (40%), Gaps = 9/184 (4%)
Query: 14 GVAVITMCNPP--VNALAIPIIRGLKNKFEEAARRNDVKAIVLTGKGGRFSGGFDISVMQ 71
G +T N P +N ++ ++ L E + + K I++ G G FS G D+ V
Sbjct: 19 GSVRLTTLNRPRQLNVISPEVVFKLAEYLELWEKDDQTKLILIKGTGRAFSAGGDLKVFY 78
Query: 72 KVHQTGDITLVPDVSVELVVNSIEDSKKPVVAAVEGLALGGGLELAMGCHARVAAPKAQL 131
++ D L + + I KK V+ V G+++GGG L + V K
Sbjct: 79 HGQESKDSCLEVVYRMYWLCYHIHTYKKTQVSLVNGISMGGGAALMVPMKFSVVTEKTVF 138
Query: 132 GLPELTLGIIPGFGGT---QRLPRLVGTSKAVEMMLTSKPITAEEGQKLGLIDAIVSPAE 188
PE + G G + RLP +G A LT + +E +G+ V +
Sbjct: 139 ATPEASFGFHTDCGFSYIHSRLPGHLGEFLA----LTGARLNGKELVAIGMATHFVPSGK 194
Query: 189 LLKL 192
L+ L
Sbjct: 195 LMDL 198
>At1g65520 hypothetical protein
Length = 240
Score = 47.4 bits (111), Expect = 3e-05
Identities = 39/155 (25%), Positives = 77/155 (49%), Gaps = 6/155 (3%)
Query: 52 IVLTGKGGRFSGGFDISVMQKVHQTGDITLVPDVSVELVVNSIEDSKKPVVAAVEGLALG 111
++ T G FS G+D+++ + +++V D + +V + P +AAV G A
Sbjct: 49 LITTSDGKFFSNGYDLALAES---NPSLSVVMDAKLRSLVADLISLPMPTIAAVTGHASA 105
Query: 112 GGLELAMGC-HARVAAPKAQLGLPELTLG-IIPGFGGTQRLPRLVGTSKAVEMMLTSKPI 169
G LAM + + + L + EL + I+P + ++ + ++MLT+ +
Sbjct: 106 AGCILAMSHDYVLMRRDRGFLYMSELDIELIVPAWFMAVIRGKIGSPAARRDVMLTAAKV 165
Query: 170 TAEEGQKLGLID-AIVSPAELLKLSRQWALEIAEQ 203
TA+ G K+G++D A S AE ++ + + EI ++
Sbjct: 166 TADVGVKMGIVDSAYGSAAETVEAAIKLGEEIVQR 200
>At4g31810 enoyl-CoA hydratase like protein
Length = 409
Score = 46.6 bits (109), Expect = 4e-05
Identities = 36/134 (26%), Positives = 64/134 (46%), Gaps = 4/134 (2%)
Query: 25 VNALAIPIIRGLKNKFEEAARRNDVKAIVLTGKGGRFSGGFDI-SVMQKVHQ--TGDITL 81
+NAL+ P++ LK +E + +++ G G F G D+ S+ +++ T + L
Sbjct: 62 LNALSAPMVGRLKRLYESWEENPAISFVLMKGSGKTFCSGADVLSLYHSINEGNTEESKL 121
Query: 82 VPDVSVELVVNSIEDSKKPVVAAVEGLALGGGLELAMGCHARVAAPKAQLGLPELTLGII 141
+ + V KP +A ++G+ +G G +++ RVA K L PE+ +G
Sbjct: 122 FFENLYKFVYLQ-GTYLKPNIAIMDGVTMGCGGGISLPGMFRVATDKTVLAHPEVQIGFH 180
Query: 142 PGFGGTQRLPRLVG 155
P G + L RL G
Sbjct: 181 PDAGASYYLSRLPG 194
>At3g60510 enoyl-CoA-hydratase - like protein
Length = 401
Score = 45.1 bits (105), Expect = 1e-04
Identities = 57/262 (21%), Positives = 104/262 (38%), Gaps = 41/262 (15%)
Query: 23 PPVNALAIPIIRGLKNKFEEAARRNDVKAIVLTGKGGRFSGGFDISVMQKVHQTGDITLV 82
P +NAL + L+ ++ ++ +++ G G F G DI + + G +
Sbjct: 56 PALNALTTHMGYRLQKLYKNWEEDPNIGFVMMKGSGRAFCAGGDIVSLYHLRTRGSPDAI 115
Query: 83 PDV--SVELVVNSIEDSKKPVVAAVEGLALGGGLELAMGCHARVAAPKAQLGLPELTLGI 140
+ S+ + + KP VA + G+ +GGG +++ RVA + PE +G
Sbjct: 116 REFFSSLYSFIYLLGTYLKPHVAILNGVTMGGGTGVSIPGTFRVATDRTIFATPETIIGF 175
Query: 141 IPGFGGTQRLPRLVGTSKAVEMMLTSKPITAEEGQKLGLIDAIVSPAELLKLSRQWALEI 200
P G + L L G G+ LGL +S AE+L A +
Sbjct: 176 HPDAGASFNLSHLPG----------------RLGEYLGLTGLKLSGAEML------ACGL 213
Query: 201 AEQRRPWIRSLHITDKLGSDAREVLRTARQHVKKTAPHLPQ-QQACIDVIEHGILHGGYS 259
A +IRS E + + +KK P ++C++ + H +
Sbjct: 214 ATH---YIRS------------EEVPVMEEQLKKLLTDDPSVVESCLEKCAE-VAHPEKT 257
Query: 260 GVLREAEVFKKLVLSETAKGLI 281
GV+R ++ +K +T + +I
Sbjct: 258 GVIRRIDLLEKCFSHDTVEEII 279
>At4g14440 carnitine racemase like protein
Length = 238
Score = 44.3 bits (103), Expect = 2e-04
Identities = 39/147 (26%), Positives = 70/147 (47%), Gaps = 7/147 (4%)
Query: 42 EAARRNDVKAIVL--TGKGGRFSGGFDISVMQKVHQTGDITLVPDV--SVELVVNSIEDS 97
E A+ K VL TG G FS GFD++ Q G I + + S + V+ ++ D
Sbjct: 36 EQAKSQSTKGSVLITTGHGKFFSNGFDLAWAQSAGH-GAIKRMHQMVKSFKPVLAALLDL 94
Query: 98 KKPVVAAVEGLALGGGLELAMG-CHARVAAPKAQLGLPELTLGI-IPGFGGTQRLPRLVG 155
P +AA+ G A GL A+ + + + L + E+ +G+ +P + + ++
Sbjct: 95 PMPTIAALNGHAAASGLMFALSHDYVFMRKDRGVLYMSEVDIGLPVPDYFSALVVAKVGS 154
Query: 156 TSKAVEMMLTSKPITAEEGQKLGLIDA 182
E++L+ K + EE LG++D+
Sbjct: 155 GIARRELLLSGKKLKGEEAVALGIVDS 181
>At1g73990 putative protease SppA (SppA)
Length = 677
Score = 31.2 bits (69), Expect = 1.9
Identities = 32/124 (25%), Positives = 54/124 (42%), Gaps = 20/124 (16%)
Query: 99 KPVVAAVEGLALGGGLELAMGCHARVAAPKAQLGLPELTLGIIPGFGGTQRLPRLVGTSK 158
KPV+A++ +A GG +AM +A VA G ++G++ +L +G +K
Sbjct: 448 KPVIASMSDVAASGGYYMAMAANAIVAENLTLTG----SIGVVTARFTLAKLYEKIGFNK 503
Query: 159 AVEMMLTSKPITAEEGQKLGLIDAIVSP-----AELLKLSRQWALEIAEQRRPWIRSLHI 213
T G+ L+ A P AEL + S Q A ++ + RS+ +
Sbjct: 504 E----------TISRGKYAELLGAEERPLKPEEAELFEKSAQHAYQLFRDKAALSRSMPV 553
Query: 214 TDKL 217
DK+
Sbjct: 554 -DKM 556
>At3g42670 putative protein
Length = 1256
Score = 30.4 bits (67), Expect = 3.3
Identities = 23/95 (24%), Positives = 45/95 (47%), Gaps = 9/95 (9%)
Query: 442 AHFFSPAHIMPLLEIVRTNKTSAQVILDLVTVGKIIKKSPVVVGNCTGFAVNRTF----- 496
A FF+P ++ + ++ K ++V+ L V +++K+ +++ C A R F
Sbjct: 1035 AKFFNPQELLEIEKLKHDAKKGSKVMFVLNLVFRVVKREKILI-FCHNIAPIRLFLELFE 1093
Query: 497 --FPYAQGAHFLANLG-VDVFRIDRLISNFGLPMG 528
F + +G L G +++F R+I F P G
Sbjct: 1094 NVFRWKRGRELLTLTGDLELFERGRVIDKFEEPGG 1128
>At4g08050
Length = 1428
Score = 30.0 bits (66), Expect = 4.3
Identities = 20/73 (27%), Positives = 37/73 (50%), Gaps = 4/73 (5%)
Query: 143 GFGGTQRLPRLVGTSKAVEMMLTSKPI-TAEEGQKLGLIDAIVSPAELLKLSRQWALEIA 201
G GG L G +++ + + A EG++L ++ A+++ EL+ +S LE+
Sbjct: 1321 GRGGPHSLDLAAGADDFCSLVVYATAVLAAAEGKRLSILRAVLAATELM-MSSTHLLELI 1379
Query: 202 EQR--RPWIRSLH 212
+ + RPW S H
Sbjct: 1380 QNKVVRPWPGSNH 1392
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.139 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,194,802
Number of Sequences: 26719
Number of extensions: 630161
Number of successful extensions: 1601
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 1572
Number of HSP's gapped (non-prelim): 30
length of query: 662
length of database: 11,318,596
effective HSP length: 106
effective length of query: 556
effective length of database: 8,486,382
effective search space: 4718428392
effective search space used: 4718428392
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC134823.6


