

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133709.13 - phase: 0
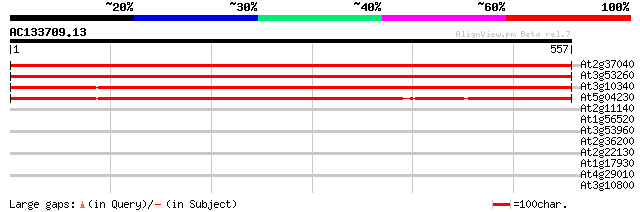
(557 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g37040 phenylalanine ammonia lyase (PAL1) 926 0.0
At3g53260 phenylalanine ammonia-lyase 918 0.0
At3g10340 putative phenylalanine ammonia-lyase 895 0.0
At5g04230 phenylalanine ammonia-lyase PAL3 786 0.0
At2g11140 putative retroelement pol polyprotein 31 2.1
At1g56520 disease resistance like protein 31 2.1
At3g53960 transporter like protein 29 6.0
At2g36200 putative kinesin-related cytokinesis protein 29 6.0
At2g22130 unknown protein 29 6.0
At1g17930 unknown protein 29 6.0
At4g29010 AIM1 protein 29 7.9
At3g10800 bZIP transcription factor AtbZip28 29 7.9
>At2g37040 phenylalanine ammonia lyase (PAL1)
Length = 725
Score = 926 bits (2393), Expect = 0.0
Identities = 449/557 (80%), Positives = 509/557 (90%)
Query: 1 MLVRINTLLQGYSGIRFEIMEAIAKFLNHNITPCLPLRGTITASGDLIPLSYVAGLLIGR 60
MLVRINTLLQG+SGIRFEI+EAI FLN+NITP LPLRGTITASGDL+PLSY+AGLL GR
Sbjct: 169 MLVRINTLLQGFSGIRFEILEAITSFLNNNITPSLPLRGTITASGDLVPLSYIAGLLTGR 228
Query: 61 PNSKSIGPNGQVLNAQEAFQLAGIETGFFELQPKEGLALVNGTAVGSGLASLVLFDTNLL 120
PNSK+ GPNG+ L A+EAF+LAGI +GFF+LQPKEGLALVNGTAVGSG+AS+VLF+TN+L
Sbjct: 229 PNSKATGPNGEALTAEEAFKLAGISSGFFDLQPKEGLALVNGTAVGSGMASMVLFETNVL 288
Query: 121 VVLSEILSAIFAEVMLGKPQFTDHLIHKVKLHPGQIEAAAIMEHILDGSYYGKAAQKVHE 180
VL+EILSA+FAEVM GKP+FTDHL H++K HPGQIEAAAIMEHILDGS Y K AQK+HE
Sbjct: 289 SVLAEILSAVFAEVMSGKPEFTDHLTHRLKHHPGQIEAAAIMEHILDGSSYMKLAQKLHE 348
Query: 181 INPLQKPKQDRYAIRTSPQWLGPQIEVIRYATKMIEREINSVNDNPLIDVSRDKALHGGN 240
++PLQKPKQDRYA+RTSPQWLGPQIEVIRYATK IEREINSVNDNPLIDVSR+KA+HGGN
Sbjct: 349 MDPLQKPKQDRYALRTSPQWLGPQIEVIRYATKSIEREINSVNDNPLIDVSRNKAIHGGN 408
Query: 241 FQGTPIGVSMDNTRLAIAAIGKLMFAQFTELVNDIYNNGLPSSLTASRNPSLDYGFKGAE 300
FQGTPIGVSMDNTRLAIAAIGKLMFAQF+ELVND YNNGLPS+LTASRNPSLDYGFKGAE
Sbjct: 409 FQGTPIGVSMDNTRLAIAAIGKLMFAQFSELVNDFYNNGLPSNLTASRNPSLDYGFKGAE 468
Query: 301 VAMASYCSELQYLASPVTTHVQSAEQHNQDVNSLGLISARKTAEAVEIWKLMSSTFLVAL 360
+AMASYCSELQYLA+PVT+HVQSAEQHNQDVNSLGLIS+RKT+EAV+I KLMS+TFLVA+
Sbjct: 469 IAMASYCSELQYLANPVTSHVQSAEQHNQDVNSLGLISSRKTSEAVDILKLMSTTFLVAI 528
Query: 361 CQAIDLRHIEENFKSVVKNTVSQVAKRILTVGVNGELHPSRFCEKDLLNVVEGEYVFTYI 420
CQA+DLRH+EEN + VKNTVSQVAK++LT GVNGELHPSRFCEKDLL VV+ E V+TY
Sbjct: 529 CQAVDLRHLEENLRQTVKNTVSQVAKKVLTTGVNGELHPSRFCEKDLLKVVDREQVYTYA 588
Query: 421 DDPCSPIYPLMQKLRYVLVDHALQNGDKEANSSTSIFQKIGAFEEELKALLPKEVENARV 480
DDPCS YPL+QKLR V+VDHAL NG+ E N+ TSIF KIGAFEEELKA+LPKEVE AR
Sbjct: 589 DDPCSATYPLIQKLRQVIVDHALINGESEKNAVTSIFHKIGAFEEELKAVLPKEVEAARA 648
Query: 481 EIENGNPAVPNMIKECRSYPLYKFMRETLGTSLLTGEKIRSPGEDCDKVFTAMCDGRFID 540
+NG A+PN IKECRSYPLY+F+RE LGT LLTGEK+ SPGE+ DKVFTA+C+G+ ID
Sbjct: 649 AYDNGTSAIPNRIKECRSYPLYRFVREELGTELLTGEKVTSPGEEFDKVFTAICEGKIID 708
Query: 541 PMLDCLKEWNGVPLPIC 557
PM++CL EWNG P+PIC
Sbjct: 709 PMMECLNEWNGAPIPIC 725
>At3g53260 phenylalanine ammonia-lyase
Length = 717
Score = 918 bits (2373), Expect = 0.0
Identities = 445/557 (79%), Positives = 503/557 (89%)
Query: 1 MLVRINTLLQGYSGIRFEIMEAIAKFLNHNITPCLPLRGTITASGDLIPLSYVAGLLIGR 60
MLVR+NTLLQGYSGIRFEI+EAI LNHNI+P LPLRGTITASGDL+PLSY+AGLL GR
Sbjct: 161 MLVRVNTLLQGYSGIRFEILEAITSLLNHNISPSLPLRGTITASGDLVPLSYIAGLLTGR 220
Query: 61 PNSKSIGPNGQVLNAQEAFQLAGIETGFFELQPKEGLALVNGTAVGSGLASLVLFDTNLL 120
PNSK+ GP+G+ L A+EAF+ AGI TGFF+LQPKEGLALVNGTAVGSG+AS+VLF+ N+
Sbjct: 221 PNSKATGPDGESLTAKEAFEKAGISTGFFDLQPKEGLALVNGTAVGSGMASMVLFEANVQ 280
Query: 121 VVLSEILSAIFAEVMLGKPQFTDHLIHKVKLHPGQIEAAAIMEHILDGSYYGKAAQKVHE 180
VL+E+LSAIFAEVM GKP+FTDHL H++K HPGQIEAAAIMEHILDGS Y K AQKVHE
Sbjct: 281 AVLAEVLSAIFAEVMSGKPEFTDHLTHRLKHHPGQIEAAAIMEHILDGSSYMKLAQKVHE 340
Query: 181 INPLQKPKQDRYAIRTSPQWLGPQIEVIRYATKMIEREINSVNDNPLIDVSRDKALHGGN 240
++PLQKPKQDRYA+RTSPQWLGPQIEVIR ATK IEREINSVNDNPLIDVSR+KA+HGGN
Sbjct: 341 MDPLQKPKQDRYALRTSPQWLGPQIEVIRQATKSIEREINSVNDNPLIDVSRNKAIHGGN 400
Query: 241 FQGTPIGVSMDNTRLAIAAIGKLMFAQFTELVNDIYNNGLPSSLTASRNPSLDYGFKGAE 300
FQGTPIGVSMDNTRLAIAAIGKLMFAQF+ELVND YNNGLPS+LTAS NPSLDYGFKGAE
Sbjct: 401 FQGTPIGVSMDNTRLAIAAIGKLMFAQFSELVNDFYNNGLPSNLTASSNPSLDYGFKGAE 460
Query: 301 VAMASYCSELQYLASPVTTHVQSAEQHNQDVNSLGLISARKTAEAVEIWKLMSSTFLVAL 360
+AMASYCSELQYLA+PVT+HVQSAEQHNQDVNSLGLIS+RKT+EAV+I KLMS+TFLV +
Sbjct: 461 IAMASYCSELQYLANPVTSHVQSAEQHNQDVNSLGLISSRKTSEAVDILKLMSTTFLVGI 520
Query: 361 CQAIDLRHIEENFKSVVKNTVSQVAKRILTVGVNGELHPSRFCEKDLLNVVEGEYVFTYI 420
CQA+DLRH+EEN + VKNTVSQVAK++LT G+NGELHPSRFCEKDLL VV+ E VFTY+
Sbjct: 521 CQAVDLRHLEENLRQTVKNTVSQVAKKVLTTGINGELHPSRFCEKDLLKVVDREQVFTYV 580
Query: 421 DDPCSPIYPLMQKLRYVLVDHALQNGDKEANSSTSIFQKIGAFEEELKALLPKEVENARV 480
DDPCS YPLMQ+LR V+VDHAL NG+ E N+ TSIFQKIGAFEEELKA+LPKEVE AR
Sbjct: 581 DDPCSATYPLMQRLRQVIVDHALSNGETEKNAVTSIFQKIGAFEEELKAVLPKEVEAARA 640
Query: 481 EIENGNPAVPNMIKECRSYPLYKFMRETLGTSLLTGEKIRSPGEDCDKVFTAMCDGRFID 540
NG +PN IKECRSYPLY+F+RE LGT LLTGEK+ SPGE+ DKVFTAMC+G+ ID
Sbjct: 641 AYGNGTAPIPNRIKECRSYPLYRFVREELGTKLLTGEKVVSPGEEFDKVFTAMCEGKLID 700
Query: 541 PMLDCLKEWNGVPLPIC 557
P++DCLKEWNG P+PIC
Sbjct: 701 PLMDCLKEWNGAPIPIC 717
>At3g10340 putative phenylalanine ammonia-lyase
Length = 707
Score = 895 bits (2314), Expect = 0.0
Identities = 437/557 (78%), Positives = 498/557 (88%), Gaps = 1/557 (0%)
Query: 1 MLVRINTLLQGYSGIRFEIMEAIAKFLNHNITPCLPLRGTITASGDLIPLSYVAGLLIGR 60
MLVR+NTLLQGYSGIRFEI+EAI K LNH ITPCLPLRGTITASGDL+PLSY+AGLL GR
Sbjct: 152 MLVRVNTLLQGYSGIRFEILEAITKLLNHEITPCLPLRGTITASGDLVPLSYIAGLLTGR 211
Query: 61 PNSKSIGPNGQVLNAQEAFQLAGIETGFFELQPKEGLALVNGTAVGSGLASLVLFDTNLL 120
PNSK++GP+G+ L A EAF+LAG+ + FFELQPKEGLALVNGTAVGSGLAS VLFD N+L
Sbjct: 212 PNSKAVGPSGETLTASEAFKLAGVSS-FFELQPKEGLALVNGTAVGSGLASTVLFDANIL 270
Query: 121 VVLSEILSAIFAEVMLGKPQFTDHLIHKVKLHPGQIEAAAIMEHILDGSYYGKAAQKVHE 180
VLSE++SA+FAEVM GKP+FTDHL HK+K HPGQIEAAAIMEHILDGS Y K AQ +HE
Sbjct: 271 AVLSEVMSAMFAEVMQGKPEFTDHLTHKLKHHPGQIEAAAIMEHILDGSSYVKEAQLLHE 330
Query: 181 INPLQKPKQDRYAIRTSPQWLGPQIEVIRYATKMIEREINSVNDNPLIDVSRDKALHGGN 240
++PLQKPKQDRYA+RTSPQWLGPQIEVIR ATKMIEREINSVNDNPLIDVSR+KALHGGN
Sbjct: 331 MDPLQKPKQDRYALRTSPQWLGPQIEVIRAATKMIEREINSVNDNPLIDVSRNKALHGGN 390
Query: 241 FQGTPIGVSMDNTRLAIAAIGKLMFAQFTELVNDIYNNGLPSSLTASRNPSLDYGFKGAE 300
FQGTPIGV+MDN+RLAIA+IGKLMFAQF+ELVND YNNGLPS+L+ RNPSLDYGFKGAE
Sbjct: 391 FQGTPIGVAMDNSRLAIASIGKLMFAQFSELVNDFYNNGLPSNLSGGRNPSLDYGFKGAE 450
Query: 301 VAMASYCSELQYLASPVTTHVQSAEQHNQDVNSLGLISARKTAEAVEIWKLMSSTFLVAL 360
+AMASYCSELQ+LA+PVT HVQSAEQHNQDVNSLGLIS+RKTAEAV+I KLMS+T+LVAL
Sbjct: 451 IAMASYCSELQFLANPVTNHVQSAEQHNQDVNSLGLISSRKTAEAVDILKLMSTTYLVAL 510
Query: 361 CQAIDLRHIEENFKSVVKNTVSQVAKRILTVGVNGELHPSRFCEKDLLNVVEGEYVFTYI 420
CQA+DLRH+EEN K VK+ VSQVAKR+LTVG NGELHPSRF E+D+L VV+ EYVF+Y
Sbjct: 511 CQAVDLRHLEENLKKAVKSAVSQVAKRVLTVGANGELHPSRFTERDVLQVVDREYVFSYA 570
Query: 421 DDPCSPIYPLMQKLRYVLVDHALQNGDKEANSSTSIFQKIGAFEEELKALLPKEVENARV 480
DDPCS YPLMQKLR++LVDHAL + ++EANS+TS+F KIGAFE ELK LLPKEVE RV
Sbjct: 571 DDPCSLTYPLMQKLRHILVDHALADPEREANSATSVFHKIGAFEAELKLLLPKEVERVRV 630
Query: 481 EIENGNPAVPNMIKECRSYPLYKFMRETLGTSLLTGEKIRSPGEDCDKVFTAMCDGRFID 540
E E G A+ N IKECRSYPLY+F+R+ L T LLTGE +RSPGE+ DKVF A+ DG+ ID
Sbjct: 631 EYEEGTSAIANRIKECRSYPLYRFVRDELNTELLTGENVRSPGEEFDKVFLAISDGKLID 690
Query: 541 PMLDCLKEWNGVPLPIC 557
P+L+CLKEWNG P+ IC
Sbjct: 691 PLLECLKEWNGAPVSIC 707
>At5g04230 phenylalanine ammonia-lyase PAL3
Length = 694
Score = 786 bits (2030), Expect = 0.0
Identities = 400/557 (71%), Positives = 458/557 (81%), Gaps = 11/557 (1%)
Query: 1 MLVRINTLLQGYSGIRFEIMEAIAKFLNHNITPCLPLRGTITASGDLIPLSYVAGLLIGR 60
ML+R+NTLLQGYSGIRFEI+EAI LN ITP LPLRGTITASGDL+PLSY+AG LIGR
Sbjct: 149 MLIRVNTLLQGYSGIRFEILEAITTLLNCKITPLLPLRGTITASGDLVPLSYIAGFLIGR 208
Query: 61 PNSKSIGPNGQVLNAQEAFQLAGIETGFFELQPKEGLALVNGTAVGSGLASLVLFDTNLL 120
PNS+S+GP+G++L A EAF+LAG+ + FFEL+PKEGLALVNGTAVGS LAS VL+D N+L
Sbjct: 209 PNSRSVGPSGEILTALEAFKLAGVSS-FFELRPKEGLALVNGTAVGSALASTVLYDANIL 267
Query: 121 VVLSEILSAIFAEVMLGKPQFTDHLIHKVKLHPGQIEAAAIMEHILDGSYYGKAAQKVHE 180
VV SE+ SA+FAEVM GKP+FTDHL HK+K HPGQIEAAAIMEHILDGS Y K A +H+
Sbjct: 268 VVFSEVASAMFAEVMQGKPEFTDHLTHKLKHHPGQIEAAAIMEHILDGSSYVKEALHLHK 327
Query: 181 INPLQKPKQDRYAIRTSPQWLGPQIEVIRYATKMIEREINSVNDNPLIDVSRDKALHGGN 240
I+PLQKPKQDRYA+RTSPQWLGPQIEVIR ATKMIEREINSVNDNPLIDVSR+KA+HGGN
Sbjct: 328 IDPLQKPKQDRYALRTSPQWLGPQIEVIRAATKMIEREINSVNDNPLIDVSRNKAIHGGN 387
Query: 241 FQGTPIGVSMDNTRLAIAAIGKLMFAQFTELVNDIYNNGLPSSLTASRNPSLDYGFKGAE 300
FQGTPIGV+MDNTRLA+A+IGKLMFAQFTELVND YNNGLPS+L+ RNPSLDYG KGAE
Sbjct: 388 FQGTPIGVAMDNTRLALASIGKLMFAQFTELVNDFYNNGLPSNLSGGRNPSLDYGLKGAE 447
Query: 301 VAMASYCSELQYLASPVTTHVQSAEQHNQDVNSLGLISARKTAEAVEIWKLMSSTFLVAL 360
VAMASYCSELQ+LA+PVT HV+SA QHNQDVNSLGLIS+R TAEAV I KLMS+T+LVAL
Sbjct: 448 VAMASYCSELQFLANPVTNHVESASQHNQDVNSLGLISSRTTAEAVVILKLMSTTYLVAL 507
Query: 361 CQAIDLRHIEENFKSVVKNTVSQVAKRILTVGVNGELHPSRFCEKDLLNVVEGEYVFTYI 420
CQA DLRH+EE K V VS AK +L + P R D+L VV EYVF+Y+
Sbjct: 508 CQAFDLRHLEEILKKAVNEVVSHTAKSVLAI------EPFR-KHDDILGVVNREYVFSYV 560
Query: 421 DDPCSPIYPLMQKLRYVLVDHALQNGDKEANSSTSIFQKIGAFEEELKALLPKEVENARV 480
DDP S PLMQKLR+VL D AL + E + ++F+KIGAFE ELK LLPKEVE R
Sbjct: 561 DDPSSLTNPLMQKLRHVLFDKALAEPEGETD---TVFRKIGAFEAELKFLLPKEVERVRT 617
Query: 481 EIENGNPAVPNMIKECRSYPLYKFMRETLGTSLLTGEKIRSPGEDCDKVFTAMCDGRFID 540
E ENG V N IK+CRSYPLY+F+R L T LLTGE +RSPGED DKVF A+ G+ ID
Sbjct: 618 EYENGTFNVANRIKKCRSYPLYRFVRNELETRLLTGEDVRSPGEDFDKVFRAISQGKLID 677
Query: 541 PMLDCLKEWNGVPLPIC 557
P+ +CLKEWNG P+ IC
Sbjct: 678 PLFECLKEWNGAPISIC 694
>At2g11140 putative retroelement pol polyprotein
Length = 411
Score = 30.8 bits (68), Expect = 2.1
Identities = 20/71 (28%), Positives = 35/71 (49%), Gaps = 1/71 (1%)
Query: 102 GTAVGSGLASLVLFDTNLLVVLSEILSAIFAEVMLGK-PQFTDHLIHKVKLHPGQIEAAA 160
G+AV S + +L D L+ + ++ A G P+ D++ + + LH +
Sbjct: 19 GSAVRSAREATILEDQKLMALKQQLTQRSDAYGGNGSNPKVPDNVDNPLILHSSDHPGLS 78
Query: 161 IMEHILDGSYY 171
I+ H+LDGS Y
Sbjct: 79 IVAHVLDGSNY 89
>At1g56520 disease resistance like protein
Length = 897
Score = 30.8 bits (68), Expect = 2.1
Identities = 21/58 (36%), Positives = 26/58 (44%), Gaps = 13/58 (22%)
Query: 272 VNDIYNNGLPSS--------LTASRNPSLDYGFKGAEVAMASYCSELQYLASPVTTHV 321
+ DIY+ G PS L+A R PS YGF +AS C L P+ HV
Sbjct: 335 IKDIYHVGFPSEGEALMIFCLSAFRQPSPPYGFLKLTYEVASICGNL-----PLGLHV 387
>At3g53960 transporter like protein
Length = 620
Score = 29.3 bits (64), Expect = 6.0
Identities = 23/76 (30%), Positives = 35/76 (45%), Gaps = 6/76 (7%)
Query: 66 IGPNGQVLNAQEAFQLAGIETGFFELQPKEGLALVNGTAVGSGLASLVLFDTNLLVVLSE 125
+ P VL +AF L G++ F++ P +L G A + F NLL+ +S+
Sbjct: 467 LAPQFLVLGVADAFTLVGLQEYFYDQVPDSMRSL--GIAFYLSVLGAASFVNNLLITVSD 524
Query: 126 ILSAIFAEVMLGKPQF 141
L AE + GK F
Sbjct: 525 HL----AEEISGKGWF 536
>At2g36200 putative kinesin-related cytokinesis protein
Length = 1056
Score = 29.3 bits (64), Expect = 6.0
Identities = 23/70 (32%), Positives = 35/70 (49%), Gaps = 4/70 (5%)
Query: 432 QKLRYVLVDHAL---QNGDKEANSSTSIFQKIGAFEEELKALLPKEVENARVEIENGNPA 488
+K VLV A N +K ++S+ QKIG E++L A K V+N +VE+
Sbjct: 493 KKSENVLVQQACILQSNLEKATKDNSSLHQKIGR-EDKLSADNRKVVDNYQVELSEQISN 551
Query: 489 VPNMIKECRS 498
+ N + C S
Sbjct: 552 LFNRVASCLS 561
>At2g22130 unknown protein
Length = 2048
Score = 29.3 bits (64), Expect = 6.0
Identities = 25/95 (26%), Positives = 44/95 (46%), Gaps = 9/95 (9%)
Query: 254 RLAIAAIGKLMFAQFTELVNDIYN-NGLPSSLTASRNPSLDYGF--------KGAEVAMA 304
R AA K + AQ E +I N NG+P + A+ PS ++ + A A+A
Sbjct: 244 RAEAAAALKSLSAQSKEAKREIANSNGIPVLINATIAPSKEFMQGEYAQALQENAMCALA 303
Query: 305 SYCSELQYLASPVTTHVQSAEQHNQDVNSLGLISA 339
+ L Y+ S + ++S Q ++LG +++
Sbjct: 304 NISGGLSYVISSLGQSLESCSSPAQTADTLGALAS 338
>At1g17930 unknown protein
Length = 478
Score = 29.3 bits (64), Expect = 6.0
Identities = 17/52 (32%), Positives = 28/52 (53%), Gaps = 5/52 (9%)
Query: 17 FEIMEAIAKFLNHNITPCLPLRGTITASGDLIPLSYVAGLLIGRPNSKSIGP 68
F+ +A+ N++ C P G + D++P S+ LL+GR +K IGP
Sbjct: 273 FKYRKALDDLDPSNVSWC-PFEGDL----DIVPQSFKDNLLLGRSRTKLIGP 319
>At4g29010 AIM1 protein
Length = 721
Score = 28.9 bits (63), Expect = 7.9
Identities = 21/77 (27%), Positives = 42/77 (54%), Gaps = 2/77 (2%)
Query: 92 QPKEGLALVNGTAVGSGLASLVLFDTNLLVVLSEILSAIFAEVMLGKPQFTDHLIHKVKL 151
+P + +A++ G +GSG+A+ +L +N+ VVL EI S + + L+ + KL
Sbjct: 307 RPIKKVAVIGGGLMGSGIATALLL-SNIRVVLKEINSEFLMKGIKSVEANMKSLVSRGKL 365
Query: 152 HPGQI-EAAAIMEHILD 167
+ +A ++ + +LD
Sbjct: 366 TQDKAGKALSLFKGVLD 382
>At3g10800 bZIP transcription factor AtbZip28
Length = 675
Score = 28.9 bits (63), Expect = 7.9
Identities = 26/91 (28%), Positives = 45/91 (48%), Gaps = 8/91 (8%)
Query: 397 LHPSRFCEKDLLNVVEGEYVFTYIDDPCSPIYPLMQKLRYVLVDHALQNGDKEANSSTSI 456
L+PS F E DL ++ + +F DP S P+ L ++L D +NGD A+ S
Sbjct: 17 LNPSMFSESDLFSIPPLDPLFLSDSDPISMDAPI-SDLDFLLDD---ENGD-FADFDFSF 71
Query: 457 FQKIGAFEEELK---ALLPKEVENARVEIEN 484
F+ +L ++P+E+ N R +++
Sbjct: 72 DNSDDFFDFDLSEPAVVIPEEIGNNRSNLDS 102
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.136 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,927,650
Number of Sequences: 26719
Number of extensions: 500797
Number of successful extensions: 1131
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 1123
Number of HSP's gapped (non-prelim): 12
length of query: 557
length of database: 11,318,596
effective HSP length: 104
effective length of query: 453
effective length of database: 8,539,820
effective search space: 3868538460
effective search space used: 3868538460
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC133709.13


