

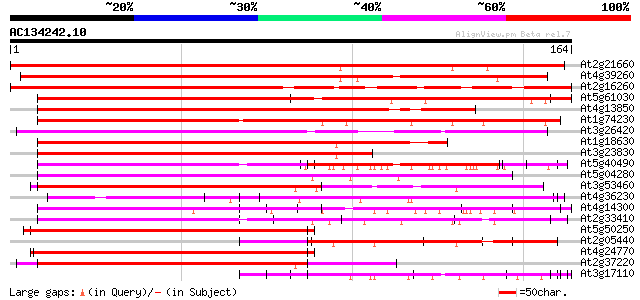
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134242.10 + phase: 0
(164 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g21660 glycine-rich RNA binding protein 248 1e-66
At4g39260 glycine-rich protein (clone AtGRP8) 224 2e-59
At2g16260 putative glycine-rich RNA-binding protein 185 8e-48
At5g61030 RNA-binding protein - like 170 3e-43
At4g13850 glycine-rich RNA-binding protein AtGRP2 - like 152 1e-37
At1g74230 putative RNA-binding protein 149 5e-37
At3g26420 RNA-binding protein, putative 127 3e-30
At1g18630 unknown protein 118 1e-27
At3g23830 glycine-rich RNA binding protein, putative 112 9e-26
At5g40490 ribonucleoprotein -like 112 1e-25
At5g04280 RNA-binding protein-like 110 3e-25
At3g53460 RNA-binding protein cp29 protein 108 1e-24
At4g36230 putative glycine-rich cell wall protein 99 1e-21
At4g14300 ribonucleoprotein like protein 93 7e-20
At2g33410 putative RNA-binding protein 93 7e-20
At5g50250 RNA-binding protein-like 92 1e-19
At2g05440 putative glycine-rich protein 92 1e-19
At4g24770 31 kDa RNA binding protein (rbp31) 90 6e-19
At2g37220 putative RNA-binding protein 89 1e-18
At3g17110 hypothetical protein 89 1e-18
>At2g21660 glycine-rich RNA binding protein
Length = 176
Score = 248 bits (632), Expect = 1e-66
Identities = 125/176 (71%), Positives = 133/176 (75%), Gaps = 14/176 (7%)
Query: 1 MASADVEYRCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEK 60
MAS DVEYRCFVGGLAWATD ALE AF+QYG++IDSKIINDRETGRSRGFGFVTF DEK
Sbjct: 1 MASGDVEYRCFVGGLAWATDDRALETAFAQYGDVIDSKIINDRETGRSRGFGFVTFKDEK 60
Query: 61 SMRDAIEGMNGQDMDGRNITVNEAQSRGSGGGGGR-----------GGGGYGGGGGGYGG 109
+M+DAIEGMNGQD+DGR+ITVNEAQSRGSGGGGG GGGGY GGGG YGG
Sbjct: 61 AMKDAIEGMNGQDLDGRSITVNEAQSRGSGGGGGHRGGGGGGYRSGGGGGYSGGGGSYGG 120
Query: 110 GGRREGGYNRSSGGGGGYG--GGGGGGYGGG-RDRGYGDDGGSRYSRGGGGDGGSW 162
GG R G SGGGGGY GGGGG YGGG R+ G G GG GG G GG W
Sbjct: 121 GGGRREGGGGYSGGGGGYSSRGGGGGSYGGGRREGGGGYGGGEGGGYGGSGGGGGW 176
>At4g39260 glycine-rich protein (clone AtGRP8)
Length = 169
Score = 224 bits (570), Expect = 2e-59
Identities = 115/169 (68%), Positives = 128/169 (75%), Gaps = 17/169 (10%)
Query: 4 ADVEYRCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSMR 63
++VEYRCFVGGLAWAT+ E L++ FSQ+G++IDSKIINDRE+GRSRGFGFVTF DEK+MR
Sbjct: 2 SEVEYRCFVGGLAWATNDEDLQRTFSQFGDVIDSKIINDRESGRSRGFGFVTFKDEKAMR 61
Query: 64 DAIEGMNGQDMDGRNITVNEAQSRGSGGGGGR-----------GGGGY-GGGGGGYGGGG 111
DAIE MNG+++DGR ITVNEAQSRGSGGGGG GGGGY GGGGGGY GGG
Sbjct: 62 DAIEEMNGKELDGRVITVNEAQSRGSGGGGGGRGGSGGGYRSGGGGGYSGGGGGGYSGGG 121
Query: 112 RREGGYNRSSGGGGGYGGGGGGGYGGGRDR---GYGDDGGSRYSRGGGG 157
GGY R SGG G GGGGG GYGGG R GYG G Y GGGG
Sbjct: 122 G--GGYERRSGGYGSGGGGGGRGYGGGGRREGGGYGGGDGGSYGGGGGG 168
>At2g16260 putative glycine-rich RNA-binding protein
Length = 185
Score = 185 bits (470), Expect = 8e-48
Identities = 101/164 (61%), Positives = 119/164 (71%), Gaps = 15/164 (9%)
Query: 1 MASADVEYRCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEK 60
+A AD EYRCFVGGLAWATD +++E+ F+++GE+ DSKII DRETGRS+GF FVTF DE
Sbjct: 37 VAYADNEYRCFVGGLAWATDEQSIERCFNEFGEVFDSKIIIDRETGRSKGFRFVTFKDED 96
Query: 61 SMRDAIEGMNGQDMDGRNITVNEAQSRGSGGGGGRGGGGYGGGGGGYGGGGRREGGYNRS 120
SMR AI+ MNGQ++DGRNIT AQ+RGSG GG GGYG GG G R +GGYNR
Sbjct: 97 SMRTAIDRMNGQELDGRNIT---AQARGSGTRGGM-VGGYGSGG---YRGRRDQGGYNR- 148
Query: 121 SGGGGGYGGGGGGGYGGGRDRGYGDDGGSRYSRGGGGDGGSWRS 164
GGGGGYGGG G G R+ GYGD G Y G +GGSWR+
Sbjct: 149 -GGGGGYGGGYG---GDRREGGYGDGG---YGGQGRSEGGSWRN 185
>At5g61030 RNA-binding protein - like
Length = 309
Score = 170 bits (431), Expect = 3e-43
Identities = 85/165 (51%), Positives = 110/165 (66%), Gaps = 11/165 (6%)
Query: 9 RCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSMRDAIEG 68
+ F+GG+A++ D ++L +AF++YGE++D+++I DRETGRSRGFGFVTF ++ AI+
Sbjct: 41 KLFIGGMAYSMDEDSLREAFTKYGEVVDTRVILDRETGRSRGFGFVTFTSSEAASSAIQA 100
Query: 69 MNGQDMDGRNITVNEAQSRGSGGGGGRGGGGYGGGGGGYGGG---GRREGGYNRS---SG 122
++G+D+ GR + VN A R S GGG GGGGYGGGGGGYGG G GGY S G
Sbjct: 101 LDGRDLHGRVVKVNYANDRTS--GGGFGGGGYGGGGGGYGGSGGYGGGAGGYGGSGGYGG 158
Query: 123 GGGGYGGGGGGGYGGGRDRGYGDDGGSRY---SRGGGGDGGSWRS 164
G GGYGG GGGYGG GYG G Y + G GG GG + S
Sbjct: 159 GAGGYGGNSGGGYGGNAAGGYGGSGAGGYGGDATGHGGAGGGYGS 203
Score = 44.3 bits (103), Expect = 3e-05
Identities = 26/78 (33%), Positives = 28/78 (35%), Gaps = 2/78 (2%)
Query: 83 EAQSRGSGGGGGRGGGGYGGGGGGYGGGGRREGGYNRSSGGGGGYGGGGGGGYGGGRDRG 142
+A G GGG GG+G G YG G G G GYG G G G
Sbjct: 190 DATGHGGAGGGYGSSGGFGSSGNTYGEGSSASAGAVGDYNGSSGYGSANTYGSSNGGFAG 249
Query: 143 YGDDGGSRYSRGG--GGD 158
GGS GGD
Sbjct: 250 DSQFGGSPVGNSSQFGGD 267
Score = 28.1 bits (61), Expect = 2.2
Identities = 28/91 (30%), Positives = 34/91 (36%), Gaps = 9/91 (9%)
Query: 80 TVNEAQSRGSGGGG---GRGGGG----YGGGGGGYGGGGRREGG-YNRSSGGGGGYGGGG 131
T E S +G G G G G YG GG+ G + G SS GG
Sbjct: 213 TYGEGSSASAGAVGDYNGSSGYGSANTYGSSNGGFAGDSQFGGSPVGNSSQFGGDNTQFT 272
Query: 132 GGGYGGGRDRGYGDDGGSRYSRGGGGDGGSW 162
GG GG D+ +G S G GG +
Sbjct: 273 AGGQFGGEDQ-FGSMEKSETKMEDGPIGGEF 302
>At4g13850 glycine-rich RNA-binding protein AtGRP2 - like
Length = 158
Score = 152 bits (383), Expect = 1e-37
Identities = 73/128 (57%), Positives = 91/128 (71%), Gaps = 6/128 (4%)
Query: 9 RCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSMRDAIEG 68
+ F+GGL+W TD +L AF+ +G+++D+K+I DRETGRSRGFGFV F DE + AI
Sbjct: 36 KLFIGGLSWGTDDASLRDAFAHFGDVVDAKVIVDRETGRSRGFGFVNFNDEGAATAAISE 95
Query: 69 MNGQDMDGRNITVNEAQSRGSGGGGGRGGGGYGGGGGGYGGGGRREGGYNRSSGGGGGYG 128
M+G++++GR+I VN A R S GGGGY GGGGGYGGGG GGY GGGGGYG
Sbjct: 96 MDGKELNGRHIRVNPANDRPSAPRAYGGGGGYSGGGGGYGGGG---GGY---GGGGGGYG 149
Query: 129 GGGGGGYG 136
GGG GG G
Sbjct: 150 GGGDGGGG 157
>At1g74230 putative RNA-binding protein
Length = 289
Score = 149 bits (377), Expect = 5e-37
Identities = 89/181 (49%), Positives = 110/181 (60%), Gaps = 29/181 (16%)
Query: 9 RCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSMRDAIEG 68
+ FVGG++++TD L +AFS+YGE++D+KII DRETGRSRGF FVTF + +A++
Sbjct: 35 KIFVGGISYSTDEFGLREAFSKYGEVVDAKIIVDRETGRSRGFAFVTFTSTEEASNAMQ- 93
Query: 69 MNGQDMDGRNITVNEAQSRGSG-GGGGRGG---------GGYGGGGGGYGGGGRREGG-- 116
++GQD+ GR I VN A RGSG GG G GG GGYG GGYGGG GG
Sbjct: 94 LDGQDLHGRRIRVNYATERGSGFGGRGFGGPGGGYGASDGGYGAPAGGYGGGAGGYGGNS 153
Query: 117 -YNRSSGGGGGYG-----GGGGGGYGG-----GRDRGYGDDGGSRYSRGG-----GGDGG 160
Y+ ++GGGGGYG GG GGYGG G G G GS + GG GG GG
Sbjct: 154 SYSGNAGGGGGYGGNSSYGGNAGGYGGNPPYSGNAVGGGGGYGSNFGGGGGYGVAGGVGG 213
Query: 161 S 161
S
Sbjct: 214 S 214
>At3g26420 RNA-binding protein, putative
Length = 245
Score = 127 bits (319), Expect = 3e-30
Identities = 68/155 (43%), Positives = 92/155 (58%), Gaps = 13/155 (8%)
Query: 3 SADVEYRCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSM 62
S D EYRCF+GGLAW T L AF +YG ++++K++ D+ +GRSRGFGF+TF ++K+M
Sbjct: 2 SEDPEYRCFIGGLAWTTSDRGLRDAFEKYGHLVEAKVVLDKFSGRSRGFGFITFDEKKAM 61
Query: 63 RDAIEGMNGQDMDGRNITVNEAQSRGSGGGGGRGGGGYGGGGGGYGGGGRREGGYNRSSG 122
+AI MNG D+DGR ITV++AQ GG GR G G R+ GY+R
Sbjct: 62 DEAIAAMNGMDLDGRTITVDKAQPH--QGGAGRDNDGDRG----------RDRGYDRDRS 109
Query: 123 GGGGYGGGGGGGYGGGRDRGYGDDGGSRYSRGGGG 157
G G GGG + G+ + + S SR GGG
Sbjct: 110 RPSG-GRGGGDCFKCGKPGHFARECPSESSRDGGG 143
>At1g18630 unknown protein
Length = 155
Score = 118 bits (296), Expect = 1e-27
Identities = 60/121 (49%), Positives = 82/121 (67%), Gaps = 6/121 (4%)
Query: 9 RCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSMRDAIEG 68
+ FVGGL+ +TD E L++AF +G+I+D+ ++ DRE+G SRGFGFVT+ + +A++
Sbjct: 37 KIFVGGLSPSTDVELLKEAFGSFGKIVDAVVVLDRESGLSRGFGFVTYDSIEVANNAMQA 96
Query: 69 MNGQDMDGRNITVNEAQSRGSGGGGG-RGGGGYGGGGGGYGGGGRREGGYNRSSGGGGGY 127
M +++DGR I V+ A S G GGGGG GGYGGG GGY GG GG+ GGGGY
Sbjct: 97 MQNKELDGRIIGVHPADSGGGGGGGGFARRGGYGGGRGGYARGGFGRGGF-----GGGGY 151
Query: 128 G 128
G
Sbjct: 152 G 152
>At3g23830 glycine-rich RNA binding protein, putative
Length = 136
Score = 112 bits (280), Expect = 9e-26
Identities = 53/99 (53%), Positives = 73/99 (73%), Gaps = 1/99 (1%)
Query: 9 RCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSMRDAIEG 68
+ FVGGL+W TD +L++AF+ +GE+ ++ +I DRETGRSRGFGFV+F+ E S +AI+
Sbjct: 36 KLFVGGLSWGTDDSSLKQAFTSFGEVTEATVIADRETGRSRGFGFVSFSCEDSANNAIKE 95
Query: 69 MNGQDMDGRNITVNEAQSRGSGGGGG-RGGGGYGGGGGG 106
M+G++++GR I VN A R S GGGGYGGGGGG
Sbjct: 96 MDGKELNGRQIRVNLATERSSAPRSSFGGGGGYGGGGGG 134
>At5g40490 ribonucleoprotein -like
Length = 423
Score = 112 bits (279), Expect = 1e-25
Identities = 68/178 (38%), Positives = 91/178 (50%), Gaps = 29/178 (16%)
Query: 9 RCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSMRDAIEG 68
+ FVGG+ + D + ++ F Q+GE+ + +I+ D TGRSRGFGFVT+ E + +
Sbjct: 131 KIFVGGIPSSVDDDEFKEFFMQFGELKEHQIMRDHSTGRSRGFGFVTYESEDMVDHLLAK 190
Query: 69 MNGQDMDGRNITVNEAQ----------------SRGSGGGG------GRGGGGYGGGGGG 106
N ++ G + + +A+ SR + GGG G GGGYGG GG
Sbjct: 191 GNRIELSGTQVEIKKAEPKKPNSVTTPSKRFGDSRSNFGGGYGDGYGGGHGGGYGGPGGP 250
Query: 107 Y----GGGGRREGGYNRSSGGGGGYGGGGGGGYGGGRDRGYGDDGGSRYSRGGGGDGG 160
Y G GG R GGY G GG +GG GGGGYGGG G+ R GGG GG
Sbjct: 251 YKSGGGYGGGRSGGY---GGYGGEFGGYGGGGYGGGVGPYRGEPALGYSGRYGGGGGG 305
Score = 77.4 bits (189), Expect = 3e-15
Identities = 47/86 (54%), Positives = 48/86 (55%), Gaps = 11/86 (12%)
Query: 86 SRGSGGGGGRG-----GGGYGGGGGGYGG-GGRREGGYNRSS--GGGGGYGGGGGGGY-- 135
S G GGG G G GG YG GGGYGG G GGY S G GGG GG GGGGY
Sbjct: 312 SMGGGGGYGGGPGDMYGGSYGEPGGGYGGPSGSYGGGYGSSGIGGYGGGMGGAGGGGYRG 371
Query: 136 GGGRDR-GYGDDGGSRYSRGGGGDGG 160
GGG D G G G Y GGGG+GG
Sbjct: 372 GGGYDMGGVGGGGAGGYGAGGGGNGG 397
Score = 76.3 bits (186), Expect = 7e-15
Identities = 46/84 (54%), Positives = 47/84 (55%), Gaps = 11/84 (13%)
Query: 88 GSGGGGGRGGGGYGGGG-GGYGGGGRREGGYNRSSGGG---GGYGGGGGGGYGGGRDRGY 143
G GG G GGGYG G GGYGGG GG GGG GG GGGG GGYG G G
Sbjct: 337 GYGGPSGSYGGGYGSSGIGGYGGGMGGAGGGGYRGGGGYDMGGVGGGGAGGYGAG---GG 393
Query: 144 GDDGGSRYSRGGG----GDGGSWR 163
G+ GGS Y GGG G GGS R
Sbjct: 394 GNGGGSFYGGGGGRGGYGGGGSGR 417
Score = 73.6 bits (179), Expect = 5e-14
Identities = 43/82 (52%), Positives = 45/82 (54%), Gaps = 11/82 (13%)
Query: 90 GGGGGRGGGGYGGGGGGYGG--GGRREGGYNRSSG--GGGGYGGGGG---GGYGGGRDRG 142
GG G GGGYGG G YGG G GGY G GGGGY GGGG GG GGG G
Sbjct: 328 GGSYGEPGGGYGGPSGSYGGGYGSSGIGGYGGGMGGAGGGGYRGGGGYDMGGVGGGGAGG 387
Query: 143 Y----GDDGGSRYSRGGGGDGG 160
Y G +GG + GGGG GG
Sbjct: 388 YGAGGGGNGGGSFYGGGGGRGG 409
Score = 68.2 bits (165), Expect = 2e-12
Identities = 40/65 (61%), Positives = 40/65 (61%), Gaps = 11/65 (16%)
Query: 88 GSGGG-GGRGGGGYGGGGG----GYGGGGRREGGYNRSSGGGGG---YGGGGG-GGYGGG 138
G GGG GG GGGGY GGGG G GGGG GGY GG GG YGGGGG GGYGGG
Sbjct: 356 GYGGGMGGAGGGGYRGGGGYDMGGVGGGGA--GGYGAGGGGNGGGSFYGGGGGRGGYGGG 413
Query: 139 RDRGY 143
Y
Sbjct: 414 GSGRY 418
Score = 67.0 bits (162), Expect = 4e-12
Identities = 41/72 (56%), Positives = 42/72 (57%), Gaps = 10/72 (13%)
Query: 90 GGGGGRGGGGYGGGGGGYGGGGRREGG-YNRSS---GGGGGYGGGGGG----G-YGGGRD 140
GG G G GGYGGG GG GGGG R GG Y+ GG GGYG GGGG YGGG
Sbjct: 347 GGYGSSGIGGYGGGMGGAGGGGYRGGGGYDMGGVGGGGAGGYGAGGGGNGGGSFYGGGGG 406
Query: 141 R-GYGDDGGSRY 151
R GYG G RY
Sbjct: 407 RGGYGGGGSGRY 418
Score = 66.2 bits (160), Expect = 7e-12
Identities = 37/63 (58%), Positives = 38/63 (59%), Gaps = 9/63 (14%)
Query: 88 GSGGGGGRGGGGY------GGGGGGYGGGGRREGGYNRSSGGGGGYGGGGGGGYGGGRDR 141
G+GGGG RGGGGY GGG GGYG GG GG GGGGG GG GGG G GR
Sbjct: 363 GAGGGGYRGGGGYDMGGVGGGGAGGYGAGGGGNGG-GSFYGGGGGRGGYGGG--GSGRYH 419
Query: 142 GYG 144
YG
Sbjct: 420 PYG 422
Score = 65.1 bits (157), Expect = 2e-11
Identities = 35/81 (43%), Positives = 48/81 (59%), Gaps = 2/81 (2%)
Query: 9 RCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSMRDAIEG 68
+ FVGGLA T S K F +YGEI DS I+ DR+TG+ RGFGFVT+AD + I+
Sbjct: 43 KIFVGGLARETTSAEFLKHFGKYGEITDSVIMKDRKTGQPRGFGFVTYADSSVVDKVIQ- 101
Query: 69 MNGQDMDGRNITVNEAQSRGS 89
+ + G+ + + RGS
Sbjct: 102 -DNHIIIGKQVEIKRTIPRGS 121
>At5g04280 RNA-binding protein-like
Length = 310
Score = 110 bits (276), Expect = 3e-25
Identities = 66/160 (41%), Positives = 88/160 (54%), Gaps = 21/160 (13%)
Query: 9 RCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSMRDAIEG 68
R FVGGL+ LE+AFS++G+I+D +I+ +R+TGRSRGFGF+TFAD ++M ++I
Sbjct: 8 RIFVGGLSPEVTDRDLERAFSRFGDILDCQIMLERDTGRSRGFGFITFADRRAMDESIRE 67
Query: 69 MNGQDMDGRNITVNEAQSR-------GSGGGGGRGGG----GYGGGGGGYGGGGR----- 112
M+G+D R I+VN A+ + G GGR G G G GGG GGGGR
Sbjct: 68 MHGRDFGDRVISVNRAEPKLGRDDGESHGSRGGRDSGYSIAGKGSFGGGGGGGGRVGEDE 127
Query: 113 -----REGGYNRSSGGGGGYGGGGGGGYGGGRDRGYGDDG 147
R G + R GG GG GG+ G DG
Sbjct: 128 CFKCGRVGHWARDCPSAGGGRGGPVGGFSSRASAYGGSDG 167
Score = 34.3 bits (77), Expect = 0.031
Identities = 27/73 (36%), Positives = 33/73 (44%), Gaps = 18/73 (24%)
Query: 103 GGGGYGGGGRR--EGGYNRS---------SGGGGGYGGGGGGG--YGGGRD-RGYGDDGG 148
GG Y G R G Y+++ S G YGGG GG GGG + RG+ G
Sbjct: 227 GGDRYSRGSDRYPPGSYDKARSFERDIAPSAGSDRYGGGRAGGPIRGGGEEGRGFRSRAG 286
Query: 149 SRYSR----GGGG 157
+ Y R GGGG
Sbjct: 287 APYERPSRSGGGG 299
Score = 32.7 bits (73), Expect = 0.090
Identities = 24/48 (50%), Positives = 24/48 (50%), Gaps = 4/48 (8%)
Query: 84 AQSRGSGG-GGGRGGGGY-GGGGGGYGGGGRREGGYNR--SSGGGGGY 127
A S GS GGGR GG GGG G G R Y R SGGGG Y
Sbjct: 254 APSAGSDRYGGGRAGGPIRGGGEEGRGFRSRAGAPYERPSRSGGGGAY 301
Score = 27.3 bits (59), Expect = 3.8
Identities = 19/46 (41%), Positives = 20/46 (43%), Gaps = 8/46 (17%)
Query: 96 GGGGYGGG--GGGYGGGGRREGGYNRSSGGGGGY----GGGGGGGY 135
G YGGG GG GGG G+ S G Y GGGG Y
Sbjct: 258 GSDRYGGGRAGGPIRGGGEEGRGFR--SRAGAPYERPSRSGGGGAY 301
>At3g53460 RNA-binding protein cp29 protein
Length = 342
Score = 108 bits (271), Expect = 1e-24
Identities = 68/162 (41%), Positives = 85/162 (51%), Gaps = 15/162 (9%)
Query: 7 EYRCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSMRDAI 66
+ + FVG L++ DS L + F G + ++I D+ TGRSRGFGFVT + + A
Sbjct: 98 DLKLFVGNLSFNVDSAQLAQLFESAGNVEMVEVIYDKVTGRSRGFGFVTMSTAAEVEAAA 157
Query: 67 EGMNGQDMDGRNITVN---------EAQSRG--SGGGGGRGGGGYGGGGGGYGGGGRREG 115
+ NG + +GR + VN E+ SRG SGG G GGGYG GG G G R G
Sbjct: 158 QQFNGYEFEGRPLRVNAGPPPPKREESFSRGPRSGGYGSERGGGYGSERGG-GYGSERGG 216
Query: 116 GYNRSSGGGGGYGG-GGGGGYGGGRDRGYGDDGGSRYSRGGG 156
GY S GGGYG GGGYGG + YG GS G G
Sbjct: 217 GYG--SERGGGYGSQRSGGGYGGSQRSSYGSGSGSGSGSGSG 256
Score = 89.0 bits (219), Expect = 1e-18
Identities = 37/83 (44%), Positives = 60/83 (71%)
Query: 9 RCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSMRDAIEG 68
R +VG L+W D ALE F++ G+++++++I DR++GRS+GFGFVT + + ++ AI
Sbjct: 258 RLYVGNLSWGVDDMALENLFNEQGKVVEARVIYDRDSGRSKGFGFVTLSSSQEVQKAINS 317
Query: 69 MNGQDMDGRNITVNEAQSRGSGG 91
+NG D+DGR I V+EA++R G
Sbjct: 318 LNGADLDGRQIRVSEAEARPPRG 340
>At4g36230 putative glycine-rich cell wall protein
Length = 221
Score = 98.6 bits (244), Expect = 1e-21
Identities = 52/97 (53%), Positives = 57/97 (58%), Gaps = 10/97 (10%)
Query: 74 MDGRNITVNE-AQSRGSGGGGGRGGGGYGGGGGGYGGGGRREGG---------YNRSSGG 123
+D +N+ E +SR GGGGG GGGG GGGG G GGGG G N+SSGG
Sbjct: 73 LDDQNVNKTEKCESRAGGGGGGGGGGGGGGGGQGSGGGGGEGNGGNGKDNSHKRNKSSGG 132
Query: 124 GGGYGGGGGGGYGGGRDRGYGDDGGSRYSRGGGGDGG 160
GGG GGGGGGG G G RG G GG GGGG GG
Sbjct: 133 GGGGGGGGGGGSGNGSGRGRGGGGGGGGGGGGGGGGG 169
Score = 87.8 bits (216), Expect = 2e-18
Identities = 61/168 (36%), Positives = 74/168 (43%), Gaps = 20/168 (11%)
Query: 12 VGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSMR--DAIEGM 69
V GL + +A E S + S +++ + T F D++++ + E
Sbjct: 31 VNGLRDLKERDASE---SNFVNATSSSVLDSKSTANISTVEFENALDDQNVNKTEKCESR 87
Query: 70 NGQDMDGRNITVNEAQSRGSGGGGGRGGGGYG--------------GGGGGYGGGGRREG 115
G G +GSGGGGG G GG G GGGGG GGGG G
Sbjct: 88 AGGGGGGGGGGGGGGGGQGSGGGGGEGNGGNGKDNSHKRNKSSGGGGGGGGGGGGGSGNG 147
Query: 116 -GYNRSSGGGGGYGGGGGGGYGGGRDRGYGDDGGSRYSRGGGGDGGSW 162
G R GGGGG GGGGGGG GGG G G GGS GG G G W
Sbjct: 148 SGRGRGGGGGGGGGGGGGGGGGGGGGGGGGGGGGSDGKGGGWGFGFGW 195
Score = 87.4 bits (215), Expect = 3e-18
Identities = 46/95 (48%), Positives = 54/95 (56%), Gaps = 1/95 (1%)
Query: 68 GMNGQDMDGRNITVNEAQSRGSGGGGGRGGGGYGGGGGGYGGGGRREGGYNRSSGGGGGY 127
G G +G++ + +S G GGGGG GGGG G G G G GG GG GGGGG
Sbjct: 111 GGEGNGGNGKDNSHKRNKSSGGGGGGGGGGGGGSGNGSGRGRGGGGGGGGGGGGGGGGGG 170
Query: 128 GGGGGGGYGGGRDRGYGDDGGSRYSRGGGGDGGSW 162
GGGGGGG GGG D G G G + GG G GG++
Sbjct: 171 GGGGGGGGGGGSD-GKGGGWGFGFGWGGDGPGGNF 204
Score = 86.3 bits (212), Expect = 7e-18
Identities = 46/92 (50%), Positives = 50/92 (54%)
Query: 68 GMNGQDMDGRNITVNEAQSRGSGGGGGRGGGGYGGGGGGYGGGGRREGGYNRSSGGGGGY 127
G G + +G N N + S GGGG GGGG GGG G G GR GG GGGGG
Sbjct: 108 GGGGGEGNGGNGKDNSHKRNKSSGGGGGGGGGGGGGSGNGSGRGRGGGGGGGGGGGGGGG 167
Query: 128 GGGGGGGYGGGRDRGYGDDGGSRYSRGGGGDG 159
GGGGGGG GGG G GG + G GGDG
Sbjct: 168 GGGGGGGGGGGGGGSDGKGGGWGFGFGWGGDG 199
Score = 71.2 bits (173), Expect = 2e-13
Identities = 45/105 (42%), Positives = 48/105 (44%), Gaps = 17/105 (16%)
Query: 58 DEKSMRDAIEGMNGQDMDGRNITVNEAQSRGSGGGGGRGGGGYGGGGGGYGGGGRREGGY 117
D R+ G G G RG GGGGG GGGG GGGGGG GGGG
Sbjct: 121 DNSHKRNKSSGGGGGGGGGGGGGSGNGSGRGRGGGGGGGGGGGGGGGGGGGGGG------ 174
Query: 118 NRSSGGGGGYGGGGGGGYGGGRDRGYGDDGGSRYSRGGGGDGGSW 162
GGGGG G GGG+G G G+G DG GG G W
Sbjct: 175 ---GGGGGGGSDGKGGGWGFG--FGWGGDG------PGGNFGRCW 208
>At4g14300 ribonucleoprotein like protein
Length = 406
Score = 92.8 bits (229), Expect = 7e-20
Identities = 64/168 (38%), Positives = 83/168 (49%), Gaps = 18/168 (10%)
Query: 9 RCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSMRDAIEG 68
+ FVGGL E + F YG + D I+ D+ T R RGFGFV+F E ++ D++
Sbjct: 106 KIFVGGLPPTLTDEEFRQYFEVYGPVTDVAIMYDQATNRPRGFGFVSFDSEDAV-DSVLH 164
Query: 69 MNGQDMDGRNITVNEAQSRGSG-GGGGRGGGGYGGGGGGYGGGGRREGGY---------- 117
D+ G+ + V A + + GGGGR G GGG GGY G G E Y
Sbjct: 165 KTFHDLSGKQVEVKRALPKDANPGGGGRSMG--GGGSGGYQGYGGNESSYDGRMDSNRFL 222
Query: 118 -NRSSGGG-GGYGGGG-GGGYGGGRD-RGYGDDGGSRYSRGGGGDGGS 161
++S G G YG G G GYG G + GYG GG S GG G G +
Sbjct: 223 QHQSVGNGLPSYGSSGYGAGYGNGSNGAGYGAYGGYTGSAGGYGAGAT 270
Score = 53.9 bits (128), Expect = 4e-08
Identities = 29/92 (31%), Positives = 46/92 (49%), Gaps = 9/92 (9%)
Query: 9 RCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFG---------FVTFADE 59
+ FVGG++W TD + L + F+ YGE+ + ++ D+ TGR RGFG + + +
Sbjct: 7 KLFVGGISWETDEDKLREHFTNYGEVSQAIVMRDKLTGRPRGFGIRKNRCIYFLLRYVLD 66
Query: 60 KSMRDAIEGMNGQDMDGRNITVNEAQSRGSGG 91
D M+ ++ T N SR SGG
Sbjct: 67 LGKVDVKRAMSREEQQVSGRTGNLNTSRSSGG 98
Score = 48.1 bits (113), Expect = 2e-06
Identities = 32/80 (40%), Positives = 35/80 (43%), Gaps = 18/80 (22%)
Query: 85 QSRGSGGGGGRGGGGYGGGGGGYGG-------GGRREGGYNRSSGGGGGYGGGGGG---- 133
QS G G GGY G GYG GGR GG ++ G GGY GGG G
Sbjct: 327 QSPSGYSNQGYGYGGYSGSDSGYGNQAAYGVVGGRPSGG-GSNNPGSGGYMGGGYGDGSW 385
Query: 134 ------GYGGGRDRGYGDDG 147
GYGGG + G G G
Sbjct: 386 RSDPSQGYGGGYNDGQGRQG 405
Score = 44.3 bits (103), Expect = 3e-05
Identities = 40/105 (38%), Positives = 44/105 (41%), Gaps = 17/105 (16%)
Query: 76 GRNITVNEAQSRGSGGGGGRGGGGYGGGGG--GYGGGGRREGGYNRS--SGGGGGYGGGG 131
G + V S + G G GYG G GYG G + S S G GYGG
Sbjct: 284 GSSTGVAPRNSWDTPASSGYGNPGYGSGAAHSGYGVPGAAPPTQSPSGYSNQGYGYGGYS 343
Query: 132 G--GGYG--------GGRDRGYGDD--GGSRYSRGGGGDGGSWRS 164
G GYG GGR G G + G Y GG GD GSWRS
Sbjct: 344 GSDSGYGNQAAYGVVGGRPSGGGSNNPGSGGYMGGGYGD-GSWRS 387
Score = 39.7 bits (91), Expect = 7e-04
Identities = 27/66 (40%), Positives = 29/66 (43%), Gaps = 5/66 (7%)
Query: 68 GMNGQDMD-GRNITVNEAQSRGSGGGGGRGGGGYGGGGGGYGGGGRREGGYNRSSGGGGG 126
G +G D G R SGGG G G G GGGYG G R + S G GGG
Sbjct: 341 GYSGSDSGYGNQAAYGVVGGRPSGGGSNNPGSG-GYMGGGYGDGSWRS---DPSQGYGGG 396
Query: 127 YGGGGG 132
Y G G
Sbjct: 397 YNDGQG 402
Score = 38.1 bits (87), Expect = 0.002
Identities = 27/69 (39%), Positives = 30/69 (43%), Gaps = 7/69 (10%)
Query: 99 GYGGGGGGYGGGGRREGGYNRSSG----GGGGYGGGGGG-GYGGGRDRGYGDDGGSRYSR 153
GY G GYGG + GY + GG GGG G GG GYGD GS S
Sbjct: 331 GYSNQGYGYGGYSGSDSGYGNQAAYGVVGGRPSGGGSNNPGSGGYMGGGYGD--GSWRSD 388
Query: 154 GGGGDGGSW 162
G GG +
Sbjct: 389 PSQGYGGGY 397
>At2g33410 putative RNA-binding protein
Length = 404
Score = 92.8 bits (229), Expect = 7e-20
Identities = 62/160 (38%), Positives = 79/160 (48%), Gaps = 13/160 (8%)
Query: 9 RCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSMRDAIEG 68
+ FVGGL A S+ F YG + D+ I+ D+ T R RGFGFV+F E S+ D +
Sbjct: 111 KIFVGGLPPALTSDEFRAYFETYGPVSDAVIMIDQTTQRPRGFGFVSFDSEDSV-DLVLH 169
Query: 69 MNGQDMDGRNITVNEAQSR----GSGGGGGRGGGGYGG--GGGGYGGGGRR----EGGYN 118
D++G+ + V A + G GGGRG GG GG G GG GG G Y
Sbjct: 170 KTFHDLNGKQVEVKRALPKDANPGIASGGGRGSGGAGGFPGYGGSGGSGYEGRVDSNRYM 229
Query: 119 RSSGGGGGYGGGGGGGYGGGRDRGYGDDGGSRYSRGGGGD 158
+ G GY GG GYG G GYG +G GG G+
Sbjct: 230 QPQNTGSGYPPYGGSGYGTG--YGYGSNGVGYGGFGGYGN 267
Score = 68.6 bits (166), Expect = 1e-12
Identities = 31/89 (34%), Positives = 51/89 (56%), Gaps = 2/89 (2%)
Query: 9 RCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSMRDAIEG 68
+ F+GG++W TD L + FS +GE++ ++ ++ TGR RGFGFV F+D + ++
Sbjct: 7 KLFIGGISWDTDENLLREYFSNFGEVLQVTVMREKATGRPRGFGFVAFSDPAVIDRVLQ- 65
Query: 69 MNGQDMDGRNITVNEAQSRGSGGGGGRGG 97
+ +D R++ V A SR GR G
Sbjct: 66 -DKHHIDNRDVDVKRAMSREEQSPAGRSG 93
Score = 49.7 bits (117), Expect = 7e-07
Identities = 36/99 (36%), Positives = 40/99 (40%), Gaps = 13/99 (13%)
Query: 78 NITVNEAQSRGSGGGGGRGGGGYGGGGGGYGGGGRREGGYNRSSGGGGGYG--GG----- 130
N+ A G GG G G G GYG G GG + S G YG GG
Sbjct: 301 NVGYGNAAPWGGSGGPGSAVMGQAGASAGYGSQGYGYGGNDSSYGTPSAYGAVGGRSGNM 360
Query: 131 ----GGGGYGGGRD--RGYGDDGGSRYSRGGGGDGGSWR 163
GGGGY D GYG+ G+ G GG GS R
Sbjct: 361 PNNHGGGGYADALDGSGGYGNHQGNNGQAGYGGGYGSGR 399
Score = 40.4 bits (93), Expect = 4e-04
Identities = 24/69 (34%), Positives = 30/69 (42%), Gaps = 7/69 (10%)
Query: 68 GMNGQDMDGRNITVNEAQSRGSGGG------GGRGGGGYGGGGGGYGGGGRREGGYNRSS 121
G G G + + + G+ GG GGGGY G GG G +G N +
Sbjct: 331 GSQGYGYGGNDSSYGTPSAYGAVGGRSGNMPNNHGGGGYADALDGSGGYGNHQGN-NGQA 389
Query: 122 GGGGGYGGG 130
G GGGYG G
Sbjct: 390 GYGGGYGSG 398
>At5g50250 RNA-binding protein-like
Length = 289
Score = 92.0 bits (227), Expect = 1e-19
Identities = 39/83 (46%), Positives = 61/83 (72%)
Query: 5 DVEYRCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSMRD 64
D +R +VG L W DS LE+ FS++G+++D+++++DRETGRSRGFGFV ++E +
Sbjct: 204 DAAFRIYVGNLPWDVDSGRLERLFSEHGKVVDARVVSDRETGRSRGFGFVQMSNENEVNV 263
Query: 65 AIEGMNGQDMDGRNITVNEAQSR 87
AI ++GQ+++GR I VN A+ R
Sbjct: 264 AIAALDGQNLEGRAIKVNVAEER 286
Score = 72.0 bits (175), Expect = 1e-13
Identities = 36/83 (43%), Positives = 52/83 (62%)
Query: 7 EYRCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSMRDAI 66
E + FVG L + DS+AL F Q G + S++I +R+T +SRGFGFVT + + A+
Sbjct: 112 EAKLFVGNLPYDVDSQALAMLFEQAGTVEISEVIYNRDTDQSRGFGFVTMSTVEEAEKAV 171
Query: 67 EGMNGQDMDGRNITVNEAQSRGS 89
E N +++GR +TVN A RGS
Sbjct: 172 EKFNSFEVNGRRLTVNRAAPRGS 194
>At2g05440 putative glycine-rich protein
Length = 154
Score = 92.0 bits (227), Expect = 1e-19
Identities = 45/73 (61%), Positives = 46/73 (62%), Gaps = 3/73 (4%)
Query: 88 GSGGGGGRGGGGYGGGGGGYGGGGRREGGYNRSSGGGGGYGGGGGGGYGGGRDRGYGDDG 147
G GGGG G GYGGGGG YGGGG GG GGGGG+ GGGGG YGGG G G G
Sbjct: 63 GHNGGGGHGLDGYGGGGGHYGGGGGHYGGGGGHYGGGGGHYGGGGGHYGGG---GGGHGG 119
Query: 148 GSRYSRGGGGDGG 160
G Y GGGG GG
Sbjct: 120 GGHYGGGGGGYGG 132
Score = 85.5 bits (210), Expect = 1e-17
Identities = 44/78 (56%), Positives = 48/78 (61%), Gaps = 9/78 (11%)
Query: 89 SGGGG------GRGGGGYGGGGGGYGGGGRREGGYNRSSGGGGGYGGGGGGGYGGGRDRG 142
+GGGG G GGG YGGGGG YGGGG GG GGGGG+ GGGGGG+GGG G
Sbjct: 65 NGGGGHGLDGYGGGGGHYGGGGGHYGGGGGHYGGGGGHYGGGGGHYGGGGGGHGGG---G 121
Query: 143 YGDDGGSRYSRGGGGDGG 160
+ GG Y GGG GG
Sbjct: 122 HYGGGGGGYGGGGGHHGG 139
Score = 82.8 bits (203), Expect = 8e-17
Identities = 45/77 (58%), Positives = 48/77 (61%), Gaps = 13/77 (16%)
Query: 88 GSGGGGGRGGGGYGGGGG----GYGGGGRREGGYNRSSGGGGGYGGGGGGGYGGGRDRGY 143
G GGGGG G GG+ GGGG GYGGGG G Y GGGGG+ GGGGG YGGG G+
Sbjct: 52 GHGGGGGHGHGGHNGGGGHGLDGYGGGG---GHY----GGGGGHYGGGGGHYGGG--GGH 102
Query: 144 GDDGGSRYSRGGGGDGG 160
GG Y GGGG GG
Sbjct: 103 YGGGGGHYGGGGGGHGG 119
Score = 77.0 bits (188), Expect = 4e-15
Identities = 37/60 (61%), Positives = 37/60 (61%)
Query: 88 GSGGGGGRGGGGYGGGGGGYGGGGRREGGYNRSSGGGGGYGGGGGGGYGGGRDRGYGDDG 147
G GG G GGG YGGGGG YGGGG GG GGGG YGGGGGG GGG G G G
Sbjct: 84 GGGGHYGGGGGHYGGGGGHYGGGGGHYGGGGGGHGGGGHYGGGGGGYGGGGGHHGGGGHG 143
Score = 74.7 bits (182), Expect = 2e-14
Identities = 37/71 (52%), Positives = 40/71 (56%)
Query: 68 GMNGQDMDGRNITVNEAQSRGSGGGGGRGGGGYGGGGGGYGGGGRREGGYNRSSGGGGGY 127
G++G G + G GG G GGG YGGGGG YGGGG GG GGGGGY
Sbjct: 71 GLDGYGGGGGHYGGGGGHYGGGGGHYGGGGGHYGGGGGHYGGGGGGHGGGGHYGGGGGGY 130
Query: 128 GGGGGGGYGGG 138
GGGGG GGG
Sbjct: 131 GGGGGHHGGGG 141
Score = 41.6 bits (96), Expect = 2e-04
Identities = 22/43 (51%), Positives = 24/43 (55%), Gaps = 4/43 (9%)
Query: 122 GGGGGYGG----GGGGGYGGGRDRGYGDDGGSRYSRGGGGDGG 160
G GGG+GG GGGGG+G G G G G Y GGG GG
Sbjct: 42 GYGGGHGGHGGHGGGGGHGHGGHNGGGGHGLDGYGGGGGHYGG 84
Score = 39.3 bits (90), Expect = 0.001
Identities = 30/69 (43%), Positives = 32/69 (45%), Gaps = 26/69 (37%)
Query: 99 GYGGGGGGYGGGGRREGGYNRSSGGGGGYGGGGGGGYGG-------GRDRGYGDDGGSRY 151
GYGGG GG+GG GGGGG G+GG G D GYG GG Y
Sbjct: 42 GYGGGHGGHGGH-----------------GGGGGHGHGGHNGGGGHGLD-GYG-GGGGHY 82
Query: 152 SRGGGGDGG 160
GGG GG
Sbjct: 83 GGGGGHYGG 91
>At4g24770 31 kDa RNA binding protein (rbp31)
Length = 329
Score = 89.7 bits (221), Expect = 6e-19
Identities = 37/80 (46%), Positives = 60/80 (74%)
Query: 8 YRCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSMRDAIE 67
+R +VG L W D+ LE+ FS++G++++++++ DRETGRSRGFGFVT +D + +AI
Sbjct: 244 FRVYVGNLPWDVDNGRLEQLFSEHGKVVEARVVYDRETGRSRGFGFVTMSDVDELNEAIS 303
Query: 68 GMNGQDMDGRNITVNEAQSR 87
++GQ+++GR I VN A+ R
Sbjct: 304 ALDGQNLEGRAIRVNVAEER 323
Score = 72.8 bits (177), Expect = 8e-14
Identities = 37/83 (44%), Positives = 53/83 (63%)
Query: 7 EYRCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSMRDAI 66
E + FVG LA+ +S+AL F Q G + +++I +RET +SRGFGFVT + A+
Sbjct: 149 EAKLFVGNLAYDVNSQALAMLFEQAGTVEIAEVIYNRETDQSRGFGFVTMSSVDEAETAV 208
Query: 67 EGMNGQDMDGRNITVNEAQSRGS 89
E N D++GR +TVN+A RGS
Sbjct: 209 EKFNRYDLNGRLLTVNKAAPRGS 231
>At2g37220 putative RNA-binding protein
Length = 289
Score = 89.0 bits (219), Expect = 1e-18
Identities = 36/79 (45%), Positives = 61/79 (76%)
Query: 9 RCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSMRDAIEG 68
R +VG L+W D ALE FS+ G+++++++I DR++GRS+GFGFVT+ + +++AI+
Sbjct: 205 RVYVGNLSWGVDDMALESLFSEQGKVVEARVIYDRDSGRSKGFGFVTYDSSQEVQNAIKS 264
Query: 69 MNGQDMDGRNITVNEAQSR 87
++G D+DGR I V+EA++R
Sbjct: 265 LDGADLDGRQIRVSEAEAR 283
Score = 83.6 bits (205), Expect = 4e-17
Identities = 46/120 (38%), Positives = 61/120 (50%), Gaps = 9/120 (7%)
Query: 3 SADVEYRCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSM 62
S + + FVG L + DS L + F G + ++I D+ TGRSRGFGFVT + +
Sbjct: 86 SFSADLKLFVGNLPFNVDSAQLAQLFESAGNVEMVEVIYDKITGRSRGFGFVTMSSVSEV 145
Query: 63 RDAIEGMNGQDMDGRNITVN---------EAQSRGSGGGGGRGGGGYGGGGGGYGGGGRR 113
A + NG ++DGR + VN + SRG G G GYGGGGG G G R
Sbjct: 146 EAAAQQFNGYELDGRPLRVNAGPPPPKREDGFSRGPRSSFGSSGSGYGGGGGSGAGSGNR 205
>At3g17110 hypothetical protein
Length = 175
Score = 88.6 bits (218), Expect = 1e-18
Identities = 49/88 (55%), Positives = 49/88 (55%), Gaps = 15/88 (17%)
Query: 88 GSGGGGGRGGGGYGGGGG------------GYGGGGRREGGYNRSSGGGGGYGGGGGGGY 135
G GGGGG GGGG GGGGG GYG G GG N GGGGG GGGGGGG
Sbjct: 48 GGGGGGGGGGGGGGGGGGGDGSGSGSGYGSGYGSGSGYGGGNNGGGGGGGGRGGGGGGGN 107
Query: 136 GG-GRDRGYGDDG--GSRYSRGGGGDGG 160
GG G GYG GS RGGGG GG
Sbjct: 108 GGSGNGSGYGSGSGYGSGAGRGGGGGGG 135
Score = 85.9 bits (211), Expect = 9e-18
Identities = 48/86 (55%), Positives = 50/86 (57%), Gaps = 5/86 (5%)
Query: 76 GRNITVNEAQSRGSGGGGGRGGGGYGGGGGGYGGGGRREGGYNRSSGGGGGYGGGGGGGY 135
GR GSGGG G GGGG GGGGGG GGGG GG SG G G G G G GY
Sbjct: 30 GRGSGSGSGYGSGSGGGNGGGGGG-GGGGGGGGGGG---GGDGSGSGSGYGSGYGSGSGY 85
Query: 136 GGGRDRGYGDDGGSRYSRGGGGDGGS 161
GGG + G G GG R GGGG+GGS
Sbjct: 86 GGGNNGG-GGGGGGRGGGGGGGNGGS 110
Score = 85.5 bits (210), Expect = 1e-17
Identities = 52/106 (49%), Positives = 52/106 (49%), Gaps = 13/106 (12%)
Query: 68 GMNGQDMDGRNITVNEAQSRGSGGGGGRGGGGYGGGGGGYGGGGRREG-----GYNRSSG 122
G G D G GSG GGG GGG GGGG G GGGG G GY SG
Sbjct: 62 GGGGGDGSGSGSGYGSGYGSGSGYGGGNNGGGGGGGGRGGGGGGGNGGSGNGSGYGSGSG 121
Query: 123 -------GGGGYGGGGGGGYGGGRDRGYGDDGGSRYSRG-GGGDGG 160
GGGG GGGGGGG GGG RG G GS Y G G G GG
Sbjct: 122 YGSGAGRGGGGGGGGGGGGGGGGGSRGNGSGYGSGYGEGYGSGYGG 167
Score = 82.0 bits (201), Expect = 1e-16
Identities = 46/79 (58%), Positives = 46/79 (58%), Gaps = 9/79 (11%)
Query: 88 GSGGGGGRGGGGYGGGGG-----GYGGGGRREGGYNRSSGGGGGYGGGGGGGYGG--GRD 140
G GGGGGRGGGG GG GG GYG G G R GGGGG GGGGGGG GG G
Sbjct: 92 GGGGGGGRGGGGGGGNGGSGNGSGYGSGSGYGSGAGRGGGGGGG-GGGGGGGGGGSRGNG 150
Query: 141 RGYGDDGGSRYSRG-GGGD 158
GYG G Y G GGGD
Sbjct: 151 SGYGSGYGEGYGSGYGGGD 169
Score = 76.6 bits (187), Expect = 5e-15
Identities = 42/81 (51%), Positives = 43/81 (52%), Gaps = 8/81 (9%)
Query: 83 EAQSRGSGGGGGRGGGGYGGGGGGYGGGGRREGGYNRSSGGGGGYGGGGGGGYGGGRDRG 142
+A RGSG G G G G GG GGG GGGG GG GGGGG GGG G G G G G
Sbjct: 27 KALGRGSGSGSGYGSGSGGGNGGGGGGGG---GG-----GGGGGGGGGDGSGSGSGYGSG 78
Query: 143 YGDDGGSRYSRGGGGDGGSWR 163
YG G GGG GG R
Sbjct: 79 YGSGSGYGGGNNGGGGGGGGR 99
Score = 76.3 bits (186), Expect = 7e-15
Identities = 45/93 (48%), Positives = 46/93 (49%), Gaps = 21/93 (22%)
Query: 88 GSGGGGGRGGG-----------------GYGGGG--GGYGGGGRREGGYNRSSGGGGGYG 128
G GGGGG GGG GYGGG GG GGGGR GG + G G G G
Sbjct: 56 GGGGGGGGGGGDGSGSGSGYGSGYGSGSGYGGGNNGGGGGGGGRGGGGGGGNGGSGNGSG 115
Query: 129 GGGGGGYGGGRDRGYGDDGGSRYSRGGGGDGGS 161
G G GYG G RG G GG GGGG GGS
Sbjct: 116 YGSGSGYGSGAGRGGGGGGGG--GGGGGGGGGS 146
Score = 65.9 bits (159), Expect = 1e-11
Identities = 39/79 (49%), Positives = 40/79 (50%), Gaps = 3/79 (3%)
Query: 68 GMNGQDMDGRNITVNEAQSRGSGGGGGRGGG-GYGGGGGGYGGGGRREGGYNRSSGGGGG 126
G G G N GSG G G G G G GGGGGG GGGG G SG G G
Sbjct: 97 GGRGGGGGGGNGGSGNGSGYGSGSGYGSGAGRGGGGGGGGGGGGGGGGGSRGNGSGYGSG 156
Query: 127 YGGGGGGGYGGGRDRGYGD 145
YG G G GYGGG + YGD
Sbjct: 157 YGEGYGSGYGGGDN--YGD 173
Score = 42.7 bits (99), Expect = 9e-05
Identities = 24/51 (47%), Positives = 25/51 (48%), Gaps = 5/51 (9%)
Query: 119 RSSGGGGGYGGG-----GGGGYGGGRDRGYGDDGGSRYSRGGGGDGGSWRS 164
R SG G GYG G GGGG GGG G G GG S G G G + S
Sbjct: 31 RGSGSGSGYGSGSGGGNGGGGGGGGGGGGGGGGGGGDGSGSGSGYGSGYGS 81
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.144 0.444
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,210,033
Number of Sequences: 26719
Number of extensions: 356566
Number of successful extensions: 26258
Number of sequences better than 10.0: 700
Number of HSP's better than 10.0 without gapping: 628
Number of HSP's successfully gapped in prelim test: 79
Number of HSP's that attempted gapping in prelim test: 2002
Number of HSP's gapped (non-prelim): 6466
length of query: 164
length of database: 11,318,596
effective HSP length: 92
effective length of query: 72
effective length of database: 8,860,448
effective search space: 637952256
effective search space used: 637952256
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC134242.10


