

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC131239.17 + phase: 0 /pseudo
(892 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
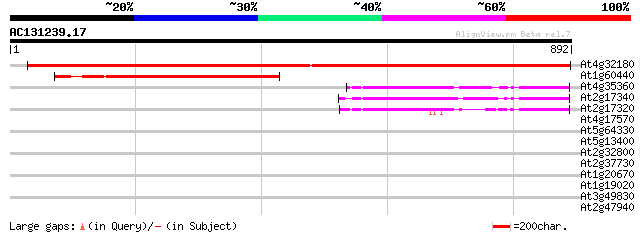
Score E
Sequences producing significant alignments: (bits) Value
At4g32180 unknown protein 1487 0.0
At1g60440 unknown protein 457 e-129
At4g35360 unknown protein 230 3e-60
At2g17340 unknown protein 228 1e-59
At2g17320 unknown protein 131 1e-30
At4g17570 unknown protein 32 1.6
At5g64330 non-phototropic hypocotyl 3 (NPH3) 31 2.7
At5g13400 peptide transporter - like protein 31 3.5
At2g32800 putative protein kinase 31 3.5
At2g37730 hypothetical protein 30 6.0
At1g20670 hypothetical protein 30 6.0
At1g19020 unknown protein 30 6.0
At3g49830 RuvB DNA helicase - like protein 30 7.9
At2g47940 DegP2 protease (DEGP2) 30 7.9
>At4g32180 unknown protein
Length = 870
Score = 1487 bits (3849), Expect = 0.0
Identities = 745/868 (85%), Positives = 804/868 (91%), Gaps = 6/868 (0%)
Query: 29 MAPT--GNPIHRSSSRPQLDVSKAEIQSNVEDKYPTILLPNQSDDLSHLALDIGGSLIKL 86
MAP+ G PIHRS SRPQLD+SKAEIQ N+E++ PTILLPNQSDD+SHLALDIGGSLIKL
Sbjct: 1 MAPSTSGTPIHRSGSRPQLDLSKAEIQGNLEERDPTILLPNQSDDISHLALDIGGSLIKL 60
Query: 87 VYFSRHEGQS-DDDKRKKTLKERLGLSNGNRRSFPILGGRLHFVKFETGKINECLDFIHS 145
+YFSRHE S DDDKRK+T+KERLG++NGN RS+P+LGGRLHFVKFET KINECLDFIHS
Sbjct: 61 LYFSRHEDYSNDDDKRKRTIKERLGITNGNLRSYPVLGGRLHFVKFETHKINECLDFIHS 120
Query: 146 KQLHRGLESRYSD-TEADRNAIIKATGGGAYKFTDLFKEKLGVSLDKEDEMNCLVAGANF 204
KQLHR +S T +IK TGGGA+KF DLFKE+LGVS++KEDEM+CLV+GANF
Sbjct: 121 KQLHRRDPYPWSSKTLPLGTGVIKVTGGGAFKFADLFKERLGVSIEKEDEMHCLVSGANF 180
Query: 205 LLKAIRHEAFTHMEGQKEFVQIDQNDLFPYLLVNVGSGVSMIKVDGDGKFERVSGTNVGG 264
LLKAIRHEAFTHMEG+KEFVQID NDL+PYLLVNVGSGVS+IKVDG+GKFERVSGTNVGG
Sbjct: 181 LLKAIRHEAFTHMEGEKEFVQIDPNDLYPYLLVNVGSGVSIIKVDGEGKFERVSGTNVGG 240
Query: 265 GTYWGLGRLLTKCKSFDELLELSQKGDNRAIDMLVGDIYGGLDYSKIGLSASTIASSFGK 324
GTYWGLGRLLTKCKSFDELLELSQKGDN AIDMLVGDIYGG+DYSKIGLSASTIASSFGK
Sbjct: 241 GTYWGLGRLLTKCKSFDELLELSQKGDNSAIDMLVGDIYGGMDYSKIGLSASTIASSFGK 300
Query: 325 TISEKKELQDYRPEDISLSLLRMISYNIGQIAYLNALRFKLKRIFFGGFFIRGHAYTMDT 384
ISE KEL DYRPEDISLSLLRMISYNIGQI+YLNALRF LKRIFFGGFFIRGHAYTMDT
Sbjct: 301 AISENKELDDYRPEDISLSLLRMISYNIGQISYLNALRFGLKRIFFGGFFIRGHAYTMDT 360
Query: 385 LSFAVQYWSNGEAQAMFLRHEGFLGALGAFMSYEKHGLDDLMVHQLVERFPMGAPYTGGK 444
+SFAV +WS GE QAMFLRHEGFLGALGAFMSYEKHGLDDLM HQLVERFPMGAPYTGG
Sbjct: 361 ISFAVHFWSKGEMQAMFLRHEGFLGALGAFMSYEKHGLDDLMSHQLVERFPMGAPYTGGN 420
Query: 445 IHGPPLGDLNEKISWMEKFLQKGTEITAPVPMTPVAGTTGLGGFEVPLSKGSALRSDASA 504
IHGPPLGDL+EKISWMEKF+++GTEITAPVPMTP + TTGLGGFEVP S+GSALRSDASA
Sbjct: 421 IHGPPLGDLDEKISWMEKFVRRGTEITAPVPMTP-SKTTGLGGFEVPSSRGSALRSDASA 479
Query: 505 LNVGVLHLVPTLEVFPLLADPKLYEPNTIDLADPSELEYWLTILSEHLPDLVDKAVASEG 564
LNVGVLHLVPTLEVFPLLADPK YEPNTIDL+D E EYWL +LSEHLPDLVD AVASEG
Sbjct: 480 LNVGVLHLVPTLEVFPLLADPKTYEPNTIDLSDQGEREYWLKVLSEHLPDLVDTAVASEG 539
Query: 565 GTDDAKRRGDAFARAFSAHLSRLMEEPSAYGKLGLANLLEMREECLREFQFVDAYRSIKQ 624
GT+DAKRRGDAFARAFSAHL+RLMEEP+AYGKLGLANLLE+REECLREFQFVDAYRSIKQ
Sbjct: 540 GTEDAKRRGDAFARAFSAHLARLMEEPAAYGKLGLANLLELREECLREFQFVDAYRSIKQ 599
Query: 625 RENEASLAVLPDLFVELDSMDEETRLLTLIEGVLAANIFDWGSRACVDLYHKGTIIEIYR 684
RENEASLAVLPDL ELDSM EE RLLTLIEGVLAANIFDWGSRACVDLYHKGTIIEIYR
Sbjct: 600 RENEASLAVLPDLLEELDSMSEEARLLTLIEGVLAANIFDWGSRACVDLYHKGTIIEIYR 659
Query: 685 MSRNKMRRPWRVDDFDLFKERMLGTGDKKKAPHRRALLFVDNSGADIVLGMLPLARELLR 744
MSRNKM+RPWRVDDFD FKERMLG+G K+ H+RALLFVDNSGAD++LGMLPLARE LR
Sbjct: 660 MSRNKMQRPWRVDDFDAFKERMLGSGGKQPHRHKRALLFVDNSGADVILGMLPLAREFLR 719
Query: 745 RGTEVVLVANSLPALNDVTAMELPDIVAEAAKHCDILRRAAEAGGLLVDAMINT-DSSKE 803
RGTEVVLVANSLPALNDVTAMELPDIVA AAKHCDILRRAAE GGLLVDAM+N D SK+
Sbjct: 720 RGTEVVLVANSLPALNDVTAMELPDIVAGAAKHCDILRRAAEMGGLLVDAMVNPGDGSKK 779
Query: 804 NSSSVPLMVVENGCGSPCIDLRQVSSELAAAAKDADLIILEGMGRSLHTNLYAQFKCDAL 863
+S+S PLMVVENGCGSPCIDLRQVSSELAAAAKDADL++LEGMGR+LHTN AQF+C+AL
Sbjct: 780 DSTSAPLMVVENGCGSPCIDLRQVSSELAAAAKDADLVVLEGMGRALHTNFNAQFQCEAL 839
Query: 864 KLAMVKNQRLAEKLIKGNIYDCICKYQP 891
KLAMVKNQRLAEKLIKGNIYDC+C+Y+P
Sbjct: 840 KLAMVKNQRLAEKLIKGNIYDCVCRYEP 867
>At1g60440 unknown protein
Length = 383
Score = 457 bits (1177), Expect = e-129
Identities = 234/357 (65%), Positives = 276/357 (76%), Gaps = 17/357 (4%)
Query: 72 LSHLALDIGGSLIKLVYFSRHEGQSDDDKRKKTLKERLGLSNGNRRSFPILGGRLHFVKF 131
+SHLALDIGG+LIKLVYFS + S++ +R ++ GRL F KF
Sbjct: 6 ISHLALDIGGTLIKLVYFSANGDYSEE----------------SRNGCSVVKGRLCFAKF 49
Query: 132 ETGKINECLDFIHSKQLHRGLESRYSDTEADRNAIIKATGGGAYKFTDLFKEKLGVSLDK 191
ET KI++CL+FI LH + + E +KATGGGA+KF DLFKEKLG+ DK
Sbjct: 50 ETRKIDDCLEFIRFNILHHS-GVQQPNGEGHDKLYVKATGGGAFKFADLFKEKLGILFDK 108
Query: 192 EDEMNCLVAGANFLLKAIRHEAFTHMEGQKEFVQIDQNDLFPYLLVNVGSGVSMIKVDGD 251
EDEM LV G NFLLK + EAFT+++GQK+FV+ID NDL+PYLLVN+GSGVSMIKVDGD
Sbjct: 109 EDEMCSLVGGVNFLLKTVPREAFTYLDGQKKFVEIDHNDLYPYLLVNIGSGVSMIKVDGD 168
Query: 252 GKFERVSGTNVGGGTYWGLGRLLTKCKSFDELLELSQKGDNRAIDMLVGDIYGGLDYSKI 311
GK+ER+SGT++GGGT+ GLG+LLTKCKSFDELLELS G+NR IDMLVGDIYGG DYSKI
Sbjct: 169 GKYERISGTSLGGGTFLGLGKLLTKCKSFDELLELSHHGNNRVIDMLVGDIYGGTDYSKI 228
Query: 312 GLSASTIASSFGKTISEKKELQDYRPEDISLSLLRMISYNIGQIAYLNALRFKLKRIFFG 371
GLS++ IASSFGK IS+ KEL+DY+PED++ SLLRMIS NIGQIAYLNALRF LKRIFFG
Sbjct: 229 GLSSTAIASSFGKAISDGKELEDYQPEDVARSLLRMISNNIGQIAYLNALRFGLKRIFFG 288
Query: 372 GFFIRGHAYTMDTLSFAVQYWSNGEAQAMFLRHEGFLGALGAFMSYEKHGLDDLMVH 428
GFFIRG YTMDT+S AV +WS GEA+AMFLRHEGFLGALGAF SY +DL H
Sbjct: 289 GFFIRGLEYTMDTISVAVHFWSRGEAKAMFLRHEGFLGALGAFTSYNDQSHNDLKPH 345
>At4g35360 unknown protein
Length = 367
Score = 230 bits (586), Expect = 3e-60
Identities = 138/356 (38%), Positives = 209/356 (57%), Gaps = 33/356 (9%)
Query: 536 ADPSELEYWLTILSEHLPDLVDKAVASEGGTDDAKRRGDAFARAFSAHLSRLMEEPSAYG 595
A P+E+ W+ + S +P ++A S+ DA R + FA+ ++ L L ++P ++G
Sbjct: 38 ATPTEIS-WIDLFSNSIPSFKERA-ESDTTVPDAPVRAEKFAKRYAEILEDLKKDPESHG 95
Query: 596 KLGLANLL-EMREECLREFQFVDAYRSIKQRENEASLAVLPDLFVELDSM-DEETRLLTL 653
LL +RE LRE F D ++ +K EN ++++ P++ D++ DE R+ L
Sbjct: 96 GPPDCILLCRIRELILRELGFRDIFKKVKDEENAKAISLFPEVVRLSDAINDEGKRIENL 155
Query: 654 IEGVLAANIFDWGSRACVDLYHKGTIIEIYRMSRNKMRRPWRVDDFDLFKERMLGTGDKK 713
+ G+ A NIFD GS +++ K + +N + RPW +DD D F+ R L
Sbjct: 156 VRGIFAGNIFDLGSAQLAEVFSKDGM-SFLASCQNLVSRPWVIDDLDNFQARWL------ 208
Query: 714 KAPHRRALLFVDNSGADIVLGMLPLARELLRRGTEVVLVANSLPALNDVTAMELPDIVAE 773
K P ++A++FVDNSGADI+LG+LP ARE+LR G +VVL AN LP++NDVT +EL
Sbjct: 209 KKPWKKAVIFVDNSGADIILGILPFAREMLRLGMQVVLAANELPSINDVTYIEL------ 262
Query: 774 AAKHCDILRRAAEAGGLLVDAMINTDSSKENSSSVPLMVVENGCGSPCIDLRQVSSELAA 833
+IL + + G L+ D+S L++ +G P IDL +VS E+A
Sbjct: 263 ----AEILSKLNDENGQLM----GVDTSN-------LLIANSGNDLPVIDLARVSQEVAY 307
Query: 834 AAKDADLIILEGMGRSLHTNLYAQFKCDALKLAMVKNQRLAEKLIKGNIYDCICKY 889
+ DADL+ILEGMGR + TNLYAQFKCD+LK+ MVK+ +A+ + G +YDC+ KY
Sbjct: 308 LSSDADLVILEGMGRGIETNLYAQFKCDSLKIGMVKHPEVAQ-FLGGRLYDCVIKY 362
>At2g17340 unknown protein
Length = 367
Score = 228 bits (581), Expect = 1e-59
Identities = 138/368 (37%), Positives = 209/368 (56%), Gaps = 41/368 (11%)
Query: 524 DPKLYEPNTIDLADPSELEYWLTILSEHLPDLVDKAVASEGGTDDAKRRGDAFARAFSAH 583
DPK PN I W+ + + +P +A S+ DA R + FA ++
Sbjct: 34 DPKKATPNEIS---------WINVFANSIPSFKKRA-ESDITVPDAPARAEKFAERYAGI 83
Query: 584 LSRLMEEPSAYGKLGLANLL-EMREECLREFQFVDAYRSIKQRENEASLAVLPDLFVELD 642
L L ++P ++G LL +RE+ LRE F D ++ +K EN ++++ P + D
Sbjct: 84 LEDLKKDPESHGGPPDGILLCRLREQVLRELGFRDIFKKVKDEENAKAISLFPQVVSLSD 143
Query: 643 SMDEE-TRLLTLIEGVLAANIFDWGSRACVDLYHKGTIIEIYRMSRNKMRRPWRVDDFDL 701
+++++ RL L+ G+ A NIFD GS +++ + + +N + RPW +DD +
Sbjct: 144 AIEDDGKRLENLVRGIFAGNIFDLGSAQLAEVFSRDGM-SFLASCQNLVPRPWVIDDLEN 202
Query: 702 FKERMLGTGDKKKAPHRRALLFVDNSGADIVLGMLPLARELLRRGTEVVLVANSLPALND 761
F+ + + KK A++FVDNSGADI+LG+LP ARELLRRG +VVL AN LP++ND
Sbjct: 203 FQAKWINKSWKK------AVIFVDNSGADIILGILPFARELLRRGAQVVLAANELPSIND 256
Query: 762 VTAMELPDIVAEAAKHCDILRRAAEAGGLLVDAMINTDSSKENSSSVPLMVVENGCGSPC 821
+T EL +I+++ E G LL D+SK L++ +G P
Sbjct: 257 ITCTELTEILSQLKD---------ENGQLL-----GVDTSK-------LLIANSGNDLPV 295
Query: 822 IDLRQVSSELAAAAKDADLIILEGMGRSLHTNLYAQFKCDALKLAMVKNQRLAEKLIKGN 881
IDL +VS ELA + DADL+I+EGMGR + TNLYAQFKCD+LK+ MVK+ +AE + G
Sbjct: 296 IDLSRVSQELAYLSSDADLVIVEGMGRGIETNLYAQFKCDSLKIGMVKHLEVAE-FLGGR 354
Query: 882 IYDCICKY 889
+YDC+ K+
Sbjct: 355 LYDCVFKF 362
>At2g17320 unknown protein
Length = 358
Score = 131 bits (330), Expect = 1e-30
Identities = 104/390 (26%), Positives = 181/390 (45%), Gaps = 82/390 (21%)
Query: 525 PKLYEPNTIDLADPSELEYWLTILSEHLPDLVDKAVASEGGTDDAKRRGDAFARAFSAHL 584
P + + + A P+E+ W+ + + +P +A S+ DA R F + ++ L
Sbjct: 21 PYRFPSDNLKKATPTEIS-WINVFANSIPSFKKRA-ESDTSVPDAPARAKIFEQRYAGFL 78
Query: 585 SRLMEEPSAYGKLGLANLL-EMREECLREFQFVDAYRSIKQRENEASLAVLPDLFVELDS 643
++P + G +L +RE LRE F D ++ +K EN ++++ P++ D+
Sbjct: 79 EGWKKDPESNGGPPDGVILGRVREHLLRELGFRDIFKKVKDEENAKAISLFPEVVSLSDA 138
Query: 644 MDEE-TRLLTLIEGVLAANIFDWGS-----------RACVDL----YHKGTIIEIYRM-- 685
++++ RL L+ G+ A NIFD GS +AC + + + E++
Sbjct: 139 IEDDGERLENLVRGIFAGNIFDLGSAQLSLFGAPTGKACPNYALFSFRILKLAEVFSRDG 198
Query: 686 ------SRNKMRRPWRVDDFDLFKERMLGTGDKKKAPHRRALLFVDNSGADIVLGMLPLA 739
S+N + RPW +DD + F+ + + K P ++
Sbjct: 199 ISFLATSQNLVPRPWVIDDLNKFQAKWV------KKPWKK-------------------- 232
Query: 740 RELLRRGTEVVLVANSLPALNDVTAMELPDIVAEAAKHCDILRRAAEAGGLLVDAMINTD 799
+ L++ VA + ++ D++ + + G L I D
Sbjct: 233 ----------------VTCLHNFGYFAFRKRVAPS-RNSDLIYQLKDGNGQL----IGVD 271
Query: 800 SSKENSSSVPLMVVENGCGSPCIDLRQVSSELAAAAKDADLIILEGMGRSLHTNLYAQFK 859
+SK L++ +G IDL +VS ELA + DADL+ILEGMGR + TNLYAQFK
Sbjct: 272 TSK-------LLIANSGNDLAVIDLSRVSHELAYLSSDADLVILEGMGRGIETNLYAQFK 324
Query: 860 CDALKLAMVKNQRLAEKLIKGNIYDCICKY 889
CD+LK+ MVK+ +A+ + G +YDC+ K+
Sbjct: 325 CDSLKIGMVKHVEVAQ-FLGGRLYDCVFKF 353
>At4g17570 unknown protein
Length = 510
Score = 32.0 bits (71), Expect = 1.6
Identities = 26/109 (23%), Positives = 47/109 (42%), Gaps = 15/109 (13%)
Query: 613 FQFVDAYRSIKQRENEASLAVLPDLFVELDSMDEETRLL----------TLIEGVLAANI 662
F F + + E + + +LP ++ D + R++ +L + ++A +
Sbjct: 297 FNFDEFIEQFTEEEQKKLMNLLPQ--IDSDDLPHSLRMMFESAQFKDNFSLFQQLIADGV 354
Query: 663 FDWGSRACVDLYHKGTIIEIYRMSRNKMRRPWRVDDFDLFKERMLGTGD 711
FD S + L T ++ NK R V+ ++L KER GTGD
Sbjct: 355 FDVSSSSGAKLEEIRTFKKLALTDFNKSRL---VESYNLLKEREKGTGD 400
>At5g64330 non-phototropic hypocotyl 3 (NPH3)
Length = 746
Score = 31.2 bits (69), Expect = 2.7
Identities = 18/56 (32%), Positives = 29/56 (51%), Gaps = 1/56 (1%)
Query: 459 WMEKFLQKGTEITAPVPMTPVAGTTGLGGFEVPLSKGSALRSDASALN-VGVLHLV 513
W+ +++G AP + + G GLGG E+ +S GS +S + G LH+V
Sbjct: 292 WLPGLIKEGGVAIAPAMSSAIGGGLGLGGDEMSISCGSNSSGGSSGPDWKGGLHMV 347
>At5g13400 peptide transporter - like protein
Length = 624
Score = 30.8 bits (68), Expect = 3.5
Identities = 15/42 (35%), Positives = 22/42 (51%), Gaps = 4/42 (9%)
Query: 433 RFPMGAPYTGG----KIHGPPLGDLNEKISWMEKFLQKGTEI 470
R +G YTGG IHG P+ +L++ W+ F G E+
Sbjct: 38 RLALGRGYTGGTTPVNIHGKPIANLSKTGGWIAAFFIFGNEM 79
>At2g32800 putative protein kinase
Length = 848
Score = 30.8 bits (68), Expect = 3.5
Identities = 23/92 (25%), Positives = 46/92 (50%), Gaps = 8/92 (8%)
Query: 569 AKRRGDAFARAFSAHLSRLMEEPSAYGKLGLANLLEMREECLREFQFVDAYRSIKQRENE 628
A+++G+ F + F+A L A +L NL+++R CL E + + Y + R +
Sbjct: 146 AEKKGEQFEKTFAAELV-------AVAQLRHRNLVKLRGWCLHEDELLLVYDYMPNRSLD 198
Query: 629 ASLAVLPDLFVELDSMDEETRLLTLIEGVLAA 660
L P++ + +D + R +++G+ AA
Sbjct: 199 RVLFRRPEVNSDFKPLDWDRR-GKIVKGLAAA 229
>At2g37730 hypothetical protein
Length = 532
Score = 30.0 bits (66), Expect = 6.0
Identities = 17/50 (34%), Positives = 24/50 (48%), Gaps = 3/50 (6%)
Query: 37 HRSSSRPQLDVSKAEIQSNVEDKYPTILLPNQSD---DLSHLALDIGGSL 83
H + +L + +N TI PNQS D+SH+A IGGS+
Sbjct: 60 HHITGLRRLGPENVNLTTNSSASSSTIRRPNQSSEATDISHIAFGIGGSI 109
>At1g20670 hypothetical protein
Length = 652
Score = 30.0 bits (66), Expect = 6.0
Identities = 30/119 (25%), Positives = 46/119 (38%), Gaps = 37/119 (31%)
Query: 5 VEDEQQQQQVLGIDESEGHNNTNNMAPTGNP---IHRSSSRPQLDVSKAEIQSNVEDKYP 61
++ +QQQ + D+ + H+N NN + + NP HRS R
Sbjct: 34 LQQQQQQHKNNHQDDDDHHHNNNNRSGSKNPNSLNHRSKRRN------------------ 75
Query: 62 TILLPNQSDDLSHLALDIGGSLIKLVYFSRHEGQSDDDKRKKTLKERLGLSNGNRRSFP 120
PN +D S D G E DD++R+K K GL++ + R P
Sbjct: 76 ----PNSNDGDSPWIKDEG------------EDNDDDERREKKHKLLHGLNSHSHRHSP 118
>At1g19020 unknown protein
Length = 86
Score = 30.0 bits (66), Expect = 6.0
Identities = 12/27 (44%), Positives = 19/27 (69%)
Query: 165 AIIKATGGGAYKFTDLFKEKLGVSLDK 191
A+++++G GA T +KEK+G LDK
Sbjct: 26 AVVRSSGSGATSNTTKYKEKMGQGLDK 52
>At3g49830 RuvB DNA helicase - like protein
Length = 473
Score = 29.6 bits (65), Expect = 7.9
Identities = 13/42 (30%), Positives = 23/42 (53%)
Query: 108 RLGLSNGNRRSFPILGGRLHFVKFETGKINECLDFIHSKQLH 149
+LG S R F ++G + FV+ G++ + + +HS LH
Sbjct: 202 KLGRSFTRSRDFDVMGSKTKFVQCPEGELEKRKEVLHSVTLH 243
>At2g47940 DegP2 protease (DEGP2)
Length = 607
Score = 29.6 bits (65), Expect = 7.9
Identities = 16/40 (40%), Positives = 26/40 (65%), Gaps = 2/40 (5%)
Query: 599 LANLLEMREECLREFQFVDAYRSIKQRE--NEASLAVLPD 636
LA+L++M ++ F+F D Y ++ +RE N ASL +L D
Sbjct: 521 LAHLIDMCKDKYLVFEFEDNYVAVLEREASNSASLCILKD 560
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.137 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,119,229
Number of Sequences: 26719
Number of extensions: 909015
Number of successful extensions: 2272
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 2247
Number of HSP's gapped (non-prelim): 15
length of query: 892
length of database: 11,318,596
effective HSP length: 108
effective length of query: 784
effective length of database: 8,432,944
effective search space: 6611428096
effective search space used: 6611428096
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Medicago: description of AC131239.17


