

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC130802.4 + phase: 0
(634 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
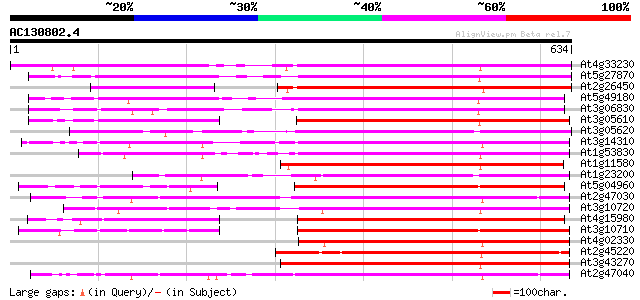
Score E
Sequences producing significant alignments: (bits) Value
At4g33230 pectinesterase - like protein 494 e-140
At5g27870 pectin methyl-esterase - like protein 399 e-111
At2g26450 putative pectinesterase 391 e-109
At5g49180 pectin methylesterase 389 e-108
At3g06830 pectin methylesterase like protein 374 e-103
At3g05610 putative pectinesterase 348 4e-96
At3g05620 putative pectinesterase 340 1e-93
At3g14310 pectin methylesterase like protein 333 2e-91
At1g53830 unknown protein 329 3e-90
At1g11580 unknown protein 326 2e-89
At1g23200 putative pectinesterase 315 6e-86
At5g04960 pectinesterase 314 8e-86
At2g47030 putative pectinesterase 313 1e-85
At3g10720 pectinesterase like protein 313 2e-85
At4g15980 pectinesterase like protein 311 9e-85
At3g10710 putative pectinesterase 311 9e-85
At4g02330 unknown protein (At4g02330) 309 3e-84
At2g45220 pectinesterase like protein 307 1e-83
At3g43270 pectinesterase like protein 306 2e-83
At2g47040 putative pectinesterase 303 2e-82
>At4g33230 pectinesterase - like protein
Length = 609
Score = 494 bits (1271), Expect = e-140
Identities = 271/661 (40%), Positives = 375/661 (55%), Gaps = 80/661 (12%)
Query: 1 MAFEDFDLVSERRKANAKQHLKKKILIGVTSVVLIACVIAAVTFVIV------KRSGPDH 54
MAF+DFD + ER A K+ +K+I++GV SV+++A I F V + G
Sbjct: 1 MAFQDFDKIQERVNAERKRKFRKRIILGVVSVLVVAAAIIGGAFAYVTYENKTQEQGKTT 60
Query: 55 NNNDK-KPVQNAPPEPE---------RVDKYSRLVTMLCSHSEYKEKCVTTLKEALKKDP 104
NN K P ++ P P+ + + +++ LC+ + YK C TLK KKD
Sbjct: 61 NNKSKDSPTKSESPSPKPPSSAAQTVKAGQVDKIIQTLCNSTLYKPTCQNTLKNETKKDT 120
Query: 105 KLKEPKGLLMVFMLVAKNEINNAFNKTANLKFASKEEKGAYEDCKQLFEDAKEEMGFSIT 164
+P+ LL ++ ++++ F + +LK +K++K A CK L ++AKEE+G S+
Sbjct: 121 PQTDPRSLLKSAIVAVNDDLDQVFKRVLSLKTENKDDKDAIAQCKLLVDEAKEELGTSMK 180
Query: 165 EVGQLDISKLASKEAELNNWLSAVISYQDTCSDGFPEGELKKKMEMIFAESRQLLSNSLA 224
+ +++ A +L++WLSAV+SYQ+TC DGF EG+LK ++ F S+ L SNSLA
Sbjct: 181 RINDSEVNNFAKIVPDLDSWLSAVMSYQETCVDGFEEGKLKTEIRKNFNSSQVLTSNSLA 240
Query: 225 VVSQVSQIVNAFQGGLSGFKLPWGKSDAHAPAPDADTDAVADDDEDLADAPDGAPDADQP 284
+ + + G LS P T + +
Sbjct: 241 M-------IKSLDGYLSS-------------VPKVKTRLLLE------------------ 262
Query: 285 IFEAPIGAPGAAPIGAPRVDAPPSWAA-------PAVDLPGSTEKPTPNVTVAKDGSGDF 337
A A D SW + AVD+ PN TVAKDGSG+F
Sbjct: 263 -----------ARSSAKETDHITSWLSNKERRMLKAVDVKALK----PNATVAKDGSGNF 307
Query: 338 KTISEALAAIPQTYKGRYVVYVKEGVYDETVTVTKKMVNLTMYGDGGLKSIITGNKNFVD 397
TI+ AL A+P Y+GRY +Y+K G+YDE+V + KK N+TM GDG K+I+TGNK+
Sbjct: 308 TTINAALKAMPAKYQGRYTIYIKHGIYDESVIIDKKKPNVTMVGDGSQKTIVTGNKSHAK 367
Query: 398 GVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQAVAARVQADCSIFVNCNFEGYQDTLY 457
+RTF TA+FV G+GF+ + MGFRNTAG QAVA RVQ+D S+F+NC FEGYQDTLY
Sbjct: 368 KIRTFLTATFVAQGEGFMAQSMGFRNTAGPEGHQAVAIRVQSDRSVFLNCRFEGYQDTLY 427
Query: 458 AQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVLRKPLDNQKNIITANGRIDSKSNTAF 517
A THRQ+YR CVI GT+DFIFG A+A+FQNC + +RK L QKN +TA GR+D T F
Sbjct: 428 AYTHRQYYRSCVIIGTVDFIFGDAAAIFQNCDIFIRKGLPGQKNTVTAQGRVDKFQTTGF 487
Query: 518 VLQKCVIKGEDDL---PSTTKNYIGRPWKEYSRTIIMESDIPALIQPEGWLPW-EGDFAL 573
V+ C + +DL + K+Y+GRPWK +SRT++MES I +I P GWL W E DFA+
Sbjct: 488 VIHNCTVAPNEDLKPVKAQFKSYLGRPWKPHSRTVVMESTIEDVIDPVGWLRWQETDFAI 547
Query: 574 KTLYYGEYDNVGAGAKTDARVKWIGRKDIKRGEALTYTVEPFLDGSWINGTGVPAHLGLY 633
TL Y EY N G T ARVKW G + + + EA+ +TV PFL G WI G P LGLY
Sbjct: 548 DTLSYAEYKNDGPSGATAARVKWPGFRVLNKEEAMKFTVGPFLQGEWIQAIGSPVKLGLY 607
Query: 634 N 634
+
Sbjct: 608 D 608
>At5g27870 pectin methyl-esterase - like protein
Length = 732
Score = 399 bits (1024), Expect = e-111
Identities = 233/622 (37%), Positives = 343/622 (54%), Gaps = 80/622 (12%)
Query: 22 KKKILIGVTSVVLIACVIAAVTFVIVKRSGPDHNNNDKKPVQNAPPEPERVDKYSRLVTM 81
K+ ++I ++SV+LI+ V+A V V +S N D E + + +
Sbjct: 14 KRYVIISISSVLLISMVVAVTIGVSVNKSD---NAGD-----------EEITTSVKAIKD 59
Query: 82 LCSHSEYKEKCVTTLKEALKKDPK-LKEPKGLLMVFMLVAKNEINNAFNKTANLKFASKE 140
+C+ ++YKE C ++ L+KD K +P L+ +I++ K+ + K+
Sbjct: 60 VCAPTDYKETC----EDTLRKDAKDTSDPLELVKTAFNATMKQISDVAKKSQTMIELQKD 115
Query: 141 EKG--AYEDCKQLFEDAKEEMGFSITEVGQLDISKLASKEAELNNWLSAVISYQDTCSDG 198
+ A + CK+L + A E+ S E+G+ + K+ +L WLSA IS++ TC DG
Sbjct: 116 PRAKMALDQCKELMDYAIGELSKSFEELGKFEFHKVDEALVKLRIWLSATISHEQTCLDG 175
Query: 199 F--PEGELKKKMEMIFAESRQLLSNSLAVVSQVSQIVNAFQGGLSGFKLPWGKSDAHAPA 256
F +G + ++ + QL N LA+V+++S + Q
Sbjct: 176 FQGTQGNAGETIKKALKTAVQLTHNGLAMVTEMSNYLGQMQ------------------I 217
Query: 257 PDADTDAVADDDEDLADAPDGAPDADQPIFEAPIGAPGAAPIGAPRVDAPPSWAAPAVDL 316
P+ ++ + + F + + A + AP + P
Sbjct: 218 PEMNSRRLLSQE-----------------FPSWMDARARRLLNAPMSEVKP--------- 251
Query: 317 PGSTEKPTPNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDETVTVTKKMVN 376
++ VA+DGSG +KTI+EAL +P+ +VV++KEG+Y E V V + M +
Sbjct: 252 ---------DIVVAQDGSGQYKTINEALNFVPKKKNTTFVVHIKEGIYKEYVQVNRSMTH 302
Query: 377 LTMYGDGGLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQAVAAR 436
L GDG K++I+G+K++ DG+ T++TA+ ++GD F+ +++ F NTAGAIK QAVA R
Sbjct: 303 LVFIGDGPDKTVISGSKSYKDGITTYKTATVAIVGDHFIAKNIAFENTAGAIKHQAVAIR 362
Query: 437 VQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVLRKPL 496
V AD SIF NC F+GYQDTLYA +HRQFYRDC ISGTIDF+FG A+AVFQNC L++RKPL
Sbjct: 363 VLADESIFYNCKFDGYQDTLYAHSHRQFYRDCTISGTIDFLFGDAAAVFQNCTLLVRKPL 422
Query: 497 DNQKNIITANGRIDSKSNTAFVLQKCVIKGEDD---LPSTTKNYIGRPWKEYSRTIIMES 553
NQ ITA+GR D + +T FVLQ C I GE D + +K Y+GRPWKEYSRTIIM +
Sbjct: 423 LNQACPITAHGRKDPRESTGFVLQGCTIVGEPDYLAVKEQSKTYLGRPWKEYSRTIIMNT 482
Query: 554 DIPALIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGRKDIKRGEALTYTVE 613
IP + PEGW PW G+F L TL+Y E N G GA RV W G K + E L +T
Sbjct: 483 FIPDFVPPEGWQPWLGEFGLNTLFYSEVQNTGPGAAITKRVTWPGIKKLSDEEILKFTPA 542
Query: 614 PFLDG-SWINGTGVPAHLGLYN 634
++ G +WI G GVP LGL++
Sbjct: 543 QYIQGDAWIPGKGVPYILGLFS 564
Score = 30.8 bits (68), Expect = 2.4
Identities = 22/85 (25%), Positives = 33/85 (37%), Gaps = 4/85 (4%)
Query: 253 HAPAPDADTDAVADDDEDLADAPDGAPDADQPIFEAPIGAPGAAPIGAPRVDAPPSWAAP 312
H +P +V L GAP A + +P +P A +G+P D P S +P
Sbjct: 603 HLGSPSDTPSSVVSPSTSLPAGQLGAPPATPSMVVSPSTSPPAGHLGSPS-DTPSSLVSP 661
Query: 313 AVDLPG---STEKPTPNVTVAKDGS 334
+ P + TP+ V S
Sbjct: 662 STSPPAGHLGSPSDTPSSVVTPSAS 686
>At2g26450 putative pectinesterase
Length = 496
Score = 391 bits (1005), Expect = e-109
Identities = 197/343 (57%), Positives = 239/343 (69%), Gaps = 15/343 (4%)
Query: 303 VDAPPSWAAP-------AVDLPGSTEKPTPNVTVAKDGSGDFKTISEALAAIPQTYKGRY 355
+D PSW + AVD+ PN TVAKDGSGDF TI++AL A+P+ Y+GRY
Sbjct: 157 LDDIPSWVSNDDRRMLRAVDVKALK----PNATVAKDGSGDFTTINDALRAMPEKYEGRY 212
Query: 356 VVYVKEGVYDETVTVTKKMVNLTMYGDGGLKSIITGNKNFVDGVRTFQTASFVVLGDGFV 415
++YVK+G+YDE VTV KK NLTM GDG K+I+TGNK+ +RTF TA+FV G+GF+
Sbjct: 213 IIYVKQGIYDEYVTVDKKKANLTMVGDGSQKTIVTGNKSHAKKIRTFLTATFVAQGEGFM 272
Query: 416 GRDMGFRNTAGAIKEQAVAARVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTID 475
+ MGFRNTAG QAVA RVQ+D SIF+NC FEGYQDTLYA THRQ+YR CVI GTID
Sbjct: 273 AQSMGFRNTAGPEGHQAVAIRVQSDRSIFLNCRFEGYQDTLYAYTHRQYYRSCVIVGTID 332
Query: 476 FIFGHASAVFQNCQLVLRKPLDNQKNIITANGRIDSKSNTAFVLQKCVIKGEDDLPSTT- 534
FIFG A+A+FQNC + +RK L QKN +TA GR+D T FV+ C I +DL
Sbjct: 333 FIFGDAAAIFQNCNIFIRKGLPGQKNTVTAQGRVDKFQTTGFVVHNCKIAANEDLKPVKE 392
Query: 535 --KNYIGRPWKEYSRTIIMESDIPALIQPEGWLPW-EGDFALKTLYYGEYDNVGAGAKTD 591
K+Y+GRPWK YSRTIIMES I +I P GWL W E DFA+ TLYY EY+N G+ T
Sbjct: 393 EYKSYLGRPWKNYSRTIIMESKIENVIDPVGWLRWQETDFAIDTLYYAEYNNKGSSGDTT 452
Query: 592 ARVKWIGRKDIKRGEALTYTVEPFLDGSWINGTGVPAHLGLYN 634
+RVKW G K I + EAL YTV PFL G WI+ +G P LGLY+
Sbjct: 453 SRVKWPGFKVINKEEALNYTVGPFLQGDWISASGSPVKLGLYD 495
Score = 92.8 bits (229), Expect = 5e-19
Identities = 49/140 (35%), Positives = 76/140 (54%)
Query: 92 CVTTLKEALKKDPKLKEPKGLLMVFMLVAKNEINNAFNKTANLKFASKEEKGAYEDCKQL 151
C TLK K L P L + +++ K +LK ++++K A E CK L
Sbjct: 4 CEKTLKNRTDKGFALDNPTTFLKSAIEAVNEDLDLVLEKVLSLKTENQDDKDAIEQCKLL 63
Query: 152 FEDAKEEMGFSITEVGQLDISKLASKEAELNNWLSAVISYQDTCSDGFPEGELKKKMEMI 211
EDAKEE S+ ++ +++ +L +WLSAV+SYQ+TC DGF EG LK +++
Sbjct: 64 VEDAKEETVASLNKINVTEVNSFEKVVPDLESWLSAVMSYQETCLDGFEEGNLKSEVKTS 123
Query: 212 FAESRQLLSNSLAVVSQVSQ 231
S+ L SNSLA++ ++
Sbjct: 124 VNSSQVLTSNSLALIKTFTE 143
>At5g49180 pectin methylesterase
Length = 571
Score = 389 bits (999), Expect = e-108
Identities = 237/618 (38%), Positives = 335/618 (53%), Gaps = 74/618 (11%)
Query: 22 KKKILIGVTSVVLIACVIAAVTFVIVKRSGPDHNNNDKKPVQNAPPEPERVDKYSRLVTM 81
KK I+ GV + +L+ V+A V + + ++ +K V P ++ + V
Sbjct: 10 KKCIIAGVITALLVLMVVA------VGITTSRNTSHSEKIV------PVQIKTATTAVEA 57
Query: 82 LCSHSEYKEKCVTTLKEALKKDPKLKEPKGLLMVFMLVAKNEINNAFNKTA---NLKFAS 138
+C+ ++YKE CV +L +K P +P L+ + V I ++ K + K A+
Sbjct: 58 VCAPTDYKETCVNSL---MKASPDSTQPLDLIKLGFNVTIRSIEDSIKKASVELTAKAAN 114
Query: 139 -KEEKGAYEDCKQLFEDAKEEMGFSITEVGQLDISKLASKEAELNNWLSAVISYQDTCSD 197
K+ KGA E C++L DA +++ + I ++ +L WLS I+YQ TC D
Sbjct: 115 DKDTKGALELCEKLMNDATDDLKKCLDNFDGFSIPQIEDFVEDLRVWLSGSIAYQQTCMD 174
Query: 198 GFPE--GELKKKMEMIFAESRQLLSNSLAVVSQVSQIVNAFQGGLSGFKLPWGKSDAHAP 255
F E +L + M+ IF SR+L SN LA+++ +S ++ F ++G GK
Sbjct: 175 TFEETNSKLSQDMQKIFKTSRELTSNGLAMITNISNLLGEFN--VTGVTGDLGKYA---- 228
Query: 256 APDADTDAVADDDEDLADAPDGAPDADQPIFEAPIGAPGAAPIGAPRVDAPPSWAAPAVD 315
L A DG P SW P
Sbjct: 229 -------------RKLLSAEDGIP----------------------------SWVGPNTR 247
Query: 316 LPGSTEKPTP-NVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDETVTVTKKM 374
+T+ NV VA DGSG +KTI+EAL A+P+ + +V+Y+K+GVY+E V VTKKM
Sbjct: 248 RLMATKGGVKANVVVAHDGSGQYKTINEALNAVPKANQKPFVIYIKQGVYNEKVDVTKKM 307
Query: 375 VNLTMYGDGGLKSIITGNKNFVDG-VRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQAV 433
++T GDG K+ ITG+ N+ G V+T+ TA+ + GD F +++GF NTAG QAV
Sbjct: 308 THVTFIGDGPTKTKITGSLNYYIGKVKTYLTATVAINGDNFTAKNIGFENTAGPEGHQAV 367
Query: 434 AARVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVLR 493
A RV AD ++F NC +GYQDTLY +HRQF+RDC +SGT+DFIFG V QNC +V+R
Sbjct: 368 ALRVSADLAVFYNCQIDGYQDTLYVHSHRQFFRDCTVSGTVDFIFGDGIVVLQNCNIVVR 427
Query: 494 KPLDNQKNIITANGRIDSKSNTAFVLQKCVIKGED---DLPSTTKNYIGRPWKEYSRTII 550
KP+ +Q +ITA GR D + +T VLQ C I GE + S K Y+GRPWKE+SRTII
Sbjct: 428 KPMKSQSCMITAQGRSDKRESTGLVLQNCHITGEPAYIPVKSINKAYLGRPWKEFSRTII 487
Query: 551 MESDIPALIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGRKDIKRGEALTY 610
M + I +I P GWLPW GDFAL TLYY EY+N G G+ RVKW G K + +AL +
Sbjct: 488 MGTTIDDVIDPAGWLPWNGDFALNTLYYAEYENNGPGSNQAQRVKWPGIKKLSPKQALRF 547
Query: 611 TVEPFLDGS-WINGTGVP 627
T FL G+ WI VP
Sbjct: 548 TPARFLRGNLWIPPNRVP 565
>At3g06830 pectin methylesterase like protein
Length = 568
Score = 374 bits (959), Expect = e-103
Identities = 233/624 (37%), Positives = 335/624 (53%), Gaps = 86/624 (13%)
Query: 22 KKKILIGVTSVVLIACVIAAVTFVIVKRSGPDHNNNDKKPVQNAPPEPERVDKYSRLVTM 81
KKK ++ + + ++ +V V K S D N+ + K ++ V
Sbjct: 8 KKKFIVAGSVSGFLVIMVVSVAVVTSKHSPKDDENH--------------IRKTTKAVQA 53
Query: 82 LCSHSEYKEKCVTTLKEALKKDPKLKEPKGLLMVFMLVAKNEINNAFNKTANLKFA---- 137
+C+ +++K+ CV +L A P +P L+ + V IN + K + A
Sbjct: 54 VCAPTDFKDTCVNSLMGA---SPDSDDPVDLIKLGFKVTIKSINESLEKASGDIKAKADK 110
Query: 138 SKEEKGAYEDCKQLFEDAKEEM------GFSITEVGQLDISKLASKEAELNNWLSAVISY 191
+ E KGA+E C++L DA +++ GFS+ ++ +L WLS I++
Sbjct: 111 NPEAKGAFELCEKLMIDAIDDLKKCMDHGFSVDQIEVF--------VEDLRVWLSGSIAF 162
Query: 192 QDTCSDGFPE--GELKKKMEMIFAESRQLLSNSLAVVSQVSQIV-NAFQGGLSGFKLPWG 248
Q TC D F E L + M IF SR+L SNSLA+V+++S ++ N+ GL+G
Sbjct: 163 QQTCMDSFGEIKSNLMQDMLKIFKTSRELSSNSLAMVTRISTLIPNSNLTGLTG------ 216
Query: 249 KSDAHAPAPDADTDAVADDDEDLADAPDGAPDADQPIFEAPIGAPGAAPIGAPRVDAPPS 308
A+A L D P P + A G P
Sbjct: 217 --------------ALAKYARKLLSTEDSIPTWVGPEARRLMAAQGGGP----------- 251
Query: 309 WAAPAVDLPGSTEKPTPNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDETV 368
G + N VA+DG+G FKTI++AL A+P+ K +++++KEG+Y E V
Sbjct: 252 ---------GPVKA---NAVVAQDGTGQFKTITDALNAVPKGNKVPFIIHIKEGIYKEKV 299
Query: 369 TVTKKMVNLTMYGDGGLKSIITGNKNFVDG-VRTFQTASFVVLGDGFVGRDMGFRNTAGA 427
TVTKKM ++T GDG K++ITG+ NF G V+TF TA+ + GD F +++G NTAG
Sbjct: 300 TVTKKMPHVTFIGDGPNKTLITGSLNFGIGKVKTFLTATITIEGDHFTAKNIGIENTAGP 359
Query: 428 IKEQAVAARVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQN 487
QAVA RV AD ++F +C +G+QDTLY +HRQFYRDC +SGT+DFIFG A + QN
Sbjct: 360 EGGQAVALRVSADYAVFHSCQIDGHQDTLYVHSHRQFYRDCTVSGTVDFIFGDAKCILQN 419
Query: 488 CQLVLRKPLDNQKNIITANGRIDSKSNTAFVLQKCVIKGED---DLPSTTKNYIGRPWKE 544
C++V+RKP Q ++TA GR + + +T VL C I G+ + S K Y+GRPWKE
Sbjct: 420 CKIVVRKPNKGQTCMVTAQGRSNVRESTGLVLHGCHITGDPAYIPMKSVNKAYLGRPWKE 479
Query: 545 YSRTIIMESDIPALIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGRKDIKR 604
+SRTIIM++ I +I P GWLPW GDFALKTLYY E+ N G G+ RVKW G K +
Sbjct: 480 FSRTIIMKTTIDDVIDPAGWLPWSGDFALKTLYYAEHMNTGPGSNQAQRVKWPGIKKLTP 539
Query: 605 GEALTYTVEPFLDG-SWINGTGVP 627
+AL YT + FL G +WI T VP
Sbjct: 540 QDALLYTGDRFLRGDTWIPQTQVP 563
>At3g05610 putative pectinesterase
Length = 669
Score = 348 bits (894), Expect = 4e-96
Identities = 169/312 (54%), Positives = 219/312 (70%), Gaps = 4/312 (1%)
Query: 325 PNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDETVTVTKKMVNLTMYGDGG 384
P++ VA+DGSG +KTI+EAL +P+ +VV++K G+Y E V V K M +L GDG
Sbjct: 254 PDIVVAQDGSGQYKTINEALQFVPKKRNTTFVVHIKAGLYKEYVQVNKTMSHLVFIGDGP 313
Query: 385 LKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQAVAARVQADCSIF 444
K+II+GNKN+ DG+ T++TA+ ++G+ F+ +++GF NTAGAIK QAVA RVQ+D SIF
Sbjct: 314 DKTIISGNKNYKDGITTYRTATVAIVGNYFIAKNIGFENTAGAIKHQAVAVRVQSDESIF 373
Query: 445 VNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVLRKPLDNQKNIIT 504
NC F+GYQDTLY +HRQF+RDC ISGTIDF+FG A+AVFQNC L++RKPL NQ IT
Sbjct: 374 FNCRFDGYQDTLYTHSHRQFFRDCTISGTIDFLFGDAAAVFQNCTLLVRKPLPNQACPIT 433
Query: 505 ANGRIDSKSNTAFVLQKCVIKGEDD---LPSTTKNYIGRPWKEYSRTIIMESDIPALIQP 561
A+GR D + +T FV Q C I GE D + T+K Y+GRPWKEYSRTIIM + IP +QP
Sbjct: 434 AHGRKDPRESTGFVFQGCTIAGEPDYLAVKETSKAYLGRPWKEYSRTIIMNTFIPDFVQP 493
Query: 562 EGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGRKDIKRGEALTYTVEPFLDG-SW 620
+GW PW GDF LKTL+Y E N G G+ RV W G K + + L +T ++ G W
Sbjct: 494 QGWQPWLGDFGLKTLFYSEVQNTGPGSALANRVTWAGIKTLSEEDILKFTPAQYIQGDDW 553
Query: 621 INGTGVPAHLGL 632
I G GVP GL
Sbjct: 554 IPGKGVPYTTGL 565
Score = 83.2 bits (204), Expect = 4e-16
Identities = 57/220 (25%), Positives = 108/220 (48%), Gaps = 19/220 (8%)
Query: 22 KKKILIGVTSVVLIACVIAAVTFVIVKRSGPDHNNNDKKPVQNAPPEPERVDKYSRLVTM 81
++ I+I ++SV+LI+ V+A V + + H+ + K + V+ + V
Sbjct: 13 RRYIVITISSVLLISMVVAVTVGVSLNK----HDGDSKGKAE--------VNASVKAVKD 60
Query: 82 LCSHSEYKEKCVTTLKEALKKDPKLKEPKGLLMVFMLVAKNEINNAFNKTANLKFASKEE 141
+C+ ++Y++ C TL +K +P L+ V +I +A K+ + K+
Sbjct: 61 VCAPTDYRKTCEDTL---IKNGKNTTDPMELVKTAFNVTMKQITDAAKKSQTIMELQKDS 117
Query: 142 KG--AYEDCKQLFEDAKEEMGFSITEVGQLDISKLASKEAELNNWLSAVISYQDTCSDGF 199
+ A + CK+L + A +E+ S E+G+ + L L WLSA IS+++TC +GF
Sbjct: 118 RTRMALDQCKELMDYALDELSNSFEELGKFEFHLLDEALINLRIWLSAAISHEETCLEGF 177
Query: 200 --PEGELKKKMEMIFAESRQLLSNSLAVVSQVSQIVNAFQ 237
+G + M+ + +L N LA++S++S V Q
Sbjct: 178 QGTQGNAGETMKKALKTAIELTHNGLAIISEMSNFVGQMQ 217
>At3g05620 putative pectinesterase
Length = 543
Score = 340 bits (872), Expect = 1e-93
Identities = 215/587 (36%), Positives = 302/587 (50%), Gaps = 89/587 (15%)
Query: 68 EPERVDKYSRLVTMLCSHSEYKEKCVTTLKEALKKDPKLKEPKGLLMVFMLVAKNEINNA 127
+P + D + LV C + E CV+ + +K+ P +L + A ++ A
Sbjct: 26 KPYKEDNFRSLVAKACQFIDAHELCVSNIWTHVKESGHGLNPHSVLRAAVKEAHDKAKLA 85
Query: 128 FNKTANLKFAS--KEEKGAYEDCKQLFEDAKEEMGFSITEVG--QLDISKL--------- 174
+ + S E+ A EDCK+L +GFS+TE+ L+++KL
Sbjct: 86 MERIPTVMMLSIRSREQVAIEDCKEL-------VGFSVTELAWSMLEMNKLHGGGGIDLD 138
Query: 175 ------ASKEAELNNWLSAVISYQDTCSDGFPEGELKKKMEMIFAESRQLLSNSLAVVSQ 228
A+ L WLSA +S QDTC +GF E K + +L+ SL V+Q
Sbjct: 139 DGSHDAAAAGGNLKTWLSAAMSNQDTCLEGFEGTERKYE---------ELIKGSLRQVTQ 189
Query: 229 VSQIVNAFQGGLSGFKLPWGKSDAHAPAPDADTDAVADDDEDLADAPDGAPDADQPIFEA 288
+ V L+ ++++ +P+ T+ DE L D
Sbjct: 190 LVSNVLDMYTQLNALPFKASRNESVIASPEWLTET----DESLMMRHD------------ 233
Query: 289 PIGAPGAAPIGAPRVDAPPSWAAPAVDLPGSTEKPTPNVTVAKDGSGDFKTISEALAAIP 348
P+V PN VA DG G ++TI+EA+ P
Sbjct: 234 -----------------------PSV--------MHPNTVVAIDGKGKYRTINEAINEAP 262
Query: 349 QTYKGRYVVYVKEGVYDETVTVTKKMVNLTMYGDGGLKSIITGNKNFVDGVRTFQTASFV 408
RYV+YVK+GVY E + + KK N+ + GDG ++IITG++NF+ G+ TF+TA+
Sbjct: 263 NHSTKRYVIYVKKGVYKENIDLKKKKTNIMLVGDGIGQTIITGDRNFMQGLTTFRTATVA 322
Query: 409 VLGDGFVGRDMGFRNTAGAIKEQAVAARVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDC 468
V G GF+ +D+ FRNTAG QAVA RV +D S F C+ EGYQDTLYA + RQFYRDC
Sbjct: 323 VSGRGFIAKDITFRNTAGPQNRQAVALRVDSDQSAFYRCSVEGYQDTLYAHSLRQFYRDC 382
Query: 469 VISGTIDFIFGHASAVFQNCQLVLRKPLDNQKNIITANGRIDSKSNTAFVLQKCVIKGED 528
I GTIDFIFG+ +AV QNC++ R PL QK ITA GR NT FV+Q +
Sbjct: 383 EIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPNQNTGFVIQNSYVL--- 439
Query: 529 DLPSTTKNYIGRPWKEYSRTIIMESDIPALIQPEGWLPWEGDFALKTLYYGEYDNVGAGA 588
+T Y+GRPWK YSRT+ M + + L+QP GWL W G+FAL TL+YGEY+N+G G
Sbjct: 440 ---ATQPTYLGRPWKLYSRTVYMNTYMSQLVQPRGWLEWFGNFALDTLWYGEYNNIGPGW 496
Query: 589 KTDARVKWIGRKDIKRGEALTYTVEPFLDG-SWINGTGVPAHLGLYN 634
++ RVKW G + + AL++TV F+DG W+ TGV GL N
Sbjct: 497 RSSGRVKWPGYHIMDKRTALSFTVGSFIDGRRWLPATGVTFTAGLAN 543
>At3g14310 pectin methylesterase like protein
Length = 592
Score = 333 bits (854), Expect = 2e-91
Identities = 217/635 (34%), Positives = 324/635 (50%), Gaps = 69/635 (10%)
Query: 14 KANAKQHLKKKILIGVTSVVLIACVIAAVTFVIVKRSGPDHNNNDKKPVQNAPPEPERVD 73
K N K++ KK +L+ +L +A ++ K N+K+ + P V
Sbjct: 11 KDNFKKN-KKLVLLSAAVALLFVAAVAGISAGASKA-------NEKRTLS---PSSHAVL 59
Query: 74 KYSRLVTMLCSHSEYKEKCVTTLKEALKKDPKLKEPKGLLMVFMLVAKNEINNAFNKTAN 133
+ S CS + Y E C++ + A +L K ++ + + + + +
Sbjct: 60 RSS------CSSTRYPELCISAVVTA--GGVELTSQKDVIEASVNLTITAVEHNYFTVKK 111
Query: 134 L----KFASKEEKGAYEDCKQLFEDAKEEMGFSITEVGQLDISKLASKEA-ELNNWLSAV 188
L K + EK A DC + ++ +E+ ++ ++ K + A +L +S+
Sbjct: 112 LIKKRKGLTPREKTALHDCLETIDETLDELHETVEDLHLYPTKKTLREHAGDLKTLISSA 171
Query: 189 ISYQDTCSDGFPEGELKKKMEMIFAESR----QLLSNSLAVVSQVSQIVNAFQGGLSGFK 244
I+ Q+TC DGF + K++ + + + SN+LA++ ++
Sbjct: 172 ITNQETCLDGFSHDDADKQVRKALLKGQIHVEHMCSNALAMIKNMT-------------- 217
Query: 245 LPWGKSDAHAPAPDADTDAVADDDEDLADAPDGAPDADQPIFEAPIGAPGAAPIGAPRVD 304
DTD + + + + + + GA + + +
Sbjct: 218 ---------------DTDIANFEQKAKITSNNRKLKEENQETTVAVDIAGAGELDS---E 259
Query: 305 APPSW--AAPAVDLPGSTEKPTPNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEG 362
P+W A L GS K + TVA DGSG FKT++ A+AA P+ RYV+++K G
Sbjct: 260 GWPTWLSAGDRRLLQGSGVKA--DATVAADGSGTFKTVAAAVAAAPENSNKRYVIHIKAG 317
Query: 363 VYDETVTVTKKMVNLTMYGDGGLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFR 422
VY E V V KK N+ GDG ++IITG++N VDG TF +A+ +G+ F+ RD+ F+
Sbjct: 318 VYRENVEVAKKKKNIMFMGDGRTRTIITGSRNVVDGSTTFHSATVAAVGERFLARDITFQ 377
Query: 423 NTAGAIKEQAVAARVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHAS 482
NTAG K QAVA RV +D S F NC+ YQDTLY ++RQF+ C+I+GT+DFIFG+A+
Sbjct: 378 NTAGPSKHQAVALRVGSDFSAFYNCDMLAYQDTLYVHSNRQFFVKCLIAGTVDFIFGNAA 437
Query: 483 AVFQNCQLVLRKPLDNQKNIITANGRIDSKSNTAFVLQKCVIKGEDDLPS---TTKNYIG 539
V Q+C + R+P QKN++TA GR D NT V+QKC I DL S + Y+G
Sbjct: 438 VVLQDCDIHARRPNSGQKNMVTAQGRTDPNQNTGIVIQKCRIGATSDLQSVKGSFPTYLG 497
Query: 540 RPWKEYSRTIIMESDIPALIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGR 599
RPWKEYS+T+IM+S I +I+PEGW W G FAL TL Y EY N GAGA T RVKW G
Sbjct: 498 RPWKEYSQTVIMQSAISDVIRPEGWSEWTGTFALNTLTYREYSNTGAGAGTANRVKWRGF 557
Query: 600 KDI-KRGEALTYTVEPFL-DGSWINGTGVPAHLGL 632
K I EA YT F+ G W++ TG P LGL
Sbjct: 558 KVITAAAEAQKYTAGQFIGGGGWLSSTGFPFSLGL 592
>At1g53830 unknown protein
Length = 587
Score = 329 bits (844), Expect = 3e-90
Identities = 212/569 (37%), Positives = 303/569 (52%), Gaps = 56/569 (9%)
Query: 78 LVTMLCSHSEYKEKCVTTLKEALKKDPKLKEPKGLLMVFM-LVAKNEINNAF---NKTAN 133
++ +CS + Y E C + + K+ L K ++ + L K +N F A
Sbjct: 61 ILKSVCSSTLYPELCFSAVAATGGKE--LTSQKEVIEASLNLTTKAVKHNYFAVKKLIAK 118
Query: 134 LKFASKEEKGAYEDCKQLFEDAKEEMGFSITEVGQLDISKLASKEAE-LNNWLSAVISYQ 192
K + E A DC + ++ +E+ ++ ++ Q K K A+ L +S+ I+ Q
Sbjct: 119 RKGLTPREVTALHDCLETIDETLDELHVAVEDLHQYPKQKSLRKHADDLKTLISSAITNQ 178
Query: 193 DTCSDGFPEGELKKKMEMIFAESR----QLLSNSLAVVSQVSQIVNAFQGGLSGFKLPWG 248
TC DGF + +K+ + + + SN+LA++ +++ ++ F+L
Sbjct: 179 GTCLDGFSYDDADRKVRKALLKGQVHVEHMCSNALAMIKNMTET------DIANFELR-- 230
Query: 249 KSDAHAPAPDADTDAVADDDEDLADAPDGAPDADQPIFEAPIGAPGAAPIGAPRVDAPPS 308
D + +++ L + G D+D G P +G R+
Sbjct: 231 ---------DKSSTFTNNNNRKLKEVT-GDLDSD--------GWPKWLSVGDRRL----- 267
Query: 309 WAAPAVDLPGSTEKPTPNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDETV 368
L GST K + TVA DGSGDF T++ A+AA P+ R+V+++K GVY E V
Sbjct: 268 -------LQGSTIKA--DATVADDGSGDFTTVAAAVAAAPEKSNKRFVIHIKAGVYRENV 318
Query: 369 TVTKKMVNLTMYGDGGLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFRNTAGAI 428
VTKK N+ GDG K+IITG++N VDG TF +A+ +G+ F+ RD+ F+NTAG
Sbjct: 319 EVTKKKTNIMFLGDGRGKTIITGSRNVVDGSTTFHSATVAAVGERFLARDITFQNTAGPS 378
Query: 429 KEQAVAARVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNC 488
K QAVA RV +D S F C+ YQDTLY ++RQF+ C I+GT+DFIFG+A+AV Q+C
Sbjct: 379 KHQAVALRVGSDFSAFYQCDMFAYQDTLYVHSNRQFFVKCHITGTVDFIFGNAAAVLQDC 438
Query: 489 QLVLRKPLDNQKNIITANGRIDSKSNTAFVLQKCVIKGEDDL---PSTTKNYIGRPWKEY 545
+ R+P QKN++TA GR D NT V+Q C I G DL T Y+GRPWKEY
Sbjct: 439 DINARRPNSGQKNMVTAQGRSDPNQNTGIVIQNCRIGGTSDLLAVKGTFPTYLGRPWKEY 498
Query: 546 SRTIIMESDIPALIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGRKDIKRG 605
SRT+IM+SDI +I+PEGW W G FAL TL Y EY N G GA T RVKW G K I
Sbjct: 499 SRTVIMQSDISDVIRPEGWHEWSGSFALDTLTYREYLNRGGGAGTANRVKWKGYKVITSD 558
Query: 606 -EALTYTVEPFL-DGSWINGTGVPAHLGL 632
EA +T F+ G W+ TG P L L
Sbjct: 559 TEAQPFTAGQFIGGGGWLASTGFPFSLSL 587
>At1g11580 unknown protein
Length = 557
Score = 326 bits (836), Expect = 2e-89
Identities = 168/329 (51%), Positives = 212/329 (64%), Gaps = 9/329 (2%)
Query: 307 PSWAAPA----VDLPGSTEKPTPNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEG 362
PSW ++ T K T NV VAKDG+G FKT++EA+AA P+ RYV+YVK+G
Sbjct: 223 PSWLTALDRKLLESSPKTLKVTANVVVAKDGTGKFKTVNEAVAAAPENSNTRYVIYVKKG 282
Query: 363 VYDETVTVTKKMVNLTMYGDGGLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFR 422
VY ET+ + KK NL + GDG +IITG+ N +DG TF++A+ GDGF+ +D+ F+
Sbjct: 283 VYKETIDIGKKKKNLMLVGDGKDATIITGSLNVIDGSTTFRSATVAANGDGFMAQDIWFQ 342
Query: 423 NTAGAIKEQAVAARVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHAS 482
NTAG K QAVA RV AD ++ C + YQDTLY T RQFYRD I+GT+DFIFG+++
Sbjct: 343 NTAGPAKHQAVALRVSADQTVINRCRIDAYQDTLYTHTLRQFYRDSYITGTVDFIFGNSA 402
Query: 483 AVFQNCQLVLRKPLDNQKNIITANGRIDSKSNTAFVLQKCVIKGEDDL---PSTTKNYIG 539
VFQNC +V R P QKN++TA GR D NTA +QKC I DL + K ++G
Sbjct: 403 VVFQNCDIVARNPGAGQKNMLTAQGREDQNQNTAISIQKCKITASSDLAPVKGSVKTFLG 462
Query: 540 RPWKEYSRTIIMESDIPALIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGR 599
RPWK YSRT+IM+S I I P GW PW+G+FAL TLYYGEY N G GA T RV W G
Sbjct: 463 RPWKLYSRTVIMQSFIDNHIDPAGWFPWDGEFALSTLYYGEYANTGPGADTSKRVNWKGF 522
Query: 600 KDIKRG-EALTYTVEPFLDGS-WINGTGV 626
K IK EA +TV + G W+ TGV
Sbjct: 523 KVIKDSKEAEQFTVAKLIQGGLWLKPTGV 551
Score = 37.0 bits (84), Expect = 0.034
Identities = 38/191 (19%), Positives = 76/191 (38%), Gaps = 26/191 (13%)
Query: 21 LKKKILIGVTSVVLIACVIAAVTFVIVKRSGPDHNNNDKKPVQNAPPEPERVDKYSRLVT 80
LK K L V S V I + +V F + + NNND + +
Sbjct: 15 LKHKNLCLVLSFVAI---LGSVAFFTAQLISVNTNNNDDSLLTTS--------------- 56
Query: 81 MLCSHSEYKEKCVTTLKEALKKDPKLKEPKGLLMVFMLVAKNEINNAFNKTANLKFASK- 139
+C + ++ C L E LL VF+ + + + + + S
Sbjct: 57 QICHGAHDQDSCQALLSEFTTLSLSKLNRLDLLHVFLKNSVWRLESTMTMVSEARIRSNG 116
Query: 140 -EEKGAYEDCKQLFEDAKEEMGFSITEV--GQLDISKLASKEAELNNWLSAVISYQDTCS 196
+K + DC+++ + +K+ M S+ E+ G ++ ++ ++ WLS+V++ TC
Sbjct: 117 VRDKAGFADCEEMMDVSKDRMMSSMEELRGGNYNLESYSN----VHTWLSSVLTNYMTCL 172
Query: 197 DGFPEGELKKK 207
+ + + K
Sbjct: 173 ESISDVSVNSK 183
>At1g23200 putative pectinesterase
Length = 554
Score = 315 bits (806), Expect = 6e-86
Identities = 191/504 (37%), Positives = 266/504 (51%), Gaps = 57/504 (11%)
Query: 139 KEEKGAYEDCKQLFEDAKEEMGFSITEVGQLDISKLASKEAELNNWLSAVISYQDTCSDG 198
K A DC +L+ED +++ S GQ S + LSA I+ QDTC +G
Sbjct: 98 KHATSALFDCLELYEDTIDQLNHSRRSYGQY------SSPHDRQTSLSAAIANQDTCRNG 151
Query: 199 FPEGELKKKMEMIFAES-----RQLLSNSLAVVSQVSQIVNAFQGGLSGFKLPWGKSDAH 253
F + +L F + +SNSLAV ++ + S + K +
Sbjct: 152 FRDFKLTSSYSKYFPVQFHRNLTKSISNSLAVTKAAAEAEAVAEKYPSTGFTKFSKQRSS 211
Query: 254 APAPDADTDAVADDDEDLADAPDGAPDADQPIFEAPIGAPGAAPIGAPRVDAPPSWAAPA 313
A + D++ P P +D+ + E
Sbjct: 212 AGGGSHRRLLLFSDEK----FPSWFPLSDRKLLE-------------------------- 241
Query: 314 VDLPGSTEKPTPNVTVAKDGSGDFKTISEAL---AAIPQTYKGRYVVYVKEGVYDETVTV 370
S ++ VAKDGSG + +I +A+ A +P+ + R V+YVK GVY E V +
Sbjct: 242 ----DSKTTAKADLVVAKDGSGHYTSIQQAVNAAAKLPRRNQ-RLVIYVKAGVYRENVVI 296
Query: 371 TKKMVNLTMYGDGGLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKE 430
K + N+ + GDG +I+TGN+N DG TF++A+F V G+GF+ + + F NTAG K
Sbjct: 297 KKSIKNVMVIGDGIDSTIVTGNRNVQDGTTTFRSATFAVSGNGFIAQGITFENTAGPEKH 356
Query: 431 QAVAARVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQL 490
QAVA R +D S+F C+F+GYQDTLY + RQF R+C I GT+DFIFG A+A+ QNC +
Sbjct: 357 QAVALRSSSDFSVFYACSFKGYQDTLYLHSSRQFLRNCNIYGTVDFIFGDATAILQNCNI 416
Query: 491 VLRKPLDNQKNIITANGRIDSKSNTAFVLQKCVIKGEDDLPSTTKNYIGRPWKEYSRTII 550
RKP+ QKN ITA R + T FV+Q + + ++ Y+GRPW+ +SRT+
Sbjct: 417 YARKPMSGQKNTITAQSRKEPDETTGFVIQSSTV------ATASETYLGRPWRSHSRTVF 470
Query: 551 MESDIPALIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGRKDIKR-GEALT 609
M+ ++ AL+ P GWLPW G FAL TLYYGEY N GAGA RVKW G IK EA
Sbjct: 471 MKCNLGALVSPAGWLPWSGSFALSTLYYGEYGNTGAGASVSGRVKWPGYHVIKTVTEAEK 530
Query: 610 YTVEPFLDGS-WINGTGVPAHLGL 632
+TVE FLDG+ WI TGVP + GL
Sbjct: 531 FTVENFLDGNYWITATGVPVNDGL 554
>At5g04960 pectinesterase
Length = 564
Score = 314 bits (805), Expect = 8e-86
Identities = 154/308 (50%), Positives = 205/308 (66%), Gaps = 3/308 (0%)
Query: 322 KPTPNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDETVTVTKKMVNLTMYG 381
K + VAKDGSG ++TI EALA + + + ++YVK+GVY E V V K N+ M G
Sbjct: 253 KKKATIVVAKDGSGKYRTIGEALAEVEEKNEKPTIIYVKKGVYLENVRVEKTKWNVVMVG 312
Query: 382 DGGLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQAVAARVQADC 441
DG K+I++ NF+DG TF+TA+F V G GF+ RDMGF NTAG K QAVA V AD
Sbjct: 313 DGQSKTIVSAGLNFIDGTPTFETATFAVFGKGFMARDMGFINTAGPAKHQAVALMVSADL 372
Query: 442 SIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVLRKPLDNQKN 501
S+F C + +QDT+YA RQFYRDCVI GT+DFIFG+A+ VFQ C+++ R+P+ Q+N
Sbjct: 373 SVFYKCTMDAFQDTMYAHAQRQFYRDCVILGTVDFIFGNAAVVFQKCEILPRRPMKGQQN 432
Query: 502 IITANGRIDSKSNTAFVLQKCVIKGEDDLPSTTKNYIGRPWKEYSRTIIMESDIPALIQP 561
ITA GR D NT + C IK D+L + + ++GRPWK++S T+IM+S + I P
Sbjct: 433 TITAQGRKDPNQNTGISIHNCTIKPLDNL-TDIQTFLGRPWKDFSTTVIMKSFMDKFINP 491
Query: 562 EGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGRK-DIKRGEALTYTVEPFLDG-S 619
+GWLPW GD A T++Y EY N G GA T RVKW G K + + EA +TV+PF+DG +
Sbjct: 492 KGWLPWTGDTAPDTIFYAEYLNSGPGASTKNRVKWQGLKTSLTKKEANKFTVKPFIDGNN 551
Query: 620 WINGTGVP 627
W+ T VP
Sbjct: 552 WLPATKVP 559
Score = 71.2 bits (173), Expect = 2e-12
Identities = 61/228 (26%), Positives = 103/228 (44%), Gaps = 26/228 (11%)
Query: 11 ERRKANAKQHLKKKI-LIGVTSVVLIACVIAAVTFVIVKRSGPDHNNNDKKPVQNAPPEP 69
E+ K A + KK+I +I ++S+VL+ V+ AV G +N KKP EP
Sbjct: 11 EQAKLEASRKTKKRIAIIAISSIVLVCIVVGAVV-------GTTARDNSKKPPTENNGEP 63
Query: 70 ERVDKYSRLVTMLCSHSEYKEKCVTTLKEALKKDPKLKEPKGLLMVFMLVAKNEINNAFN 129
V V LC + +KEKC TL A + P+ L + V E++ +
Sbjct: 64 ISVS-----VKALCDVTLHKEKCFETLGSA--PNASRSSPEELFKYAVKVTITELSKVLD 116
Query: 130 KTANLKFASKEEKGAYEDCKQLFEDAKEEMGFSITEVGQLDISKLASKEAELNNWLSAVI 189
+N + A C +L +G ++ ++ + S L + + +L WLS+V
Sbjct: 117 GFSNGEHMDNATSAAMGACVEL-------IGLAVDQLNETMTSSLKNFD-DLRTWLSSVG 168
Query: 190 SYQDTCSDGFPEG---ELKKKMEMIFAESRQLLSNSLAVVSQVSQIVN 234
+YQ+TC D E L E S ++ SN+LA+++ + +I +
Sbjct: 169 TYQETCMDALVEANKPSLTTFGENHLKNSTEMTSNALAIITWLGKIAD 216
>At2g47030 putative pectinesterase
Length = 588
Score = 313 bits (803), Expect = 1e-85
Identities = 202/621 (32%), Positives = 305/621 (48%), Gaps = 48/621 (7%)
Query: 24 KILIGVTSVVLIACVIAAVTFVIVKRSGPDHNNNDKKPVQNAPPEPERVDKYSRLVTMLC 83
K+++ V S++LI V V + ++G + + K VQ +C
Sbjct: 4 KVVVSVASILLIVGVAIGVV-AFINKNGDANLSPQMKAVQG-----------------IC 45
Query: 84 SHSEYKEKCVTTLKEALKKDPKLKEPKGLLMVFMLVAKNEINNAFNKTANLKF-----AS 138
+ K CV TL+ K ++P L+ FML K+E+ + N T + S
Sbjct: 46 QSTSDKASCVKTLEPV-----KSEDPNKLIKAFMLATKDELTKSSNFTGQTEVNMGSSIS 100
Query: 139 KEEKGAYEDCKQLFEDAKEEMGFSITEVGQLDISKLASKEAELNNWLSAVISYQDTCSDG 198
K + CK++F A E++ I E+G+ D+S++ SK +L WL V +YQ C D
Sbjct: 101 PNNKAVLDYCKRVFMYALEDLATIIEEMGE-DLSQIGSKIDQLKQWLIGVYNYQTDCLDD 159
Query: 199 FPEGELKKKMEMIFAESRQLLSNSLAVVSQVSQIVNAFQGGLSGFKLPWGKSDAHAPAPD 258
E +L+K + A S+ L +N++ + V + + K G P
Sbjct: 160 IEEDDLRKAIGEGIANSKILTTNAIDIFHTVVSAMAKINNKVDDLKNMTGGIPTPGAPPV 219
Query: 259 ADTDAVADDDEDLADAPDGAPDADQPIFEAPIGAPGAAPIGAPRVDAPPSWAAPAVDLPG 318
D VAD D + + P + + A G R G
Sbjct: 220 VDESPVADPDGPARRLLEDIDETGIPTWVSGADRKLMAKAGRGRRGGR-----------G 268
Query: 319 STEKPTPNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDETVTVTKKMVNLT 378
+ N VAKDGSG FKT+ +A+ A P+ +GR ++Y+K G+Y E V + KK N+
Sbjct: 269 GGARVRTNFVVAKDGSGQFKTVQQAVDACPENNRGRCIIYIKAGLYREQVIIPKKKNNIF 328
Query: 379 MYGDGGLKSIITGNKNFV--DGVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQAVAAR 436
M+GDG K++I+ N++ G T +A+ V +GF+ + MGF+NTAG + QA A R
Sbjct: 329 MFGDGARKTVISYNRSVALSRGTTTSLSATVQVESEGFMAKWMGFKNTAGPMGHQAAAIR 388
Query: 437 VQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVLRKPL 496
V D ++ NC F+GYQDTLY RQFYR+CV+SGT+DFIFG ++ V QN +V+RK
Sbjct: 389 VNGDRAVIFNCRFDGYQDTLYVNNGRQFYRNCVVSGTVDFIFGKSATVIQNTLIVVRKGS 448
Query: 497 DNQKNIITANG-RIDSKSNTAFVLQKCVIKGEDDLPS---TTKNYIGRPWKEYSRTIIME 552
Q N +TA+G + VLQ C I + L T Y+GRPWK++S T+IM
Sbjct: 449 KGQYNTVTADGNELGLGMKIGIVLQNCRIVPDRKLTPERLTVATYLGRPWKKFSTTVIMS 508
Query: 553 SDIPALIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGRKDIKRGEALTYTV 612
+++ LI+PEGW W+G+ K+ Y EY+N G GA + RV W + E +T
Sbjct: 509 TEMGDLIRPEGWKIWDGESFHKSCRYVEYNNRGPGAFANRRVNW-AKVARSAAEVNGFTA 567
Query: 613 EPFLDG-SWINGTGVPAHLGL 632
+L +WI VP +GL
Sbjct: 568 ANWLGPINWIQEANVPVTIGL 588
>At3g10720 pectinesterase like protein
Length = 619
Score = 313 bits (802), Expect = 2e-85
Identities = 209/590 (35%), Positives = 296/590 (49%), Gaps = 52/590 (8%)
Query: 61 PVQNAPPEPERVD-KYSRLVTMLCSHSEYKEKCVTTLKEALKKDPKLKEPKGLLMVFMLV 119
P+ P +P + S+ ++ C + Y + C T L A+K P G + +
Sbjct: 60 PIFVPPSQPPSLPPSQSQSPSLACKSTPYPKLCRTILN-AVKSSPSDPYRYGKFTIKQCL 118
Query: 120 AK----NEINNAFNKTANLK--FASKEEKGAYEDCKQLFEDAKEEMGFSITEVGQLDISK 173
+ +++ ++ + K A+ EE GA DC +L E + + TE+ +
Sbjct: 119 KQASRLSKVITSYARRVESKPGSATAEEIGAVADCGELSELSVNYLETVTTELKTAQVMT 178
Query: 174 LASKEAELNNWLSAVISYQDTCSDGFPEGE--LKKKMEMIFAESRQLLSNSLAVVSQVSQ 231
A E +N+ LS V++ Q TC DG E + + +L S SL +VS
Sbjct: 179 AALVE-HVNSLLSGVVTNQQTCLDGLVEAKSGFAAAIGSPMGNLTRLYSISLGLVS---- 233
Query: 232 IVNAFQGGLSGFKLPWGKSDAHAPAPDADTDAVADDDEDLADAPDGAPDADQPIFEAP-- 289
+A L FK GK + E L D D+ +
Sbjct: 234 --HALNRNLKRFKASKGK------ILGGGNSTYREPLETLIKGLRKTCDNDKDCRKTSRN 285
Query: 290 IGAPGAAPIGAPRVDAPPSWAAPAVDLPGSTEKPTPNVTVAKDGSGDFKTISEALAAIPQ 349
+G G G+ V V V S +F TI++A+AA P
Sbjct: 286 LGELGETSGGSILVSKA--------------------VIVGPFKSDNFTTITDAIAAAPN 325
Query: 350 TYK---GRYVVYVKEGVYDETVTVTKKMVNLTMYGDGGLKSIITGNKNFVDGVRTFQTAS 406
+ G +V+Y +EGVY+E + V NL + GDG K+IITGN N VDG T+ +S
Sbjct: 326 NTRPEDGYFVIYAREGVYEEYIVVPINKKNLMLMGDGINKTIITGNHNVVDGWTTYNCSS 385
Query: 407 FVVLGDGFVGRDMGFRNTAGAIKEQAVAARVQADCSIFVNCNFEGYQDTLYAQTHRQFYR 466
F V+G+ F+ D+ FRNTAG K QAVA R A+ S F C+FEGYQDTLY + RQFYR
Sbjct: 386 FAVVGERFMAVDVTFRNTAGPEKHQAVALRNNAEGSSFYRCSFEGYQDTLYVHSLRQFYR 445
Query: 467 DCVISGTIDFIFGHASAVFQNCQLVLRKPLDNQKNIITANGRIDSKSNTAFVLQKCVIKG 526
+C I GT+DFIFG+A+A+FQNC + RKP+ QKN ITA+GR+D NT + C IK
Sbjct: 446 ECDIYGTVDFIFGNAAAIFQNCNIYARKPMAKQKNAITAHGRLDPNQNTGISIINCTIKA 505
Query: 527 EDDL---PSTTKNYIGRPWKEYSRTIIMESDIPALIQPEGWLPWEGDFALKTLYYGEYDN 583
DL P + ++GRPWK YSRT+ M+S I ++QP GWL W G L T+YYGEY N
Sbjct: 506 APDLAAEPKSAMTFLGRPWKPYSRTVFMQSYISDIVQPVGWLEWNGTIGLDTIYYGEYSN 565
Query: 584 VGAGAKTDARVKWIGRKDIKRGEALTYTVEPFLDG-SWINGTGVPAHLGL 632
G GA T+ RV+W+G + EA+ +TV F G +W+ T +P + GL
Sbjct: 566 FGPGANTNQRVQWLGYNLLNLAEAMNFTVYNFTMGDTWLPQTDIPFYGGL 615
>At4g15980 pectinesterase like protein
Length = 701
Score = 311 bits (796), Expect = 9e-85
Identities = 159/307 (51%), Positives = 209/307 (67%), Gaps = 5/307 (1%)
Query: 326 NVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDETVTVTKKMVNLTMYGDGGL 385
NV VAKDGSG KTI++ALA +P ++V+++KEGVY E V VTKKM+++ GDG
Sbjct: 391 NVVVAKDGSGKCKTIAQALAMVPMKNTKKFVIHIKEGVYKEKVEVTKKMLHVMFVGDGPT 450
Query: 386 KSIITGNKNFV-DGVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQAVAARVQADCSIF 444
K++ITG+ F+ D V T++TAS V GD F+ +D+GF NTAGA + QAVA RV AD ++F
Sbjct: 451 KTVITGDIAFLPDQVGTYRTASVAVNGDYFMAKDIGFENTAGAARHQAVALRVSADFAVF 510
Query: 445 VNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVLRKPLDNQKNIIT 504
NC+ GYQDTLY THRQFYR+C +SGTIDF+FG A AVFQNC+ V+R+P+++Q+ I+T
Sbjct: 511 FNCHMNGYQDTLYVHTHRQFYRNCRVSGTIDFVFGDAKAVFQNCEFVIRRPMEHQQCIVT 570
Query: 505 ANGRIDSKSNTAFVLQKCVIKGEDD-LPSTTKN--YIGRPWKEYSRTIIMESDIPALIQP 561
A GR D + T V+ I G+ LP KN ++GRPWKE+SRTIIM ++I +I P
Sbjct: 571 AQGRKDRRETTGIVIHNSRITGDASYLPVKAKNRAFLGRPWKEFSRTIIMNTEIDDVIDP 630
Query: 562 EGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGRKDIKRGEALTYTVEPFLDG-SW 620
EGWL W FAL TL+Y EY N G G+ RV+W G K I A + FL G +W
Sbjct: 631 EGWLKWNETFALNTLFYTEYRNRGRGSGQGRRVRWRGIKRISDRAAREFAPGNFLRGNTW 690
Query: 621 INGTGVP 627
I T +P
Sbjct: 691 IPQTRIP 697
Score = 72.0 bits (175), Expect = 9e-13
Identities = 54/227 (23%), Positives = 108/227 (46%), Gaps = 30/227 (13%)
Query: 21 LKKKILIGVTSVVL--IACVIAAVTFVIVKRSGPDHNNNDKKPVQNAPPEPERVDKYSRL 78
+ K +L+GVT++++ + CV A D N+ +K E V + S+L
Sbjct: 1 MNKYVLLGVTALIMAMVICVEAN-----------DGNSPSRKM--------EEVHRESKL 41
Query: 79 ------VTMLCSHSEYKEKCVTTLKEALKKDPKLKEPKGLLMVFMLVAKNEINNAFNKTA 132
V+++C+ ++YK+ C T+L DP+ + + ++ ++ I+
Sbjct: 42 MITKTTVSIICASTDYKQDCTTSLATVRSPDPR-NLIRSAFDLAIISIRSGIDRGMIDLK 100
Query: 133 NLKFASKEEKGAYEDCKQLFEDAKEEMGFSITEVGQLDISKLASKEAELNNWLSAVISYQ 192
+ A + A C++L +DA +++ + + ++L+ +L WLS I+YQ
Sbjct: 101 SRADADMHTREALNTCRELMDDAIDDLRKTRDKFRGFLFTRLSDFVEDLCVWLSGSITYQ 160
Query: 193 DTCSDGFP--EGELKKKMEMIFAESRQLLSNSLAVVSQVSQIVNAFQ 237
TC DGF + E ME + + + L SN LA+ + + +++ AF+
Sbjct: 161 QTCIDGFEGIDSEAAVMMERVMRKGQHLTSNGLAIAANLDKLLKAFR 207
>At3g10710 putative pectinesterase
Length = 561
Score = 311 bits (796), Expect = 9e-85
Identities = 151/308 (49%), Positives = 201/308 (65%), Gaps = 2/308 (0%)
Query: 326 NVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDETVTVTKKMVNLTMYGDGGL 385
++ VAKDGSG ++TI AL +P+ + R ++YVK+GVY E V V KKM N+ + GDG
Sbjct: 255 DIVVAKDGSGKYRTIKRALQDVPEKSEKRTIIYVKKGVYFENVKVEKKMWNVIVVGDGES 314
Query: 386 KSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQAVAARVQADCSIFV 445
KSI++G N +DG TF+TA+F V G GF+ RDMGF NTAG K QAVA V AD + F
Sbjct: 315 KSIVSGRLNVIDGTPTFKTATFAVFGKGFMARDMGFINTAGPSKHQAVALMVSADLTAFY 374
Query: 446 NCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVLRKPLDNQKNIITA 505
C YQDTLY RQFYR+C I GT+DFIFG++++V Q+C+++ R+P+ Q+N ITA
Sbjct: 375 RCTMNAYQDTLYVHAQRQFYRECTIIGTVDFIFGNSASVLQSCRILPRRPMKGQQNTITA 434
Query: 506 NGRIDSKSNTAFVLQKCVIKGEDDLPSTTKNYIGRPWKEYSRTIIMESDIPALIQPEGWL 565
GR D NT + +C I DL + ++GRPWK +S T+IM+S + I +GWL
Sbjct: 435 QGRTDPNMNTGISIHRCNISPLGDL-TDVMTFLGRPWKNFSTTVIMDSYLHGFIDRKGWL 493
Query: 566 PWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGRKDIKRGEALTYTVEPFLDGS-WINGT 624
PW GD A T++YGEY N G GA T RVKW G + + EA +TV+PF+DG W+ T
Sbjct: 494 PWTGDSAPDTIFYGEYKNTGPGASTKNRVKWKGLRFLSTKEANRFTVKPFIDGGRWLPAT 553
Query: 625 GVPAHLGL 632
VP GL
Sbjct: 554 KVPFRSGL 561
Score = 57.0 bits (136), Expect = 3e-08
Identities = 53/231 (22%), Positives = 102/231 (43%), Gaps = 23/231 (9%)
Query: 11 ERRKANAKQHLKKKI-LIGVTSVVLIACVIAAVTFVIVKRSGPDH---NNNDKKPVQNAP 66
E + A++ +K I +I V+ V+L VI AV + + P+ NNN
Sbjct: 12 EHVRLEARRKTRKNIAIIAVSLVILAGIVIGAVFGTMAHKKSPETVETNNNG-------- 63
Query: 67 PEPERVDKYSRLVTMLCSHSEYKEKCVTTLKEALKKDPKLKEPKGLLMVFMLVAKNEINN 126
D S V +C + +KEKC TL A + P+ L + + E++
Sbjct: 64 ------DSISVSVKAVCDVTLHKEKCFETLGSA--PNASSLNPEELFRYAVKITIAEVSK 115
Query: 127 AFNKTANLKFASKEEKGAYEDCKQLFEDAKEEMGFSITEVGQLDISKLASKEAELNNWLS 186
A N ++ ++ C +L + + + ++T D++ + +L WLS
Sbjct: 116 AINAFSS-SLGDEKNNITMNACAELLDLTIDNLNNTLTSSSNGDVT-VPELVDDLRTWLS 173
Query: 187 AVISYQDTCSDGFPEGELKKKMEMIFAESRQLLSNSLAVVSQVSQIVNAFQ 237
+ +YQ TC + +++ E S +L SN+LA+++ + +I ++F+
Sbjct: 174 SAGTYQRTCVETLAP-DMRPFGESHLKNSTELTSNALAIITWLGKIADSFK 223
>At4g02330 unknown protein (At4g02330)
Length = 573
Score = 309 bits (792), Expect = 3e-84
Identities = 160/313 (51%), Positives = 203/313 (64%), Gaps = 7/313 (2%)
Query: 327 VTVAKDGSGDFKTISEALAAIPQTYKGR---YVVYVKEGVYDETVTVTKKMVNLTMYGDG 383
VTV ++G+G+F TI+EA+ + P G +V+YV GVY+E V + K L M GDG
Sbjct: 259 VTVNQNGTGNFTTITEAVNSAPNKTDGTAGYFVIYVTSGVYEENVVIAKNKRYLMMIGDG 318
Query: 384 GLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFRNTAGAIKEQAVAARVQADCSI 443
++++TGN+N VDG TF +A+F V FV +M FRNTAG K QAVA R AD SI
Sbjct: 319 INRTVVTGNRNVVDGWTTFNSATFAVTSPNFVAVNMTFRNTAGPEKHQAVAMRSSADLSI 378
Query: 444 FVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQLVLRKPLDNQKNII 503
F +C+FE YQDTLY + RQFYR+C I GT+DFIFG+A+ VFQ+C L R+P+ NQ N I
Sbjct: 379 FYSCSFEAYQDTLYTHSLRQFYRECDIYGTVDFIFGNAAVVFQDCNLYPRQPMQNQFNAI 438
Query: 504 TANGRIDSKSNTAFVLQKCVIKGEDDLPS---TTKNYIGRPWKEYSRTIIMESDIPALIQ 560
TA GR D NT + C IK DDL S T K Y+GRPWKEYSRT+ M+S I +++
Sbjct: 439 TAQGRTDPNQNTGISIHNCTIKPADDLVSSNYTVKTYLGRPWKEYSRTVFMQSYIDEVVE 498
Query: 561 PEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGRKDIKRGEALTYTVEPFLDG-S 619
P GW W GDFAL TLYY EY+N G+G+ T RV W G I +A +TVE FL G
Sbjct: 499 PVGWREWNGDFALSTLYYAEYNNTGSGSSTTDRVVWPGYHVINSTDANNFTVENFLLGDG 558
Query: 620 WINGTGVPAHLGL 632
W+ +GVP GL
Sbjct: 559 WMVQSGVPYISGL 571
Score = 30.0 bits (66), Expect = 4.1
Identities = 21/68 (30%), Positives = 30/68 (43%), Gaps = 2/68 (2%)
Query: 133 NLKFASKEEKGAYEDCKQLFEDAKEEMGFSITEVGQLDISKLAS--KEAELNNWLSAVIS 190
N S+ GA +DC+ L + + S V SK S K E+ LSA ++
Sbjct: 85 NNALLSQSAVGALQDCRYLASLTTDYLITSFETVNITTSSKTLSFSKADEIQTLLSAALT 144
Query: 191 YQDTCSDG 198
+ TC DG
Sbjct: 145 NEQTCLDG 152
>At2g45220 pectinesterase like protein
Length = 511
Score = 307 bits (787), Expect = 1e-83
Identities = 162/337 (48%), Positives = 214/337 (63%), Gaps = 11/337 (3%)
Query: 301 PRVDAPPSWAAPAVDLPGSTEKPTPNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVK 360
P D PSW P + P N VAKDGSG+FKTI EA+ A + GR+V+YVK
Sbjct: 181 PEKDGFPSWVKPGDRKLLQSSTPKDNAVVAKDGSGNFKTIKEAIDAASGS--GRFVIYVK 238
Query: 361 EGVYDETVTVTKKMVNLTMYGDGGLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMG 420
+GVY E + + KK N+ + GDG K+IITG+K+ G TF +A+ +GDGF+ R +
Sbjct: 239 QGVYSENLEIRKK--NVMLRGDGIGKTIITGSKSVGGGTTTFNSATVAAVGDGFIARGIT 296
Query: 421 FRNTAGAIKEQAVAARVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGH 480
FRNTAGA EQAVA R +D S+F C+FE YQDTLY ++RQFYRDC + GT+DFIFG+
Sbjct: 297 FRNTAGASNEQAVALRSGSDLSVFYQCSFEAYQDTLYVHSNRQFYRDCDVYGTVDFIFGN 356
Query: 481 ASAVFQNCQLVLRKPLDNQKNIITANGRIDSKSNTAFVLQKCVIKGEDDLP---STTKNY 537
A+AV QNC + R+P ++ N ITA GR D NT ++ + DL +TK Y
Sbjct: 357 AAAVLQNCNIFARRP-RSKTNTITAQGRSDPNQNTGIIIHNSRVTAASDLRPVLGSTKTY 415
Query: 538 IGRPWKEYSRTIIMESDIPALIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWI 597
+GRPW++YSRT+ M++ + +LI P GWL W+G+FALKTL+Y E+ N G GA T RV W
Sbjct: 416 LGRPWRQYSRTVFMKTSLDSLIDPRGWLEWDGNFALKTLFYAEFQNTGPGASTSGRVTWP 475
Query: 598 GRKDI-KRGEALTYTVEPFL-DGSWINGTGVPAHLGL 632
G + + EA +TV FL GSWI + VP GL
Sbjct: 476 GFRVLGSASEASKFTVGTFLAGGSWI-PSSVPFTSGL 511
Score = 42.0 bits (97), Expect = 0.001
Identities = 36/147 (24%), Positives = 62/147 (41%), Gaps = 13/147 (8%)
Query: 83 CSHSEYKEKCVTTLKEALKKDPKLKEPKGLLMVFMLVAKNEI---NNAFNKTANLKFASK 139
CS + + C L +P E + L + LV I +AF T K
Sbjct: 34 CSQTPNPKPCEYFLTHNSNNEPIKSESEFLKISMKLVLDRAILAKTHAF--TLGPKCRDT 91
Query: 140 EEKGAYEDCKQLFEDAKEEMGFSITEVGQLDISKLASKEAELNNWLSAVISYQDTCSDGF 199
EK A+EDC +L++ +++++ + + + + WLS ++ DTC GF
Sbjct: 92 REKAAWEDCIKLYD-------LTVSKINETMDPNVKCSKLDAQTWLSTALTNLDTCRAGF 144
Query: 200 PE-GELKKKMEMIFAESRQLLSNSLAV 225
E G + ++ LL N+LA+
Sbjct: 145 LELGVTDIVLPLMSNNVSNLLCNTLAI 171
>At3g43270 pectinesterase like protein
Length = 527
Score = 306 bits (784), Expect = 2e-83
Identities = 161/332 (48%), Positives = 209/332 (62%), Gaps = 6/332 (1%)
Query: 307 PSWAAPAVDLPGSTEKPT-PNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYD 365
PSW P T+ T + VA DG+G+F TIS+A+ A P RYV++VK GVY
Sbjct: 194 PSWVKPGDRKLLQTDNITVADAVVAADGTGNFTTISDAVLAAPDYSTKRYVIHVKRGVYV 253
Query: 366 ETVTVTKKMVNLTMYGDGGLKSIITGNKNFVDGVRTFQTASFVVLGDGFVGRDMGFRNTA 425
E V + KK N+ M GDG ++ITGN++F+DG TF++A+F V G GF+ RD+ F+NTA
Sbjct: 254 ENVEIKKKKWNIMMVGDGIDATVITGNRSFIDGWTTFRSATFAVSGRGFIARDITFQNTA 313
Query: 426 GAIKEQAVAARVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVF 485
G K QAVA R D +F C GYQDTLYA + RQF+R+C+I+GT+DFIFG A+AVF
Sbjct: 314 GPEKHQAVAIRSDTDLGVFYRCAMRGYQDTLYAHSMRQFFRECIITGTVDFIFGDATAVF 373
Query: 486 QNCQLVLRKPLDNQKNIITANGRIDSKSNTAFVLQKCVIKGEDDL---PSTTKNYIGRPW 542
Q+CQ+ ++ L NQKN ITA GR D T F +Q I + DL +TT Y+GRPW
Sbjct: 374 QSCQIKAKQGLPNQKNSITAQGRKDPNEPTGFTIQFSNIAADTDLLLNLNTTATYLGRPW 433
Query: 543 KEYSRTIIMESDIPALIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGRKDI 602
K YSRT+ M++ + I P GWL W G+FAL TLYYGEY N G GA D RVKW G +
Sbjct: 434 KLYSRTVFMQNYMSDAINPVGWLEWNGNFALDTLYYGEYMNSGPGASLDRRVKWPGYHVL 493
Query: 603 K-RGEALTYTVEPFLDGS-WINGTGVPAHLGL 632
EA +TV + G+ W+ TG+ GL
Sbjct: 494 NTSAEANNFTVSQLIQGNLWLPSTGITFIAGL 525
Score = 40.4 bits (93), Expect = 0.003
Identities = 37/134 (27%), Positives = 57/134 (41%), Gaps = 9/134 (6%)
Query: 78 LVTMLCS-HSEYKEKCVTTLKEALKKDPKLKEPKGLLMVFMLVAKNEINNAFNKTANLKF 136
++ LCS H E E L+ P +V + I + F+K A
Sbjct: 17 IIISLCSAHKEAFSSTDLVQMECLRVPPLEFAEAAKTVVDAITKAVAIVSKFDKKAGKSR 76
Query: 137 ASKEEKGAYEDCKQLFEDAKEEMGFSITEVGQLDISKLASKE--AELNNWLSAVISYQDT 194
S A DC L + A EE+ + I+ + ++ + ++L W+SA +S QDT
Sbjct: 77 VSN----AIVDCVDLLDSAAEELSWIISASQSPNGKDNSTGDVGSDLRTWISAALSNQDT 132
Query: 195 CSDGF--PEGELKK 206
C DGF G +KK
Sbjct: 133 CLDGFEGTNGIIKK 146
>At2g47040 putative pectinesterase
Length = 595
Score = 303 bits (776), Expect = 2e-82
Identities = 213/628 (33%), Positives = 318/628 (49%), Gaps = 55/628 (8%)
Query: 24 KILIGVTSVVLIACVIAAVTFVIVKRSGPDHNNNDKKPVQNAPPEPERVDKYSRLVTMLC 83
K+++ V S++LI V V I K N D N P+ + V +C
Sbjct: 4 KVVVSVASILLIVGVAIGVVAYINK-------NGDA----NLSPQ-------MKAVRGIC 45
Query: 84 SHSEYKEKCVTTLKEALKKDPKLKEPKGLLMVFMLVAKNEINNAFNKTANLKF-----AS 138
+ K CV TL E +K D +P L+ FML ++ I + N T + S
Sbjct: 46 EATSDKASCVKTL-EPVKSD----DPNKLIKAFMLATRDAITQSSNFTGKTEGNLGSGIS 100
Query: 139 KEEKGAYEDCKQLFEDAKEEMGFSITEVGQLDISKLASKEAELNNWLSAVISYQDTCSDG 198
K + CK++F A E++ + E+G+ D++++ SK +L WL+ V +YQ C D
Sbjct: 101 PNNKAVLDYCKKVFMYALEDLSTIVEEMGE-DLNQIGSKIDQLKQWLTGVYNYQTDCLDD 159
Query: 199 FPEGELKKKMEMIFAESRQLLSNSL----AVVSQVSQI---VNAFQGGLSGFKLPWGKSD 251
E +L+K + A S+ L SN++ VVS ++++ V F+ G P SD
Sbjct: 160 IEEDDLRKTIGEGIASSKILTSNAIDIFHTVVSAMAKLNLKVEDFKNMTGGIFAP---SD 216
Query: 252 AHAPAPDADTDAVADDDEDLADAPDGAPDADQPIFEAPIGAPGAAPIGAPRVDAPPSWAA 311
A + T VADD +P PD I G P D A
Sbjct: 217 KGAAPVNKGTPPVADD------SPVADPDGPARRLLEDIDETGI-PTWVSGADRKLMTKA 269
Query: 312 PAVDLPGSTEKPTPNVTVAKDGSGDFKTISEALAAIPQTYKGRYVVYVKEGVYDETVTVT 371
G + VAKDGSG FKT+ +A+ A P+ GR ++++K G+Y E V +
Sbjct: 270 GRGSNDGGA-RIRATFVVAKDGSGQFKTVQQAVNACPEKNPGRCIIHIKAGIYREQVIIP 328
Query: 372 KKMVNLTMYGDGGLKSIITGNKN--FVDGVRTFQTASFVVLGDGFVGRDMGFRNTAGAIK 429
KK N+ M+GDG K++I+ N++ G T + + V +GF+ + +GF+NTAG +
Sbjct: 329 KKKNNIFMFGDGARKTVISYNRSVKLSPGTTTSLSGTVQVESEGFMAKWIGFKNTAGPMG 388
Query: 430 EQAVAARVQADCSIFVNCNFEGYQDTLYAQTHRQFYRDCVISGTIDFIFGHASAVFQNCQ 489
QAVA RV D ++ NC F+GYQDTLY RQFYR+ V+SGT+DFIFG ++ V QN
Sbjct: 389 HQAVAIRVNGDRAVIFNCRFDGYQDTLYVNNGRQFYRNIVVSGTVDFIFGKSATVIQNSL 448
Query: 490 LVLRKPLDNQKNIITANGRIDSKS-NTAFVLQKCVIKGEDDLPS---TTKNYIGRPWKEY 545
+V+RK Q N +TA+G + VLQ C I + L + ++Y+GRPWK++
Sbjct: 449 IVVRKGNKGQFNTVTADGNEKGLAMKIGIVLQNCRIVPDKKLAAERLIVESYLGRPWKKF 508
Query: 546 SRTIIMESDIPALIQPEGWLPWEGDFALKTLYYGEYDNVGAGAKTDARVKWIGRKDIKRG 605
S T+I+ S+I +I+PEGW W+G+ K+ Y EY+N G GA T+ RV W+ +
Sbjct: 509 STTVIINSEIGDVIRPEGWKIWDGESFHKSCRYVEYNNRGPGAITNRRVNWV-KIARSAA 567
Query: 606 EALTYTVEPFLDG-SWINGTGVPAHLGL 632
E +TV +L +WI VP LGL
Sbjct: 568 EVNDFTVANWLGPINWIQEANVPVTLGL 595
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.134 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,691,569
Number of Sequences: 26719
Number of extensions: 666061
Number of successful extensions: 2429
Number of sequences better than 10.0: 132
Number of HSP's better than 10.0 without gapping: 84
Number of HSP's successfully gapped in prelim test: 48
Number of HSP's that attempted gapping in prelim test: 2040
Number of HSP's gapped (non-prelim): 215
length of query: 634
length of database: 11,318,596
effective HSP length: 105
effective length of query: 529
effective length of database: 8,513,101
effective search space: 4503430429
effective search space used: 4503430429
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC130802.4


