

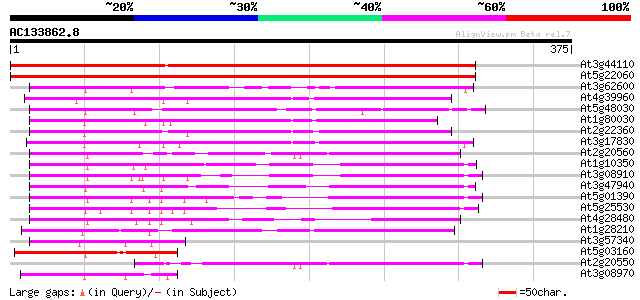
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133862.8 + phase: 0
(375 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g44110 dnaJ protein homolog atj3 566 e-162
At5g22060 DNAJ PROTEIN HOMOLOG ATJ 562 e-160
At3g62600 unknown protein 183 1e-46
At4g39960 DnaJ like protein 177 8e-45
At5g48030 DnaJ protein-like 176 1e-44
At1g80030 unknown protein 174 7e-44
At2g22360 DnaJ like protein 172 4e-43
At3g17830 DnaJ, putative 162 2e-40
At2g20560 putative heat shock protein 156 2e-38
At1g10350 putative heat-shock protein 144 1e-34
At3g08910 putative heat shock protein 137 1e-32
At3g47940 heat shock protein-like protein 137 1e-32
At5g01390 heat shock protein 40-like 134 8e-32
At5g25530 heat-shock protein - like 130 9e-31
At4g28480 heat-shock protein 129 3e-30
At1g28210 AtJ1 (atj) 119 3e-27
At3g57340 dnaJ-like protein 93 2e-19
At5g03160 unknown protein 93 3e-19
At2g20550 putative heat shock protein 83 2e-16
At3g08970 putative DnaJ protein 82 4e-16
>At3g44110 dnaJ protein homolog atj3
Length = 420
Score = 566 bits (1458), Expect = e-162
Identities = 274/312 (87%), Positives = 291/312 (92%), Gaps = 2/312 (0%)
Query: 1 MFGRAP-KKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYE 59
MFGR P KKSDNTK+YEILGV K+ASP+DLKKAYKKAAIKNHPDKGGDPEKFKELAQAYE
Sbjct: 1 MFGRGPSKKSDNTKFYEILGVPKSASPEDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYE 60
Query: 60 VLSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGE 119
VLSDPEKRE+YDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGG F GG +SR RRQRRGE
Sbjct: 61 VLSDPEKREIYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGPF-GGNTSRQRRQRRGE 119
Query: 120 DVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHL 179
DVVHPLKVSLED+YLGT KKLSLSRN LCSKCNGKGSKSGAS+ C GCQGSGMK+S+R L
Sbjct: 120 DVVHPLKVSLEDVYLGTMKKLSLSRNALCSKCNGKGSKSGASLKCGGCQGSGMKVSIRQL 179
Query: 180 GANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFP 239
G MIQQMQH CNECKGTGETI+D+DRCPQCKG+KV+ +KKVLEV+VEKGMQ+ QKITF
Sbjct: 180 GPGMIQQMQHACNECKGTGETINDRDRCPQCKGDKVIPEKKVLEVNVEKGMQHSQKITFE 239
Query: 240 GEADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQL 299
G+ADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQF LTHLD R L
Sbjct: 240 GQADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFVLTHLDGRSL 299
Query: 300 LIKSNPGEVVKP 311
LIKSNPGEVVKP
Sbjct: 300 LIKSNPGEVVKP 311
>At5g22060 DNAJ PROTEIN HOMOLOG ATJ
Length = 419
Score = 562 bits (1448), Expect = e-160
Identities = 269/312 (86%), Positives = 288/312 (92%), Gaps = 1/312 (0%)
Query: 1 MFGRAP-KKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYE 59
MFGR P +KSDNTK+YEILGV K A+P+DLKKAYKKAAIKNHPDKGGDPEKFKELAQAYE
Sbjct: 1 MFGRGPSRKSDNTKFYEILGVPKTAAPEDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYE 60
Query: 60 VLSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGE 119
VLSDPEKRE+YDQYGEDALKEGMGGGGGGHDPFDIFSSFFG GG P G SRGRRQRRGE
Sbjct: 61 VLSDPEKREIYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGSGGHPFGSHSRGRRQRRGE 120
Query: 120 DVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHL 179
DVVHPLKVSLED+YLGT+KKLSLSR LCSKCNGKGSKSGASM C GCQGSGMKIS+R
Sbjct: 121 DVVHPLKVSLEDVYLGTTKKLSLSRKALCSKCNGKGSKSGASMKCGGCQGSGMKISIRQF 180
Query: 180 GANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFP 239
G M+QQ+QH CN+CKGTGETI+D+DRCPQCKGEKVV +KKVLEV+VEKGMQ+ QKITF
Sbjct: 181 GPGMMQQVQHACNDCKGTGETINDRDRCPQCKGEKVVSEKKVLEVNVEKGMQHNQKITFS 240
Query: 240 GEADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQL 299
G+ADEAPDTVTGDIVFV+QQKEHPKFKRKGEDLFVEHT+SLTEALCGFQF LTHLD RQL
Sbjct: 241 GQADEAPDTVTGDIVFVIQQKEHPKFKRKGEDLFVEHTISLTEALCGFQFVLTHLDKRQL 300
Query: 300 LIKSNPGEVVKP 311
LIKS PGEVVKP
Sbjct: 301 LIKSKPGEVVKP 312
>At3g62600 unknown protein
Length = 346
Score = 183 bits (465), Expect = 1e-46
Identities = 116/310 (37%), Positives = 169/310 (54%), Gaps = 45/310 (14%)
Query: 14 YYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPE----KFKELAQAYEVLSDPEKREV 69
YY++L V K AS + +K+AY+K A+K HPDK E KF E+ AYEVLSD EKRE+
Sbjct: 27 YYDVLQVPKGASDEQIKRAYRKLALKYHPDKNQGNEEATRKFAEINNAYEVLSDEEKREI 86
Query: 70 YDQYGEDALKE-----GMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGEDVVHP 124
Y++YGE+ LK+ G GGGGGG + DIFSSFFGGG + +G+DV+
Sbjct: 87 YNKYGEEGLKQFSANGGRGGGGGGMNMQDIFSSFFGGGSM-----EEEEKVVKGDDVIVE 141
Query: 125 LKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHLGANMI 184
L+ +LEDLY+G S K+ +NV+ K C+ ++ R +G M
Sbjct: 142 LEATLEDLYMGGSMKVWREKNVI---------KPAPGKRKCNCRN---EVYHRQIGPGMF 189
Query: 185 QQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPGEADE 244
QQM E + DK CP K E+ + + V +EKGM++G++++F + +
Sbjct: 190 QQMT----------EQVCDK--CPNVKYER---EGYFVTVDIEKGMKDGEEVSFYEDGEP 234
Query: 245 APDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQLLIKS- 303
D GD+ F ++ H +F+R G DL + ++L EAL GF+ + HLD ++ I S
Sbjct: 235 ILDGDPGDLKFRIRTAPHARFRRDGNDLHMNVNITLVEALVGFEKSFKHLDDHEVDISSK 294
Query: 304 ---NPGEVVK 310
P EV K
Sbjct: 295 GITKPKEVKK 304
>At4g39960 DnaJ like protein
Length = 447
Score = 177 bits (449), Expect = 8e-45
Identities = 106/297 (35%), Positives = 157/297 (52%), Gaps = 16/297 (5%)
Query: 11 NTKYYEILGVSKNASPDDLKKAYKKAAIKNHPD---KGGDPEKFKELAQAYEVLSDPEKR 67
+T +Y +LGVSKNA+ ++K AY+K A HPD G +KFKE++ AYE+LSD EKR
Sbjct: 83 DTDFYSVLGVSKNATKAEIKSAYRKLARSYHPDVNKDAGAEDKFKEISNAYEILSDDEKR 142
Query: 68 EVYDQYGEDALK-EGMGGGGGGHDPFDIFSSFFGG----GGFPGGGSSRGRRQRR--GED 120
+YD+YGE +K GMGG G +PFD+F S F G GG GG SRG R R GED
Sbjct: 143 SLYDRYGEAGVKGAGMGGMGDYSNPFDLFESLFEGMGGMGGMGGGMGSRGSRSRAIDGED 202
Query: 121 VVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMT-CAGCQGSGMKISMRHL 179
+ L ++ ++ G K++ +SR C CNG G+K+G T C C G G ++
Sbjct: 203 EYYSLILNFKEAVFGIEKEIEISRLESCGTCNGSGAKAGTKPTKCKTCGGQGQVVASTRT 262
Query: 180 GANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFP 239
+ QQ+ C+ C GTGE C C G+ V++ K + + V G+ +G ++
Sbjct: 263 PLGVFQQVM-TCSPCNGTGEI---SKPCGACSGDGRVRRTKRISLKVPAGVDSGSRLRVR 318
Query: 240 GEADEAP-DTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLD 295
GE + GD+ V++ P KR ++ +S +A+ G + +D
Sbjct: 319 GEGNAGKRGGSPGDLFAVIEVIPDPVLKRDDTNILYTCKISYVDAILGTTLKVPTVD 375
>At5g48030 DnaJ protein-like
Length = 456
Score = 176 bits (447), Expect = 1e-44
Identities = 116/323 (35%), Positives = 170/323 (51%), Gaps = 35/323 (10%)
Query: 14 YYEILGVSKNASPDDLKKAYKKAAIKNHPDKG-GDPE---KFKELAQAYEVLSDPEKREV 69
YY +LGVSKNA ++KKAY A K HPD DPE KF+E+++AYE+L D EKR++
Sbjct: 95 YYSVLGVSKNAQEGEIKKAYYGLAKKLHPDMNKDDPEAETKFQEVSKAYEILKDKEKRDL 154
Query: 70 YDQYGEDALKEGM-----------GGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRG 118
YDQ G +A ++ GGGGGG +PFDIF G F G + R+ G
Sbjct: 155 YDQVGHEAFEQNASGGFPNDQGFGGGGGGGFNPFDIF------GSFNGDIFNMYRQDIGG 208
Query: 119 EDVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASM-TCAGCQGSGMKISMR 177
+DV L +S + G SK ++ + C+ C G+G G C C GSGM R
Sbjct: 209 QDVKVLLDLSFMEAVQGCSKTVTFQTEMACNTCGGQGVPPGTKREKCKACNGSGMTSLRR 268
Query: 178 HLGANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQ--K 235
+ +Q C +C G G+T S C C+G +VV+ +K ++V ++ G+ N K
Sbjct: 269 GM-----LSIQTTCQKCGGAGQTFS--SICKSCRGARVVRGQKSVKVTIDPGVDNSDTLK 321
Query: 236 ITFPGEADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLD 295
+ G AD D GD+ L+ +E P F+R+G D+ V+ LS+T+A+ G + L
Sbjct: 322 VARVGGADPEGDQ-PGDLYVTLKVREDPVFRREGSDIHVDAVLSVTQAILGGTIQVPTL- 379
Query: 296 SRQLLIKSNPGEVVKPGWMSVRR 318
+ +++K PG +PG V R
Sbjct: 380 TGDVVVKVRPG--TQPGHKVVLR 400
>At1g80030 unknown protein
Length = 500
Score = 174 bits (441), Expect = 7e-44
Identities = 105/287 (36%), Positives = 156/287 (53%), Gaps = 17/287 (5%)
Query: 14 YYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDP---EKFKELAQAYEVLSDPEKREVY 70
YY LGVSK+A+ ++K AY++ A + HPD +P EKFKE++ AYEVLSD +KR +Y
Sbjct: 76 YYATLGVSKSANNKEIKAAYRRLARQYHPDVNKEPGATEKFKEISAAYEVLSDEQKRALY 135
Query: 71 DQYGEDALKEGMGGGGGGH--DPFDIFSSFFGG--GGFPG-----GGSSRGRRQRRGEDV 121
DQYGE +K +GG G + +PFD+F +FFG GGFPG G +R R +GED+
Sbjct: 136 DQYGEAGVKSTVGGASGPYTSNPFDLFETFFGASMGGFPGMDQADFGRTRRSRVTKGEDL 195
Query: 122 VHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASM-TCAGCQGSGMKISMRHLG 180
+ + + L + G+ K+ L+ C C G G+K+G+ M C+ C G G +
Sbjct: 196 RYDITLELSEAIFGSEKEFDLTHLETCEACAGTGAKAGSKMRICSTCGGRGQVMRTEQTP 255
Query: 181 ANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPG 240
M Q+ C C G GE IS + C +C GE V+ KK ++V + G+ G + G
Sbjct: 256 FGMFSQVS-ICPNCGGDGEVIS--ENCRKCSGEGRVRIKKSIKVKIPPGVSAGSILRVAG 312
Query: 241 EADEAP-DTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCG 286
E D P GD+ L ++ +R G +L ++S +A+ G
Sbjct: 313 EGDSGPRGGPPGDLYVYLDVEDVRGIERDGINLLSTLSISYLDAILG 359
>At2g22360 DnaJ like protein
Length = 442
Score = 172 bits (435), Expect = 4e-43
Identities = 104/290 (35%), Positives = 151/290 (51%), Gaps = 15/290 (5%)
Query: 14 YYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDP---EKFKELAQAYEVLSDPEKREVY 70
YY +LGVSKNA+ ++K AY+K A HPD DP EKFKE++ AYEVLSD EK+ +Y
Sbjct: 87 YYSVLGVSKNATKAEIKSAYRKLARNYHPDVNKDPGAEEKFKEISNAYEVLSDDEKKSLY 146
Query: 71 DQYGEDALKEGMG-GGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRR--GEDVVHPLKV 127
D+YGE LK G G G +PFD+F S F G GGG RG R R G+D + L +
Sbjct: 147 DRYGEAGLKGAAGFGNGDFSNPFDLFDSLFEGF---GGGMGRGSRSRAVDGQDEYYTLIL 203
Query: 128 SLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMT-CAGCQGSGMKISMRHLGANMIQQ 186
+ ++ G K++ +SR C C G G+K G T C C G G +S + QQ
Sbjct: 204 NFKEAVFGMEKEIEISRLESCGTCEGSGAKPGTKPTKCTTCGGQGQVVSAARTPLGVFQQ 263
Query: 187 MQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPGEADEAP 246
+ C+ C GTGE + C C G+ V++ K + + V G+ +G ++ GE +
Sbjct: 264 VM-TCSSCNGTGEISTP---CGTCSGDGRVRKTKRISLKVPAGVDSGSRLRVRGEGNAGK 319
Query: 247 -DTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLD 295
GD+ V++ P KR ++ +S +A+ G + +D
Sbjct: 320 RGGSPGDLFVVIEVIPDPILKRDDTNILYTCKISYIDAILGTTLKVPTVD 369
>At3g17830 DnaJ, putative
Length = 517
Score = 162 bits (411), Expect = 2e-40
Identities = 104/318 (32%), Positives = 171/318 (53%), Gaps = 22/318 (6%)
Query: 12 TKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDP---EKFKELAQAYEVLSDPEKRE 68
T +Y L V++NA+ ++K +Y+K A K HPD +P +KFK+++ AYEVLSD EKR
Sbjct: 62 TDHYSTLNVNRNATLQEIKSSYRKLARKYHPDMNKNPGAEDKFKQISAAYEVLSDEEKRS 121
Query: 69 VYDQYGEDALKEGMGGG---GGGHDPFDIFSSFFGG--GGFPGGGSSRG------RRQRR 117
YD++GE L+ G G DPFD++S+FFGG G F G G S G ++
Sbjct: 122 AYDRFGEAGLEGDFNGSQDTSPGVDPFDLYSAFFGGSDGFFGGMGESGGMGFDFMNKRSL 181
Query: 118 GEDVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASM-TCAGCQGSGMKISM 176
D+ + L++S E+ G +++ +S C C G G+KS S+ C+ C G G ++
Sbjct: 182 DLDIRYDLRLSFEEAVFGVKREIEVSYLETCDGCGGTGAKSSNSIKQCSSCDGKGRVMNS 241
Query: 177 RHLGANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKI 236
+ ++ Q+ C++C G G+TI+DK C +C G ++ +K ++V V G+ + +
Sbjct: 242 QRTPFGIMSQVS-TCSKCGGEGKTITDK--CRKCIGNGRLRARKKMDVVVPPGVSDRATM 298
Query: 237 TFPGEAD-EAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLD 295
GE + + GD+ VLQ E +R+G +L+ + T+A+ G + ++
Sbjct: 299 RIQGEGNMDKRSGRAGDLFIVLQVDEKRGIRREGLNLYSNINIDFTDAILGATTKVETVE 358
Query: 296 -SRQLLIK--SNPGEVVK 310
S L I + PG+ VK
Sbjct: 359 GSMDLRIPPGTQPGDTVK 376
>At2g20560 putative heat shock protein
Length = 337
Score = 156 bits (394), Expect = 2e-38
Identities = 106/298 (35%), Positives = 154/298 (51%), Gaps = 31/298 (10%)
Query: 14 YYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEK-----FKELAQAYEVLSDPEKRE 68
YY++L V ++AS DDLKKAY+K A+K HPDK + +K FK++++AYEVLSDP+K+
Sbjct: 5 YYKVLQVDRSASDDDLKKAYRKLAMKWHPDKNPNNKKDAEAMFKQISEAYEVLSDPQKKA 64
Query: 69 VYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGEDVVHPLKVS 128
VYDQYGE+ LK + G +++F G P +S R +D+
Sbjct: 65 VYDQYGEEGLKGNVPPPDAGG------ATYFSTGDGP---TSFRFNPRNADDIFAE---- 111
Query: 129 LEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHLGANMIQQMQ 188
+ G S R + G AS G G G SM H GA ++
Sbjct: 112 ----FFGFSSPFGGGRGGTRFSSSMFGDNMFASFGEGGGGGGG---SMHHGGARKAAPIE 164
Query: 189 H--PCNE---CKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPGEAD 243
+ PC+ KGT + + G K +Q +++L + V+ G + G KITFP + +
Sbjct: 165 NKLPCSLEDLYKGTTKKMRISREIADVSG-KTMQVEEILTIDVKPGWKKGTKITFPEKGN 223
Query: 244 EAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQLLI 301
E P + D+VF++ +K HP F R+G DL V +SL EAL G+ LT LD R+L I
Sbjct: 224 EQPGVIPADLVFIIDEKPHPVFTREGNDLIVTQKISLVEALTGYTVNLTTLDGRRLTI 281
>At1g10350 putative heat-shock protein
Length = 349
Score = 144 bits (362), Expect = 1e-34
Identities = 99/325 (30%), Positives = 153/325 (46%), Gaps = 54/325 (16%)
Query: 14 YYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEK-----FKELAQAYEVLSDPEKRE 68
YY +L V++NA+ DDLKK+Y++ A+K HPDK +K FK++++AY+VLSDP++R+
Sbjct: 5 YYNVLKVNRNANEDDLKKSYRRMAMKWHPDKNPTSKKEAEAKFKQISEAYDVLSDPQRRQ 64
Query: 69 VYDQYGEDALKEG---MGGGGGGH------------------DPFDIFSSFFGGGGFPGG 107
+YDQYGE+ LK H D DIF+ FFG G G
Sbjct: 65 IYDQYGEEGLKSTDLPTAAETAAHQQQRSYSSSNSEFRYYPRDAEDIFAEFFGESGDAFG 124
Query: 108 GSSRGRRQRRGEDVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGC 167
G S GR + G D S E ++K + N K + + C
Sbjct: 125 GGSSGRTRGDGGDGGGRRFKSAE-AGSQANRKTPPTNKKTTPPANRKAPAIESKLACT-- 181
Query: 168 QGSGMKISMRHLGANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVE 227
+ + GA ++ + G +T+ + +L++ ++
Sbjct: 182 ------LEELYKGAKKKMRISRVVPDDFGKPKTVQE-----------------ILKIDIK 218
Query: 228 KGMQNGQKITFPGEADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGF 287
G + G KITFP + ++ P D++FV+ +K H FKR G DL +E +SL +AL G
Sbjct: 219 PGWKKGTKITFPEKGNQEPGVTPADLIFVVDEKPHSVFKRDGNDLILEKKVSLIDALTGL 278
Query: 288 QFALTHLDSRQLLIKSNPGEVVKPG 312
++T LD R L I ++VKPG
Sbjct: 279 TISVTTLDGRSLTIPVL--DIVKPG 301
>At3g08910 putative heat shock protein
Length = 323
Score = 137 bits (345), Expect = 1e-32
Identities = 101/329 (30%), Positives = 149/329 (44%), Gaps = 81/329 (24%)
Query: 14 YYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEK-----FKELAQAYEVLSDPEKRE 68
YY++L V +NA DDLKKAY+K A+K HPDK + +K FK++++AY+VLSDP+KR
Sbjct: 5 YYKVLQVDRNAKDDDLKKAYRKLAMKWHPDKNPNNKKDAEAKFKQISEAYDVLSDPQKRA 64
Query: 69 VYDQYGEDALKE-----GMGGG---GG------GHDPFDIFSSFFGG----GGFPGGGSS 110
+YDQYGE+ L G GGG GG G DIFS FFG G G G S
Sbjct: 65 IYDQYGEEGLTSQAPPPGAGGGFSDGGASFRFNGRSADDIFSEFFGFTRPFGDSRGAGPS 124
Query: 111 RGRRQRR---GEDVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGC 167
G R +VV P K + + R + CS
Sbjct: 125 NGFRFAEDVFSSNVVPPRKAA------------PIERQLPCS------------------ 154
Query: 168 QGSGMKISMRHLGANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVE 227
+ + G + ++ + G T+ + +L + ++
Sbjct: 155 ------LEDLYKGVSKKMKISRDVLDSSGRPTTVEE-----------------ILTIEIK 191
Query: 228 KGMQNGQKITFPGEADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGF 287
G + G KITFP + +E + D+VF++ +K H FKR G DL + + L EAL G+
Sbjct: 192 PGWKKGTKITFPEKGNEQRGIIPSDLVFIVDEKPHAVFKRDGNDLVMTQKIPLVEALTGY 251
Query: 288 QFALTHLDSRQLLIKSNPGEVVKPGWMSV 316
++ LD R + + N V+ P + V
Sbjct: 252 TAQVSTLDGRSVTVPIN--NVISPSYEEV 278
>At3g47940 heat shock protein-like protein
Length = 350
Score = 137 bits (344), Expect = 1e-32
Identities = 100/342 (29%), Positives = 148/342 (43%), Gaps = 91/342 (26%)
Query: 14 YYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPE------KFKELAQAYEVLSDPEKR 67
YY IL V+ NA+ DDLKKAYK+ A+ HPDK KFK +++AY+VLSDP+KR
Sbjct: 5 YYNILKVNHNATEDDLKKAYKRLAMIWHPDKNPSTRRDEAEAKFKRISEAYDVLSDPQKR 64
Query: 68 EVYDQYGEDALKEGMGGGGGG-----------------------------------HDPF 92
++YD YGE+ LK G D
Sbjct: 65 QIYDLYGEEGLKSGKIPNSSSSEASSSSSSSSSRYPHFHQHRPQHPPNASSFRFNPRDAE 124
Query: 93 DIFSSFFG---GGGFPGGGSSRGRRQRRGEDVVHPLKVSLEDLYLGTSKKLSLSRNVLCS 149
DI++ FFG GGG G R R G + Y G +K+ N L
Sbjct: 125 DIYAEFFGSENGGGSNNAGGRGNRAFRNGH-----FNTGGANGYSGEMRKVPAMENPL-- 177
Query: 150 KCNGKGSKSGASMTCAGCQGSGMKISMRHLGANMIQQMQHPCNECKGTGETISDKDRCPQ 209
+S+ L ++++M+ N +G
Sbjct: 178 -----------------------PVSLEDLYKGVVKKMRITRNVYDASG----------- 203
Query: 210 CKGEKVVQQKKVLEVHVEKGMQNGQKITFPGEADEAPDTVTGDIVFVLQQKEHPKFKRKG 269
+++ + ++L + ++ G + G K+TFP + +E P + DIVFV+++K HP +KR G
Sbjct: 204 ----RMMVEAEILPIEIKPGWKKGTKLTFPKKGNEEPGIIPADIVFVVEEKPHPVYKRDG 259
Query: 270 EDLFVEHTLSLTEALCGFQFALTHLDSRQLLIKSNPGEVVKP 311
DL V ++L EAL G L LD R L+I E++KP
Sbjct: 260 NDLLVSQEITLLEALTGKTVNLITLDGRTLMIPLT--EIIKP 299
>At5g01390 heat shock protein 40-like
Length = 335
Score = 134 bits (337), Expect = 8e-32
Identities = 102/330 (30%), Positives = 157/330 (46%), Gaps = 70/330 (21%)
Query: 14 YYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEK-----FKELAQAYEVLSDPEKRE 68
+Y++L V ++A+ D+LKKAY+K A+K HPDK + +K FK++++AY+VLSDP+KR
Sbjct: 5 FYKVLEVDRSANDDELKKAYRKLAMKWHPDKNPNNKKEAEAKFKQISEAYDVLSDPQKRA 64
Query: 69 VYDQYGEDALKE----GMGGG-GGGHDPF-----------DIFSSFFG--GGGFPGGGSS 110
+Y+QYGE+ L + G GGG GG D DIFS FFG F G S
Sbjct: 65 IYEQYGEEGLNQAPPPGAGGGYPGGSDAGASFRFNPRSADDIFSEFFGFTRPSFGTGSDS 124
Query: 111 RGRRQ--RRGEDVVHPLKVSLE--DLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAG 166
R R G+D+ + + + + + K + R + CS + G S
Sbjct: 125 RAGPSGFRYGDDIFASFRAATTGGEASIPSRKSAPIERQLPCSL---EDLYKGVSKK--- 178
Query: 167 CQGSGMKISMRHLGANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHV 226
MKIS L ++ + +++L + +
Sbjct: 179 -----MKISRDVLDSS------------------------------GRPTPVEEILTIEI 203
Query: 227 EKGMQNGQKITFPGEADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCG 286
+ G + G KITF + +E + D+VF++ +K HP FKR G DL V +SL +AL G
Sbjct: 204 KPGWKKGTKITFLEKGNEHRGVIPSDLVFIVDEKPHPVFKRDGNDLVVMQKISLVDALTG 263
Query: 287 FQFALTHLDSRQLLIKSNPGEVVKPGWMSV 316
+ +T LD R L + N V+ P + V
Sbjct: 264 YTAQVTTLDGRTLTVPVN--NVISPSYEEV 291
>At5g25530 heat-shock protein - like
Length = 347
Score = 130 bits (328), Expect = 9e-31
Identities = 108/360 (30%), Positives = 154/360 (42%), Gaps = 124/360 (34%)
Query: 14 YYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPE-----KFKELAQAYE--------V 60
YY+IL V++NA+ DDLKK+Y+K A+K HPDK + + KFK++++AYE V
Sbjct: 5 YYDILKVNRNATEDDLKKSYRKLAMKWHPDKNPNTKTEAEAKFKQISEAYEAKYEVMFQV 64
Query: 61 LSDPEKREVYDQYGEDALKE-------GMGGGGGGHDPF---DIFSSFFG---------- 100
LSDP+KR VYDQYGE+ L + G G GG +P DIF+ FFG
Sbjct: 65 LSDPQKRAVYDQYGEEGLSDMPPPGSTGNNGRAGGFNPRNAEDIFAEFFGSSPFGFGSAG 124
Query: 101 ------------GGGFPGG--GSSRGRRQ-------------RRGEDVVHPLKVSLEDLY 133
GGG GG G + G ++ V L SLE+LY
Sbjct: 125 GPGRSMRFQSDGGGGMFGGFGGGNNGSENNIFRTYSEGTPAPKKPPPVESKLPCSLEELY 184
Query: 134 LGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHLGANMIQQMQHPCNE 193
G+++K MKIS + AN Q
Sbjct: 185 SGSTRK--------------------------------MKISRSIVDANGRQ-------- 204
Query: 194 CKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPGEADEAPDTVTGDI 253
Q+ ++L + V+ G + G KI FP + +E + + D+
Sbjct: 205 ----------------------AQETEILTIVVKPGWKKGTKIKFPDKGNEQVNQLPADL 242
Query: 254 VFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQLLIKSNPGEVVKPGW 313
VFV+ +K H F R G DL ++L EA+ G + LD R L + E+V PG+
Sbjct: 243 VFVIDEKPHDLFTRDGNDLITSRRVTLAEAIGGTTVNINTLDGRNLPV--GVAEIVSPGY 300
>At4g28480 heat-shock protein
Length = 348
Score = 129 bits (323), Expect = 3e-30
Identities = 104/350 (29%), Positives = 146/350 (41%), Gaps = 124/350 (35%)
Query: 14 YYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEK-----FKELAQAYEVLSDPEKRE 68
YY++L V ++A+ DDLKKAY+K A+K HPDK + +K FK++++AY+VLSDP+KR
Sbjct: 5 YYKVLQVDRSANDDDLKKAYRKLAMKWHPDKNPNNKKDAEAKFKQISEAYDVLSDPQKRA 64
Query: 69 VYDQYGEDALKEGMG------------GGGGGHDPF--------DIFSSFFG-----GGG 103
VYDQYGE+ LK + G G F DIF+ FFG GGG
Sbjct: 65 VYDQYGEEGLKGNVPPPNAATSGASYFSTGDGSSSFRFNPRSADDIFAEFFGFSTPFGGG 124
Query: 104 FPGGGSSRGRRQRRGED--------------------------------VVHPLKVSLED 131
G G R + G+D + + L SLED
Sbjct: 125 GGGTGGQRFASRMFGDDMYASFGEGAGGGGAMHHHHHHHHHAAARKVAPIENKLPCSLED 184
Query: 132 LYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHLGANMIQQMQHPC 191
LY GT+KK+ +SR ++ SG +M G+K +
Sbjct: 185 LYKGTTKKMKISREIV--------DVSGKAMQVEEILTIGVKPGWK-------------- 222
Query: 192 NECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPGEADEAPDTVTG 251
KGT T +K +E P +
Sbjct: 223 ---KGTKITFPEK-------------------------------------GNEHPGVIPA 242
Query: 252 DIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQLLI 301
D+VF++ +K HP F R+G DL V +SL +AL G+ + LD R L I
Sbjct: 243 DLVFIIDEKPHPVFTREGNDLIVTQKVSLADALTGYTANIATLDGRTLTI 292
>At1g28210 AtJ1 (atj)
Length = 427
Score = 119 bits (298), Expect = 3e-27
Identities = 86/304 (28%), Positives = 140/304 (45%), Gaps = 36/304 (11%)
Query: 9 SDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGD----PEKFKELAQAYEVLSDP 64
S YY++LGVS A+ +++KK++ + A K HPD + KF+E+ +AYE L +
Sbjct: 44 SSARNYYDVLGVSPKATREEIKKSFHELAKKFHPDTNRNNPSAKRKFQEIREAYETLGNS 103
Query: 65 EKREVYDQYGEDALKEGMGGGGGGHDPF---------DIFSSFFGGGGFPGGGSSRGRRQ 115
E+RE YD+ + + + GG + F D F F S
Sbjct: 104 ERREEYDKL-QYRNSDYVNNDGGDSERFRRAYQSNFSDTFHKIF---------SEIFENN 153
Query: 116 RRGEDVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMT-CAGCQGSGMKI 174
+ D+ L +SL + G +K+LS V C C+G G S A+M+ C C+G
Sbjct: 154 QIKPDIRVELSLSLSEAAEGCTKRLSFDAYVFCDSCDGLGHPSDAAMSICPTCRG----- 208
Query: 175 SMRHLGANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQ 234
+G I C CKGTG I K+ C C+G +V+ K E+ + G+++
Sbjct: 209 ----VGRVTIPPFTASCQTCKGTGHII--KEYCMSCRGSGIVEGTKTAELVIPGGVESEA 262
Query: 235 KITFPGEADEAPDT-VTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTH 293
IT G + + T G++ L+ F R G D++V+ +S T+A+ G + +
Sbjct: 263 TITIVGAGNVSSRTSQPGNLYIKLKVANDSTFTRDGSDIYVDANISFTQAILGGKVVVPT 322
Query: 294 LDSR 297
L +
Sbjct: 323 LSGK 326
>At3g57340 dnaJ-like protein
Length = 367
Score = 93.2 bits (230), Expect = 2e-19
Identities = 54/119 (45%), Positives = 67/119 (55%), Gaps = 15/119 (12%)
Query: 14 YYEILGVSKNASPDDLKKAYKKAAIKNHPDKG---GDPEKFKELAQAYEVLSDPEKREVY 70
YYEILG+ N S DD++KAY+K ++K HPDK G E FK +++A++ LS+ E R+ Y
Sbjct: 114 YYEILGLESNCSVDDVRKAYRKLSLKVHPDKNQAPGSEEAFKSVSKAFQCLSNDEARKKY 173
Query: 71 DQYGED---------ALKEGMGGGGGGHDPFD---IFSSFFGGGGFPGGGSSRGRRQRR 117
D G D A G GG D FD IF SFFGGGGF GGG Q R
Sbjct: 174 DVSGSDEPIYQPRRSARSNGFNGGYYYEDEFDPNEIFRSFFGGGGFGGGGMPPATAQFR 232
>At5g03160 unknown protein
Length = 482
Score = 92.8 bits (229), Expect = 3e-19
Identities = 56/120 (46%), Positives = 77/120 (63%), Gaps = 13/120 (10%)
Query: 4 RAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKG-GDPE----KFKELAQAY 58
+A K S +Y+ILG+S+ AS ++KKAYKK A++ HPDK G+ E KF+E+A AY
Sbjct: 361 KALKMSKRKDWYKILGISRTASISEIKKAYKKLALQWHPDKNVGNREEAENKFREIAAAY 420
Query: 59 EVLSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFS------SFFGGGGFPGGGSSRG 112
E+L D +KR +D+ GED L++ GGGGGG++PF +F GGFPGGG G
Sbjct: 421 EILGDDDKRARFDR-GED-LEDMGGGGGGGYNPFHGGGGGGQQYTFHFEGGFPGGGGGFG 478
>At2g20550 putative heat shock protein
Length = 284
Score = 83.2 bits (204), Expect = 2e-16
Identities = 68/238 (28%), Positives = 109/238 (45%), Gaps = 23/238 (9%)
Query: 84 GGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGEDVVHPLKVSLEDLYLGTSKKLSLS 143
G G + DI+S FFG P G R +D+ + + ++
Sbjct: 22 GPSGPRNADDIYSEFFGVSS-PSG-------PRNKDDI-------FSEFFEVSNPSGPRD 66
Query: 144 RNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHLGANMIQQMQH--PCNE---CKGTG 198
++ + ++ G S SG+ + G G +M H GA ++ PC+ KGT
Sbjct: 67 KDDISAEYFGVPSPSGSGSSGGREGGGGGGGTMHHGGARKAAPVEKKLPCSLEDLYKGTT 126
Query: 199 ETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPGEADEAPDTVTGDIVFVLQ 258
+ + G K Q +++L V V+ G + G KITF + +E P + D+VF++
Sbjct: 127 KKMKISREIAGVFG-KTTQVQEILTVDVKPGWKTGTKITFSEKGNEQPGVIPADLVFIID 185
Query: 259 QKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQLLIKSNPGEVVKPGWMSV 316
+K HP F R+G DL V +S+ EA G+ LT LD R+L I N V+ P ++ V
Sbjct: 186 EKPHPVFTREGNDLVVTQKISVLEAFTGYTVNLTTLDGRRLTIPVN--TVIHPEYVEV 241
Score = 40.0 bits (92), Expect = 0.002
Identities = 37/141 (26%), Positives = 58/141 (40%), Gaps = 31/141 (21%)
Query: 6 PKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEVLSDPE 65
P+ +D+ Y E GVS + P + + + ++P D + +++ Y + P
Sbjct: 26 PRNADDI-YSEFFGVSSPSGPRNKDDIFSEFFEVSNPSGPRDKD---DISAEYFGVPSPS 81
Query: 66 KREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGEDVVHPL 125
G +EG GGGGG + GG R+ V L
Sbjct: 82 GS------GSSGGREGGGGGGG--------TMHHGGA-------------RKAAPVEKKL 114
Query: 126 KVSLEDLYLGTSKKLSLSRNV 146
SLEDLY GT+KK+ +SR +
Sbjct: 115 PCSLEDLYKGTTKKMKISREI 135
>At3g08970 putative DnaJ protein
Length = 572
Score = 82.4 bits (202), Expect = 4e-16
Identities = 48/114 (42%), Positives = 67/114 (58%), Gaps = 13/114 (11%)
Query: 8 KSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDP---EKFKELAQAYEVLSDP 64
K+ + Y++LGVSK+A +++KA+ K ++K HPDK D EKF E+ AYE+LSD
Sbjct: 22 KAKSVDPYKVLGVSKDAKQREIQKAFHKQSLKYHPDKNKDKGAQEKFAEINNAYEILSDE 81
Query: 65 EKREVYDQYGED----ALKEGMGGGGGGHDPFDIFSSFFGGGGF--PGGGSSRG 112
EKR+ YD YG++ G GG GG+ +SS GG F PGG + G
Sbjct: 82 EKRKNYDLYGDEKGQPGFDSGFPGGNGGYS----YSSSGGGFNFGGPGGWQNMG 131
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,199,059
Number of Sequences: 26719
Number of extensions: 442467
Number of successful extensions: 2852
Number of sequences better than 10.0: 188
Number of HSP's better than 10.0 without gapping: 115
Number of HSP's successfully gapped in prelim test: 78
Number of HSP's that attempted gapping in prelim test: 1835
Number of HSP's gapped (non-prelim): 666
length of query: 375
length of database: 11,318,596
effective HSP length: 101
effective length of query: 274
effective length of database: 8,619,977
effective search space: 2361873698
effective search space used: 2361873698
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC133862.8


