

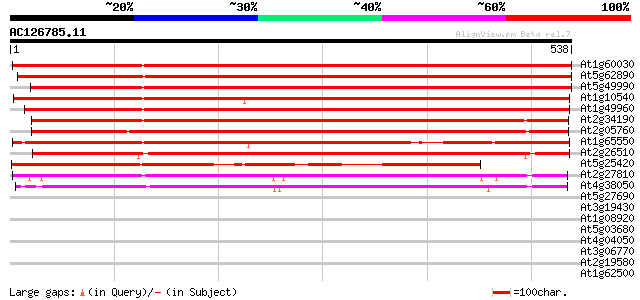
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126785.11 + phase: 0
(538 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g60030 unknown protein 855 0.0
At5g62890 permease 1 - like protein 840 0.0
At5g49990 permease 806 0.0
At1g10540 putative permease 776 0.0
At1g49960 permease, putative 636 0.0
At2g34190 putative membrane transporter 606 e-174
At2g05760 putative membrane transporter 575 e-164
At1g65550 hypothetical protein 540 e-153
At2g26510 putative membrane transporter 496 e-140
At5g25420 permease 1-like protein 398 e-111
At2g27810 putative membrane transporter 312 3e-85
At4g38050 unknown protein 309 3e-84
At5g27690 unknown protein (At5g27690) 35 0.14
At3g19430 putative late embryogenesis abundant protein 32 0.89
At1g08920 putative sugar transport protein 30 4.4
At5g03680 GT2 -like protein 29 7.5
At4g04050 putative transposon protein 29 7.5
At3g06770 unknown protein 29 7.5
At2g19580 putative senescence-associated protein 5 29 7.5
At1g62500 putative proline-rich cell wall protein (pir|IS52985);... 29 7.5
>At1g60030 unknown protein
Length = 538
Score = 855 bits (2210), Expect = 0.0
Identities = 410/536 (76%), Positives = 469/536 (87%), Gaps = 1/536 (0%)
Query: 3 GGGGGGGATPPPKQDEFQPHPVKDQLPNVSYCITSPPPWPEAIMLGFQHYLVMLGTTVLI 62
GGGGGGG PP K D +PHPVKDQL ++SYCITSPPPWPEAI+LGFQHYLVMLGTTVLI
Sbjct: 4 GGGGGGGVAPPLKHDGLEPHPVKDQLSSISYCITSPPPWPEAILLGFQHYLVMLGTTVLI 63
Query: 63 PTALVSQMGGGNEEKAMLIQNHLFVAGINTLIQTLFGTRLPAVIGGSFTFVPTTISIILA 122
PT LV QMGGGNEEKA ++Q LFV+G+NTL+Q+ FGTRLPAVIGGS+T+VPTT+SIILA
Sbjct: 64 PTYLVPQMGGGNEEKAKMVQTLLFVSGLNTLLQSFFGTRLPAVIGGSYTYVPTTLSIILA 123
Query: 123 SRYDDDIMHPREKFKRIMRGTQGALIVASSLQIIVGFSGLWCHVVRFISPLSAVPLVALT 182
RY D I+ P+EKFKRIMRG QGALIVAS LQI+VGFSGLW +VVR +SPLSAVPLVAL
Sbjct: 124 GRYSD-ILDPQEKFKRIMRGIQGALIVASILQIVVGFSGLWRNVVRLLSPLSAVPLVALA 182
Query: 183 GFGLYELGFPMLAKCIEIGLPEIVILVFLSQFMPHMMKGGRHIFARFAVIFSVIIVWVYA 242
GFGLYE GFP+LAKCIEIGLPEI++L+ SQ++PH+++G R +F RFAVIFSV+IVW+YA
Sbjct: 183 GFGLYEHGFPLLAKCIEIGLPEIILLLLFSQYIPHLIRGERQVFHRFAVIFSVVIVWIYA 242
Query: 243 IILTGCGAYKNAEHETQDTCRTDRAGLIHGASWISPPIPFRWGAPTFDAGEAFAMMAASF 302
+LT GAYKN TQ +CRTDR+GLI G+ WI P PF+WG PTF AGEAFAMMA SF
Sbjct: 243 HLLTVGGAYKNTGVNTQTSCRTDRSGLISGSPWIRVPYPFQWGPPTFHAGEAFAMMAVSF 302
Query: 303 VAQIESTGGFIAVARFASATPVPPSVLSRGIGWQGVGILLSGIFGTGNGSSVSIENAGLL 362
V+ IESTG +I V+RFASATP PPSVLSRG+GWQGVG+LL G+FG GNG+SVS+ENAGLL
Sbjct: 303 VSLIESTGTYIVVSRFASATPPPPSVLSRGVGWQGVGVLLCGLFGAGNGASVSVENAGLL 362
Query: 363 ALTRVGSRRVVQISAGFMIFFSILGKFGAVFASIPMPIVAALYCLLFSQVGSAGLSFLQF 422
ALTRVGSRRVVQISAGFMIFFSILGKFGA+FASIP P+VAAL+CL F+ VG+ GLS LQF
Sbjct: 363 ALTRVGSRRVVQISAGFMIFFSILGKFGAIFASIPAPVVAALHCLFFAYVGAGGLSLLQF 422
Query: 423 CNLNSFRTKFIIGFSIFMGFSVPQYFKEYTAIKQYGPVHTNARWFNDMINVPFSSGAFVA 482
CNLNSFRTKFI+GFS+FMG S+PQYF +YTA+ +YGPVHT+ARWFNDMINVPFSS AFVA
Sbjct: 423 CNLNSFRTKFILGFSVFMGLSIPQYFNQYTAVNKYGPVHTHARWFNDMINVPFSSKAFVA 482
Query: 483 GILALFFDVTLHKSDNQTRKDRGMHWWDRFSSFKTDTRSEEFYSLPFNLNKFFPSV 538
GILA F DVT+ D+ TRKDRGM WWDRF SFK+DTRSEEFYSLPFNLNK+FPSV
Sbjct: 483 GILAFFLDVTMSSKDSATRKDRGMFWWDRFMSFKSDTRSEEFYSLPFNLNKYFPSV 538
>At5g62890 permease 1 - like protein
Length = 532
Score = 840 bits (2169), Expect = 0.0
Identities = 405/531 (76%), Positives = 464/531 (87%), Gaps = 1/531 (0%)
Query: 8 GGATPPPKQDEFQPHPVKDQLPNVSYCITSPPPWPEAIMLGFQHYLVMLGTTVLIPTALV 67
GG P PK DE QPHP KDQLPN+SYCITSPPPWPEAI+LGFQHYLVMLGTTVLIPTALV
Sbjct: 3 GGGAPAPKADEPQPHPPKDQLPNISYCITSPPPWPEAILLGFQHYLVMLGTTVLIPTALV 62
Query: 68 SQMGGGNEEKAMLIQNHLFVAGINTLIQTLFGTRLPAVIGGSFTFVPTTISIILASRYDD 127
QMGGG EEKA +IQ LFVAGINTL+QTLFGTRLPAV+G S+TFVPTTISIIL+ R+ D
Sbjct: 63 PQMGGGYEEKAKVIQTILFVAGINTLLQTLFGTRLPAVVGASYTFVPTTISIILSGRFSD 122
Query: 128 DIMHPREKFKRIMRGTQGALIVASSLQIIVGFSGLWCHVVRFISPLSAVPLVALTGFGLY 187
+P ++F+RIMR TQGALIVAS+LQ+I+GFSGLW +VVRF+SP+SAVPLV L GFGLY
Sbjct: 123 T-SNPIDRFERIMRATQGALIVASTLQMILGFSGLWRNVVRFLSPISAVPLVGLVGFGLY 181
Query: 188 ELGFPMLAKCIEIGLPEIVILVFLSQFMPHMMKGGRHIFARFAVIFSVIIVWVYAIILTG 247
E GFP +AKCIEIGLPE++ILVF+SQ++PH++K G+++F RFAVIF+V+IVW+YA +LT
Sbjct: 182 EFGFPGVAKCIEIGLPELLILVFVSQYLPHVIKSGKNVFDRFAVIFAVVIVWIYAHLLTV 241
Query: 248 CGAYKNAEHETQDTCRTDRAGLIHGASWISPPIPFRWGAPTFDAGEAFAMMAASFVAQIE 307
GAY A TQ +CRTDRAG+I A WI P PF+WGAP+FDAGEAFAMM ASFVA +E
Sbjct: 242 GGAYNGAAPTTQTSCRTDRAGIIGAAPWIRVPWPFQWGAPSFDAGEAFAMMMASFVALVE 301
Query: 308 STGGFIAVARFASATPVPPSVLSRGIGWQGVGILLSGIFGTGNGSSVSIENAGLLALTRV 367
STG F+AV+R+ASAT +PPS+LSRGIGWQGV IL+SG+FGTG GSSVS+ENAGLLALTRV
Sbjct: 302 STGAFVAVSRYASATMLPPSILSRGIGWQGVAILISGLFGTGAGSSVSVENAGLLALTRV 361
Query: 368 GSRRVVQISAGFMIFFSILGKFGAVFASIPMPIVAALYCLLFSQVGSAGLSFLQFCNLNS 427
GSRRVVQI+AGFMIFFSILGKFGAVFASIP PI+AALYCL F+ VG+ GLSFLQFCNLNS
Sbjct: 362 GSRRVVQIAAGFMIFFSILGKFGAVFASIPAPIIAALYCLFFAYVGAGGLSFLQFCNLNS 421
Query: 428 FRTKFIIGFSIFMGFSVPQYFKEYTAIKQYGPVHTNARWFNDMINVPFSSGAFVAGILAL 487
FRTKFI+GFS+F+G S+PQYF EYTAIK YGPVHT ARWFNDM+NVPFSS FVAG +A
Sbjct: 422 FRTKFILGFSVFLGLSIPQYFNEYTAIKGYGPVHTGARWFNDMVNVPFSSEPFVAGSVAF 481
Query: 488 FFDVTLHKSDNQTRKDRGMHWWDRFSSFKTDTRSEEFYSLPFNLNKFFPSV 538
F D TLHK D+ RKDRG HWWD+F SFK DTRSEEFYSLPFNLNK+FPSV
Sbjct: 482 FLDNTLHKKDSSIRKDRGKHWWDKFRSFKGDTRSEEFYSLPFNLNKYFPSV 532
>At5g49990 permease
Length = 528
Score = 806 bits (2081), Expect = 0.0
Identities = 388/518 (74%), Positives = 447/518 (85%), Gaps = 1/518 (0%)
Query: 21 PHPVKDQLPNVSYCITSPPPWPEAIMLGFQHYLVMLGTTVLIPTALVSQMGGGNEEKAML 80
PHP K+QLP++SYCITSPPPWPEA++LGFQHYLVMLGTTVLIP+ALV QMGG NEEKA L
Sbjct: 12 PHPPKEQLPDISYCITSPPPWPEAVLLGFQHYLVMLGTTVLIPSALVPQMGGRNEEKAKL 71
Query: 81 IQNHLFVAGINTLIQTLFGTRLPAVIGGSFTFVPTTISIILASRYDDDIMHPREKFKRIM 140
IQ LFVAG+NTL+QT+FGTRLPAVIG S+TFVP TISI+L+ R++D + P E+FKRI+
Sbjct: 72 IQTILFVAGLNTLLQTVFGTRLPAVIGASYTFVPVTISIMLSGRFND-VADPVERFKRII 130
Query: 141 RGTQGALIVASSLQIIVGFSGLWCHVVRFISPLSAVPLVALTGFGLYELGFPMLAKCIEI 200
R TQGALIVAS+LQII+GFSGLW +VVRF+SPLSA PLV L G+GLYELGFP +AKCIEI
Sbjct: 131 RATQGALIVASTLQIILGFSGLWRNVVRFLSPLSAAPLVGLVGYGLYELGFPGVAKCIEI 190
Query: 201 GLPEIVILVFLSQFMPHMMKGGRHIFARFAVIFSVIIVWVYAIILTGCGAYKNAEHETQD 260
GLP ++IL+ +SQ+MPH++KGG+H+FARFAVIFSV IVW+YA LT GAY +TQ
Sbjct: 191 GLPGLIILILISQYMPHVIKGGKHVFARFAVIFSVAIVWLYAFFLTLGGAYNGVGTDTQR 250
Query: 261 TCRTDRAGLIHGASWISPPIPFRWGAPTFDAGEAFAMMAASFVAQIESTGGFIAVARFAS 320
+CRTDRAGLI A WI P PF+WGAP FDAGEAFAMM ASFVA +ESTG FIAV+R+AS
Sbjct: 251 SCRTDRAGLISAAPWIRVPWPFQWGAPLFDAGEAFAMMMASFVALVESTGAFIAVSRYAS 310
Query: 321 ATPVPPSVLSRGIGWQGVGILLSGIFGTGNGSSVSIENAGLLALTRVGSRRVVQISAGFM 380
AT PPSV+SRG+GWQGV IL+SG+FGTG GSSVS+ENAGLLALT++GSRRVVQISAGFM
Sbjct: 311 ATMPPPSVISRGVGWQGVAILISGLFGTGIGSSVSVENAGLLALTKIGSRRVVQISAGFM 370
Query: 381 IFFSILGKFGAVFASIPMPIVAALYCLLFSQVGSAGLSFLQFCNLNSFRTKFIIGFSIFM 440
IFFSILGKFGAVFASIP PI+AALYCL F+ VG+ GLS LQFCNLNSFRT FI+GFSIF+
Sbjct: 371 IFFSILGKFGAVFASIPSPIIAALYCLFFAYVGAGGLSLLQFCNLNSFRTLFILGFSIFL 430
Query: 441 GFSVPQYFKEYTAIKQYGPVHTNARWFNDMINVPFSSGAFVAGILALFFDVTLHKSDNQT 500
G S+PQYF E+TAIK YGPVHT ARWFNDM+NVPFSS AFV G +A D TLHK D
Sbjct: 431 GLSIPQYFNEHTAIKGYGPVHTGARWFNDMVNVPFSSKAFVGGCVAYLLDTTLHKKDGSI 490
Query: 501 RKDRGMHWWDRFSSFKTDTRSEEFYSLPFNLNKFFPSV 538
RKDRG HWWDRF +FK D R+EEFY+LPFNLNK+FPSV
Sbjct: 491 RKDRGKHWWDRFWTFKNDPRTEEFYALPFNLNKYFPSV 528
>At1g10540 putative permease
Length = 539
Score = 776 bits (2005), Expect = 0.0
Identities = 377/537 (70%), Positives = 451/537 (83%), Gaps = 4/537 (0%)
Query: 4 GGGGGGATPPPKQDEFQPHPVKDQLPNVSYCITSPPPWPEAIMLGFQHYLVMLGTTVLIP 63
G G A PP KQ++ QPHPVKDQL ++YC+TSPPPWPE I+LGFQHYLVMLGTTVLIP
Sbjct: 3 GDGVENAKPPQKQEDLQPHPVKDQLYGITYCLTSPPPWPETILLGFQHYLVMLGTTVLIP 62
Query: 64 TALVSQMGGGNEEKAMLIQNHLFVAGINTLIQTLFGTRLPAVIGGSFTFVPTTISIILAS 123
T LVS++ NE+K LIQ LFV+GINTL Q+ FGTRLPAVIG S+++VPTT+SI+LA+
Sbjct: 63 TMLVSKIDARNEDKVKLIQTLLFVSGINTLFQSFFGTRLPAVIGASYSYVPTTMSIVLAA 122
Query: 124 RYDDDIMHPREKFKRIMRGTQGALIVASSLQIIVGFSGLWCHVVRFISPLSAVPLVALTG 183
RY+D IM P+++F++IMRG QGALI+AS L I+VGFSGLW +V RF+SPLSAVPLVA +G
Sbjct: 123 RYND-IMDPQKRFEQIMRGIQGALIIASFLHILVGFSGLWRNVTRFLSPLSAVPLVAFSG 181
Query: 184 FGLYELGFPMLAKCIEIGLPEIVILVFLSQFMPHMMKGGR--HIFARFAVIFSVIIVWVY 241
FGLYE GFPMLAKCIEIGLPEI++LV SQ++PH+M+G + F RFAVIFSV+IVW+Y
Sbjct: 182 FGLYEQGFPMLAKCIEIGLPEIILLVIFSQYIPHLMQGETCSNFFHRFAVIFSVVIVWLY 241
Query: 242 AIILTGCGAYKNAEHETQDTCRTDRAGLIHGASWISPPIPFRWG-APTFDAGEAFAMMAA 300
A ILT GAY N E TQ +CRTDRAG+I + WI P P +WG APTF+AG+ FAMMAA
Sbjct: 242 AYILTIGGAYSNTEINTQISCRTDRAGIISASPWIRVPHPIQWGGAPTFNAGDIFAMMAA 301
Query: 301 SFVAQIESTGGFIAVARFASATPVPPSVLSRGIGWQGVGILLSGIFGTGNGSSVSIENAG 360
SFV+ +ESTG +IAV+R+ASATP+PPSVLSRGIGWQG GILL G+FG GN +SVS+ENAG
Sbjct: 302 SFVSLVESTGTYIAVSRYASATPIPPSVLSRGIGWQGFGILLCGLFGAGNATSVSVENAG 361
Query: 361 LLALTRVGSRRVVQISAGFMIFFSILGKFGAVFASIPMPIVAALYCLLFSQVGSAGLSFL 420
LLA+TRVGSRRV+Q++AGFMIFFSILGKFGA+FASIP PIVAALYCL FS VG+ GLS +
Sbjct: 362 LLAVTRVGSRRVIQVAAGFMIFFSILGKFGAIFASIPAPIVAALYCLFFSYVGAGGLSLI 421
Query: 421 QFCNLNSFRTKFIIGFSIFMGFSVPQYFKEYTAIKQYGPVHTNARWFNDMINVPFSSGAF 480
QFCNLNSFRTKFI+GFSIFMG S+PQYF +YT ++ YGPV T+A WFN++INVPFSS AF
Sbjct: 422 QFCNLNSFRTKFILGFSIFMGLSIPQYFYQYTTLETYGPVRTSATWFNNIINVPFSSKAF 481
Query: 481 VAGILALFFDVTLHKSDNQTRKDRGMHWWDRFSSFKTDTRSEEFYSLPFNLNKFFPS 537
V+GILA F D TL D T+KDRG+ WW RF SF++D RSEEFYSLP NL+K+FPS
Sbjct: 482 VSGILAFFLDTTLPPKDKTTKKDRGLVWWKRFKSFQSDNRSEEFYSLPLNLSKYFPS 538
>At1g49960 permease, putative
Length = 526
Score = 636 bits (1640), Expect = 0.0
Identities = 310/523 (59%), Positives = 386/523 (73%), Gaps = 1/523 (0%)
Query: 15 KQDEFQPHPVKDQLPNVSYCITSPPPWPEAIMLGFQHYLVMLGTTVLIPTALVSQMGGGN 74
K D+F P PVKDQLP V +C++S P WPE I+LGFQHY+VMLGTTV+IP+ LV MGGG+
Sbjct: 4 KTDDFAPFPVKDQLPGVEFCVSSSPNWPEGIVLGFQHYIVMLGTTVIIPSILVPLMGGGD 63
Query: 75 EEKAMLIQNHLFVAGINTLIQTLFGTRLPAVIGGSFTFVPTTISIILASRYDDDIMHPRE 134
EKA +I LFV+GINTL+Q+LFG+RLP V+G S+ ++ + I + R+ +HP
Sbjct: 64 VEKAEVINTVLFVSGINTLLQSLFGSRLPVVMGASYAYLIPALYITFSYRFTY-YLHPHL 122
Query: 135 KFKRIMRGTQGALIVASSLQIIVGFSGLWCHVVRFISPLSAVPLVALTGFGLYELGFPML 194
+F+ MR QGALI+AS +I+GF GLW +VRF+SPLSA PLV LTG GL FP L
Sbjct: 123 RFEETMRAIQGALIIASISHMIMGFFGLWRILVRFLSPLSAAPLVILTGVGLLAFAFPQL 182
Query: 195 AKCIEIGLPEIVILVFLSQFMPHMMKGGRHIFARFAVIFSVIIVWVYAIILTGCGAYKNA 254
A+CIEIGLP ++IL+ LSQ++PH+ K R I +FAV+F++ IVW YA ILT GAY
Sbjct: 183 ARCIEIGLPALIILIILSQYLPHLFKCKRSICEQFAVLFTIAIVWAYAEILTAAGAYDKR 242
Query: 255 EHETQDTCRTDRAGLIHGASWISPPIPFRWGAPTFDAGEAFAMMAASFVAQIESTGGFIA 314
TQ +CRTDR+GLI + W+ P P +WG P+F +AFAMMAA++VA +E+TG FIA
Sbjct: 243 PDNTQLSCRTDRSGLISASPWVRIPYPLQWGRPSFHGSDAFAMMAATYVAIVETTGSFIA 302
Query: 315 VARFASATPVPPSVLSRGIGWQGVGILLSGIFGTGNGSSVSIENAGLLALTRVGSRRVVQ 374
+RF SAT +PPSVLSRGIGWQG+G+LL+G+FGT GS+ +EN GLL LT+VGSRRVVQ
Sbjct: 303 ASRFGSATHIPPSVLSRGIGWQGIGVLLNGLFGTATGSTALVENTGLLGLTKVGSRRVVQ 362
Query: 375 ISAGFMIFFSILGKFGAVFASIPMPIVAALYCLLFSQVGSAGLSFLQFCNLNSFRTKFII 434
ISAGFMIFFSI GKFGAV ASIP+PI AALYC+LF+ V SAGL LQFCNLNSFR KFI+
Sbjct: 363 ISAGFMIFFSIFGKFGAVLASIPLPIFAALYCVLFAYVASAGLGLLQFCNLNSFRNKFIL 422
Query: 435 GFSIFMGFSVPQYFKEYTAIKQYGPVHTNARWFNDMINVPFSSGAFVAGILALFFDVTLH 494
GFSIF+G SV QYF EY I GPVHT FN ++ V FSS A V + A D T
Sbjct: 423 GFSIFIGLSVAQYFTEYLFISGRGPVHTRTSAFNVIMQVIFSSAATVGIMAAFLLDCTHS 482
Query: 495 KSDNQTRKDRGMHWWDRFSSFKTDTRSEEFYSLPFNLNKFFPS 537
R+D G HWW++F + TDTR+EEFY+LP+NLN+FFPS
Sbjct: 483 YGHASVRRDSGRHWWEKFRVYHTDTRTEEFYALPYNLNRFFPS 525
>At2g34190 putative membrane transporter
Length = 524
Score = 606 bits (1563), Expect = e-174
Identities = 298/515 (57%), Positives = 377/515 (72%), Gaps = 3/515 (0%)
Query: 22 HPVKDQLPNVSYCITSPPPWPEAIMLGFQHYLVMLGTTVLIPTALVSQMGGGNEEKAMLI 81
HP DQL + YCI S PPW EAI LGF+HY++ LGT V+IP+ LV MGG + +K ++
Sbjct: 11 HPPMDQLQGLEYCIDSNPPWGEAIALGFEHYILALGTAVMIPSILVPMMGGDDGDKVRVV 70
Query: 82 QNHLFVAGINTLIQTLFGTRLPAVIGGSFTFVPTTISIILASRYDDDIMHPREKFKRIMR 141
Q LF+ G+NTL+QTLFGTRLP VIGGS+ F+ ISII S I P+ +F MR
Sbjct: 71 QTLLFLQGVNTLLQTLFGTRLPTVIGGSYAFMVPIISIIHDSSLTR-IEDPQLRFLSTMR 129
Query: 142 GTQGALIVASSLQIIVGFSGLWCHVVRFISPLSAVPLVALTGFGLYELGFPMLAKCIEIG 201
QGA+IVASS+QII+GFS +W RF SP+ VP++ALTGFGL+ GFP++ C+EIG
Sbjct: 130 AVQGAIIVASSVQIILGFSQMWAICSRFFSPIGMVPVIALTGFGLFNRGFPVVGNCVEIG 189
Query: 202 LPEIVILVFLSQFMPHMMKGGRHIFARFAVIFSVIIVWVYAIILTGCGAYKNAEHETQDT 261
LP +++ V SQ++ + + RFA+I ++IIVW YA +LT GAYK+ H+TQ
Sbjct: 190 LPMLILFVIFSQYLKNFQFRQFPVVERFALIIALIIVWAYAHVLTASGAYKHRPHQTQLN 249
Query: 262 CRTDRAGLIHGASWISPPIPFRWGAPTFDAGEAFAMMAASFVAQIESTGGFIAVARFASA 321
CRTD + LI A WI P P +WGAP+FDAG AFAMMAA V+ IESTG F A AR ASA
Sbjct: 250 CRTDMSNLISSAPWIKIPYPLQWGAPSFDAGHAFAMMAAVLVSLIESTGAFKAAARLASA 309
Query: 322 TPVPPSVLSRGIGWQGVGILLSGIFGTGNGSSVSIENAGLLALTRVGSRRVVQISAGFMI 381
TP PP VLSRGIGWQG+GILL+G+FGT +GSSVS+EN GLL TRVGSRRV+QISAGFMI
Sbjct: 310 TPPPPHVLSRGIGWQGIGILLNGLFGTLSGSSVSVENIGLLGSTRVGSRRVIQISAGFMI 369
Query: 382 FFSILGKFGAVFASIPMPIVAALYCLLFSQVGSAGLSFLQFCNLNSFRTKFIIGFSIFMG 441
FFS+LGKFGA+FASIP I AA+YC+LF V S GLSFLQF N+NS R FI+G S+F+G
Sbjct: 370 FFSMLGKFGALFASIPFTIFAAVYCVLFGLVASVGLSFLQFTNMNSLRNLFIVGVSLFLG 429
Query: 442 FSVPQYFKEYTAIKQYGPVHTNARWFNDMINVPFSSGAFVAGILALFFDVTLHKSDNQTR 501
S+P+YF++++ +GP HTNA WFND +N F S VA ++A+F D TL +T
Sbjct: 430 LSIPEYFRDFSMKALHGPAHTNAGWFNDFLNTIFLSSPMVALMVAVFLDNTL--DYKETA 487
Query: 502 KDRGMHWWDRFSSFKTDTRSEEFYSLPFNLNKFFP 536
+DRG+ WW +F +FK D+R+EEFY+LPFNLN+FFP
Sbjct: 488 RDRGLPWWAKFRTFKGDSRNEEFYTLPFNLNRFFP 522
>At2g05760 putative membrane transporter
Length = 520
Score = 575 bits (1481), Expect = e-164
Identities = 283/516 (54%), Positives = 364/516 (69%), Gaps = 4/516 (0%)
Query: 22 HPVKDQLPNVSYCITSPPPWPEAIMLGFQHYLVMLGTTVLIPTALVSQMGGGNEEKAMLI 81
HP +QL ++ YCI S PPWPE ++L FQ+Y++MLGT+ IP LV MGG + ++A +I
Sbjct: 6 HPPMEQLQDLEYCIDSNPPWPETVLLAFQNYILMLGTSAFIPALLVPAMGGSDGDRARVI 65
Query: 82 QNHLFVAGINTLIQTLFGTRLPAVIGGSFTFVPTTISIILASRYDDDIMHPREKFKRIMR 141
Q LFVAGI TL+Q LFGTRLPAV+GGS +V I+ I+ I + E+F MR
Sbjct: 66 QTLLFVAGIKTLLQALFGTRLPAVVGGSLAYV-VPIAYIINDSSLQKISNDHERFIHTMR 124
Query: 142 GTQGALIVASSLQIIVGFSGLWCHVVRFISPLSAVPLVALTGFGLYELGFPMLAKCIEIG 201
QGALIVASS+QII+G+S +W RF SPL P+V L G G+++ GFP L CIEIG
Sbjct: 125 AIQGALIVASSIQIILGYSQVWGLFSRFFSPLGMAPVVGLVGLGMFQRGFPQLGNCIEIG 184
Query: 202 LPEIVILVFLSQFMPHMMK-GGRHIFARFAVIFSVIIVWVYAIILTGCGAYKNAEHETQD 260
LP +++++ L+Q++ H+ IF RF ++ V IVW+YA+ILT GAY+ TQ
Sbjct: 185 LPMLLLVIGLTQYLKHVRPFKDVPIFERFPILICVTIVWIYAVILTASGAYRGKPSLTQH 244
Query: 261 TCRTDRAGLIHGASWISPPIPFRWGAPTFDAGEAFAMMAASFVAQIESTGGFIAVARFAS 320
+CRTD+A LI A W P P +WG PTF G +FAMM+A V+ +ESTG +IA +R A
Sbjct: 245 SCRTDKANLISTAPWFKFPYPLQWGPPTFSVGHSFAMMSAVLVSMVESTGAYIAASRLAI 304
Query: 321 ATPVPPSVLSRGIGWQGVGILLSGIFGTGNGSSVSIENAGLLALTRVGSRRVVQISAGFM 380
ATP P VLSRGIGWQG+G+LL G+FGTG GS+V +EN GLL LTRVGSRRVVQ+SAGFM
Sbjct: 305 ATPPPAYVLSRGIGWQGIGVLLDGLFGTGTGSTVLVENVGLLGLTRVGSRRVVQVSAGFM 364
Query: 381 IFFSILGKFGAVFASIPMPIVAALYCLLFSQVGSAGLSFLQFCNLNSFRTKFIIGFSIFM 440
I FS LGKFGAVFASIP+PI AAL+C+LF V + GLSFLQF N+NS R I G S+F+
Sbjct: 365 IVFSTLGKFGAVFASIPVPIYAALHCILFGLVAAVGLSFLQFTNMNSMRNLMITGLSLFL 424
Query: 441 GFSVPQYFKEYTAIKQYGPVHTNARWFNDMINVPFSSGAFVAGILALFFDVTLHKSDNQT 500
G S+PQ+F +Y + YG VHTNA WFN +N F S A V I+A+F D T+ ++
Sbjct: 425 GISIPQFFAQYWDARHYGLVHTNAGWFNAFLNTLFMSPATVGLIIAVFMDNTMEV--ERS 482
Query: 501 RKDRGMHWWDRFSSFKTDTRSEEFYSLPFNLNKFFP 536
+KDRGM WW +F +F+ D R+EEFY+LPFNLN+FFP
Sbjct: 483 KKDRGMPWWVKFRTFRGDNRNEEFYTLPFNLNRFFP 518
>At1g65550 hypothetical protein
Length = 515
Score = 540 bits (1390), Expect = e-153
Identities = 270/542 (49%), Positives = 355/542 (64%), Gaps = 38/542 (7%)
Query: 3 GGGGGGGATPPPKQDEFQPHPVKDQLPNVSYCITSPPPWPEAIMLGFQHYLVMLGTTVLI 62
GGGG GA + +E QPHPVK+QLP + YC+ SPPPW EA++LGFQHYL+ LG TVLI
Sbjct: 4 GGGGNNGAAN--RTEELQPHPVKEQLPGIQYCVNSPPPWLEAVVLGFQHYLLSLGITVLI 61
Query: 63 PTALVSQMGGGNEEKAMLIQNHLFVAGINTLIQTLFGTRLPAVIGGSFTFVPTTISIILA 122
P+ LV MGGG EK +IQ LFV+G+ TL Q+ FGTRLP + S+ ++ SII +
Sbjct: 62 PSVLVPLMGGGYAEKVKVIQTLLFVSGLTTLFQSFFGTRLPVIAVASYAYIIPITSIIYS 121
Query: 123 SRYDDDIMHPREKFKRIMRGTQGALIVASSLQIIVGFSGLWCHVVRFISPLSAVPLVALT 182
+R+ P E+F R MR QGALI+ Q+++ G+W ++VRF+SPLS PL T
Sbjct: 122 TRFTY-YTDPFERFVRTMRSIQGALIITGCFQVLICILGVWRNIVRFLSPLSIAPLATFT 180
Query: 183 GFGLYELGFPMLAKCIEIGLPEIVILVFLSQFMPHMMKGGRHIFA-------RFAVIFSV 235
G GLY +GFP+LA+C+E+GLP +++L+F++Q++P +K + + R+ +I +
Sbjct: 181 GLGLYHIGFPLLARCVEVGLPGLILLIFVTQYLPRFLKMKKGVMILDGSRCDRYGMILCI 240
Query: 236 IIVWVYAIILTGCGAYKNAEHETQDTCRTDRAGLIHGASWISPPIPFRWGAPTFDAGEAF 295
+VW++A +LT G Y + H TQ +CRTDR GLI WI P PF+WG+PTFD ++F
Sbjct: 241 PLVWLFAQLLTSSGVYDHKSHTTQTSCRTDRTGLITNTPWIYIPYPFQWGSPTFDITDSF 300
Query: 296 AMMAASFVAQIESTGGFIAVARFASATPVPPSVLSRGIGWQGVGILLSGIFGTGNGSSVS 355
AMMAASFV ESTG F A AR+ SATP+PPSV+SRG W GVG+LL+G+ G G + S
Sbjct: 301 AMMAASFVTLFESTGLFYASARYGSATPIPPSVVSRGTCWLGVGVLLNGMLGGITGITTS 360
Query: 356 IENAGLLALTRVGSRRVVQISAGFMIFFSILGKFGAVFASIPMPIVAALYCLLFSQVGSA 415
EN GLLA+T++GSRRV+QISA FMIFFSI FAS+
Sbjct: 361 TENVGLLAMTKIGSRRVIQISAAFMIFFSI-------FASV------------------- 394
Query: 416 GLSFLQFCNLNSFRTKFIIGFSIFMGFSVPQYFKEYTAIKQYGPVHTNARWFNDMINVPF 475
GLS+LQFCNLNSF KFI+GFS FM S+PQYF+EY H+N W DMI V F
Sbjct: 395 GLSYLQFCNLNSFNIKFILGFSFFMAISIPQYFREYYNGGWRSDHHSN--WLEDMIRVIF 452
Query: 476 SSGAFVAGILALFFDVTLHKSDNQTRKDRGMHWWDRFSSFKTDTRSEEFYSLPFNLNKFF 535
S VA I+A+ D TL + ++ +KD GM WWD+F + D R++EFY LP LNKFF
Sbjct: 453 MSHTTVAAIIAIVLDCTLCRDSDEAKKDCGMKWWDKFRLYNLDVRNDEFYGLPCRLNKFF 512
Query: 536 PS 537
PS
Sbjct: 513 PS 514
>At2g26510 putative membrane transporter
Length = 551
Score = 496 bits (1276), Expect = e-140
Identities = 250/521 (47%), Positives = 338/521 (63%), Gaps = 14/521 (2%)
Query: 23 PVKDQLPNVSYCITSPPPWPEAIMLGFQHYLVMLGTTVLIPTALVSQMGGGNEEKAMLIQ 82
P +QL ++ YCI S P W E ++L FQHY+VMLGTTVLI LVS MGG +KA +IQ
Sbjct: 36 PPAEQLHHLQYCIHSNPSWHETVVLAFQHYIVMLGTTVLIANTLVSPMGGDPGDKARVIQ 95
Query: 83 NHLFVAGINTLIQTLFGTRLPAVIGGSFTFVPTTISIILA---SRYDDDIMHPREKFKRI 139
LF++GINTL+QTL GTRLP V+G SF +V +SII ++D + +++F+
Sbjct: 96 TILFMSGINTLLQTLIGTRLPTVMGVSFAYVLPVLSIIRDYNNGQFDSE----KQRFRHT 151
Query: 140 MRGTQGALIVASSLQIIVGFSGLWCHVVRFISPLSAVPLVALTGFGLYELGFPMLAKCIE 199
MR QG+LI++S + II+G+ W +++R SP+ VP+V++ GL+ GFP+LA C+E
Sbjct: 152 MRTVQGSLIISSFVNIIIGYGQAWGNLIRIFSPIIVVPVVSVVSLGLFLRGFPLLANCVE 211
Query: 200 IGLPEIVILVFLSQFMPHMMKGGRHIFARFAVIFSVIIVWVYAIILTGCGAYKNAEHETQ 259
IGLP +++L+ Q++ H I R+A++ + I+W +A ILT GAY N T+
Sbjct: 212 IGLPMLILLIITQQYLKHAFSRISMILERYALLVCLAIIWAFAAILTVSGAYNNVSTATK 271
Query: 260 DTCRTDRAGLIHGASWISPPIPFRWGAPTFDAGEAFAMMAASFVAQIESTGGFIAVARFA 319
+CRTDRA L+ A WI P PF+WG P F A F M A+ VA ESTG F A +R A
Sbjct: 272 QSCRTDRAFLMSSAPWIRIPYPFQWGTPIFKASHVFGMFGAAIVASAESTGVFFAASRLA 331
Query: 320 SATPVPPSVLSRGIGWQGVGILLSGIFGTGNGSSVSIENAGLLALTRVGSRRVVQISAGF 379
AT P V+SR IG QG+G+LL GIFG+ G++ S+EN GLL LTR+GSRRVVQ+S F
Sbjct: 332 GATAPPAHVVSRSIGLQGIGVLLEGIFGSITGNTASVENVGLLGLTRIGSRRVVQVSTFF 391
Query: 380 MIFFSILGKFGAVFASIPMPIVAALYCLLFSQVGSAGLSFLQFCNLNSFRTKFIIGFSIF 439
MIFFSI GKFGA FASIP+PI A +YC+L V + G+SF+QF + NS R ++IG S+F
Sbjct: 392 MIFFSIFGKFGAFFASIPLPIFAGVYCILLGIVVAVGISFIQFTDTNSMRNMYVIGVSLF 451
Query: 440 MGFSVPQYFKEYTAIKQYGPVHTNARWFNDMINVPFSSGAFVAGILALFFDVTL---HKS 496
+ S+ QYF T+ YGPV T WFND++N F+S VA ILA D TL H S
Sbjct: 452 LSLSIAQYFLANTSRAGYGPVRTAGGWFNDILNTIFASAPLVATILATILDNTLEARHAS 511
Query: 497 DNQTRKDRGMHWWDRFSSFKTDTRSEEFYSLPFNLNKFFPS 537
D+ RG+ WW F D R++EFYS+P +N+ P+
Sbjct: 512 DDA----RGIPWWKPFQHRNGDGRNDEFYSMPLRINELMPT 548
>At5g25420 permease 1-like protein
Length = 419
Score = 398 bits (1022), Expect = e-111
Identities = 215/451 (47%), Positives = 274/451 (60%), Gaps = 70/451 (15%)
Query: 2 AGGGGGGGAT-PPPKQDEFQPHPVKDQLPNVSYCITSPPPWPEAIMLGFQHYLVMLGTTV 60
AGG GGGG + +E QPHPVK+QLP + YC+ SPPPW EA++LGFQHYL+ LG TV
Sbjct: 12 AGGNGGGGNNGAGNRAEELQPHPVKEQLPGIQYCVNSPPPWLEAVVLGFQHYLLSLGITV 71
Query: 61 LIPTALVSQMGGGNEEKAMLIQNHLFVAGINTLIQTLFGTRLPAVIGGSFTFVPTTISII 120
LIP+ LV MGGG+ EK +IQ LFV+G+ TL Q+ FGTRLP + S+ ++ SII
Sbjct: 72 LIPSLLVPLMGGGDAEKVKVIQTLLFVSGLTTLFQSFFGTRLPVIASASYAYIIPITSII 131
Query: 121 LASRYDDDIMHPREKFKRIMRGTQGALIVASSLQIIVGFSGLWCHVVRFISPLSAVPLVA 180
++R+ P E+F R MR QGALI+ Q++V F G+W ++VRF+SPLS PLV
Sbjct: 132 YSTRF-TYYTDPFERFVRTMRSIQGALIITGCFQVLVCFLGVWRNIVRFLSPLSIAPLVT 190
Query: 181 LTGFGLYELGFPMLAKCIEIGLPEIVILVFLSQFMPHMMKGGRHIFARFAVIFSVIIVWV 240
TG GLY +GFP++ K P + G R R+ ++ + +VW+
Sbjct: 191 FTGLGLYHIGFPLVKK------------------GPMIWDGNR--CDRYGMMLCIPVVWL 230
Query: 241 YAIILTGCGAYKNAEHETQDTCRTDRAGLIHGASWISPPIPFRWGAPTFDAGEAFAMMAA 300
+A +LT G Y + TQ +CRTDR GLI PTFD ++FAMMAA
Sbjct: 231 FAQLLTSSGVYDHKPQTTQTSCRTDRTGLITNTP-----------CPTFDITDSFAMMAA 279
Query: 301 SFVAQIESTGGFIAVARFASATPVPPSVLSRGIGWQGVGILLSGIFGTGNGSSVSIENAG 360
SFV ESTG F A AR+ +N G
Sbjct: 280 SFVTLFESTGLFYASARYG-------------------------------------KNVG 302
Query: 361 LLALTRVGSRRVVQISAGFMIFFSILGKFGAVFASIPMPIVAALYCLLFSQVGSAGLSFL 420
LLA+T+VGSRRV+QISA FM+FFSI GKFGA FASIP+PI+A+LYC++ V SAGLSFL
Sbjct: 303 LLAMTKVGSRRVIQISAAFMLFFSIFGKFGAFFASIPLPIMASLYCIVLCFVSSAGLSFL 362
Query: 421 QFCNLNSFRTKFIIGFSIFMGFSVPQYFKEY 451
QFCNLNSF TKFI+GFS FM S+PQYF+EY
Sbjct: 363 QFCNLNSFNTKFILGFSFFMAISIPQYFREY 393
>At2g27810 putative membrane transporter
Length = 721
Score = 312 bits (799), Expect = 3e-85
Identities = 184/590 (31%), Positives = 298/590 (50%), Gaps = 63/590 (10%)
Query: 3 GGGGGGGATPPPKQD-------EFQPHPVKDQLP----NVSYCITSPPPWPEAIMLGFQH 51
G G G+ P ++ E P + D L ++ Y + P G QH
Sbjct: 130 GPDGANGSGDPVRRPGRIEETVEVLPQSMDDDLVARNLHMKYGLRDTPGLVPIGFYGLQH 189
Query: 52 YLVMLGTTVLIPTALVSQMGGGNEEKAMLIQNHLFVAGINTLIQTLFGTRLPAVIGGSFT 111
YL MLG+ +L+P +V MGG +EE A ++ LFV+GI TL+ T FG+RLP + G SF
Sbjct: 190 YLSMLGSLILVPLVIVPAMGGSHEEVANVVSTVLFVSGITTLLHTSFGSRLPLIQGPSFV 249
Query: 112 FVPTTISIILASRYDDDIMHPREKFKRIMRGTQGALIVASSLQIIVGFSGLWCHVVRFIS 171
F+ ++II + + ++ FK IMR QGA+I+ S+ Q ++G+SGL ++R ++
Sbjct: 250 FLAPALAIINSPEFQG--LNGNNNFKHIMRELQGAIIIGSAFQAVLGYSGLMSLILRLVN 307
Query: 172 PLSAVPLVALTGFGLYELGFPMLAKCIEIGLPEIVILVFLSQFMPHMMKGGRHIFARFAV 231
P+ P VA G Y GFP++ KC+EIG+ +I++++ + ++ + IF +AV
Sbjct: 308 PVVVAPTVAAVGLSFYSYGFPLVGKCLEIGVVQILLVIIFALYLRKISVLSHRIFLIYAV 367
Query: 232 IFSVIIVWVYAIILTGCGAY--KNAEHETQDT----------------CRTDRAGLIHGA 273
S+ I W A +LT GAY K + + CR D + + A
Sbjct: 368 PLSLAITWAAAFLLTETGAYTYKGCDPNVPVSNVVSTHCRKYMTRMKYCRVDTSHALSSA 427
Query: 274 SWISPPIPFRWGAPTFDAGEAFAMMAASFVAQIESTGGFIAVARFASATPVPPSVLSRGI 333
W P P +WG P F+ AF M S +A ++S G + A + ++ P V+SR I
Sbjct: 428 PWFRFPYPLQWGVPLFNWKMAFVMCVVSVIASVDSVGSYHASSLLVASRPPTRGVVSRAI 487
Query: 334 GWQGVGILLSGIFGTGNGSSVSIENAGLLALTRVGSRRVVQISAGFMIFFSILGKFGAVF 393
G +G +L+G++GTG GS+ EN +A+T++GSRRVV++ A ++ FS++GK G
Sbjct: 488 GLEGFTSVLAGLWGTGTGSTTLTENVHTIAVTKMGSRRVVELGACVLVIFSLVGKVGGFL 547
Query: 394 ASIPMPIVAALYCLLFSQVGSAGLSFLQFCNLNSFRTKFIIGFSIFMGFSVPQYFKEY-- 451
ASIP +VA+L C +++ + GLS L++ S R I+G S+F SVP YF++Y
Sbjct: 548 ASIPQVMVASLLCFMWAMFTALGLSNLRYSEAGSSRNIIIVGLSLFFSLSVPAYFQQYGI 607
Query: 452 --------------TAIKQYGPVHTNAR------------WFNDMINVPFSSGAFVAGIL 485
+ +GP + + N ++N S +A I+
Sbjct: 608 SPNSNLSVPSYYQPYIVSSHGPFKSQYKGDLQFSYLLVYLQMNYVMNTLLSMSMVIAFIM 667
Query: 486 ALFFDVTLHKSDNQTRKDRGMHWWDRFSSFKTDTRSEEFYSLPFNLNKFF 535
A+ D T+ S +++RG++ W + + + Y LPF + +FF
Sbjct: 668 AVILDNTVPGS----KQERGVYVWSDSETATREPALAKDYELPFRVGRFF 713
>At4g38050 unknown protein
Length = 709
Score = 309 bits (791), Expect = 3e-84
Identities = 184/562 (32%), Positives = 283/562 (49%), Gaps = 47/562 (8%)
Query: 6 GGGGATPPPKQDEFQPHPVKDQLPNVSYCITSPPPWPEAIMLGFQHYLVMLGTTVLIPTA 65
GGGG + +D P P+ + + + P + I G QHYL ++G+ V IP
Sbjct: 158 GGGGES---SEDGQWPKPIL-----MKFGLRDNPGFVPLIYYGLQHYLSLVGSLVFIPLV 209
Query: 66 LVSQMGGGNEEKAMLIQNHLFVAGINTLIQTLFGTRLPAVIGGSFTFVPTTISIILASRY 125
+V M G +++ A +I L + G+ T++ FGTRLP V G SF ++ + +I + +
Sbjct: 210 IVPAMDGSDKDTASVISTMLLLTGVTTILHCYFGTRLPLVQGSSFVYLAPVLVVINSEEF 269
Query: 126 DDDIMHPREKFKRIMRGTQGALIVASSLQIIVGFSGLWCHVVRFISPLSAVPLVALTGFG 185
+ H KF+ MR QGA+IV S Q I+GFSGL ++RFI+P+ P VA G
Sbjct: 270 RNLTEH---KFRDTMRELQGAIIVGSLFQCILGFSGLMSLLLRFINPVVVAPTVAAVGLA 326
Query: 186 LYELGFPMLAKCIEIGLPEIVILVFLSQFMPHMMKGGRHIFARFAVIFSVIIVWVYAIIL 245
+ GFP C+EI +P I++L+ + ++ + G +F +AV S +++W YA L
Sbjct: 327 FFSYGFPQAGTCVEISVPLILLLLIFTLYLRGVSLFGHRLFRIYAVPLSALLIWTYAFFL 386
Query: 246 TGCGAYK----NAEHE--------------TQDTCRTDRAGLIHGASWISPPIPFRWGAP 287
T GAY NA+ T CRTD + ASW+ P PF+WG P
Sbjct: 387 TVGGAYDYRGCNADIPSSNILIDECKKHVYTMKHCRTDASNAWRTASWVRIPYPFQWGFP 446
Query: 288 TFDAGEAFAMMAASFVAQIESTGGFIAVARFASATPVPPSVLSRGIGWQGVGILLSGIFG 347
F + M+ S VA ++S G + + + +A ++SRGI +G LL+GI+G
Sbjct: 447 NFHMRTSIIMIFVSLVASVDSVGTYHSASMIVNAKRPTRGIVSRGIALEGFCSLLAGIWG 506
Query: 348 TGNGSSVSIENAGLLALTRVGSRRVVQISAGFMIFFSILGKFGAVFASIPMPIVAALYCL 407
+G GS+ EN + +T+V SRR + I A F+I S LGK GA+ ASIP + A++ C
Sbjct: 507 SGTGSTTLTENIHTINITKVASRRALVIGAMFLIVLSFLGKLGAILASIPQALAASVLCF 566
Query: 408 LFSQVGSAGLSFLQFCNLNSFRTKFIIGFSIFMGFSVPQYFKEYTAIKQY---------- 457
+++ S GLS L++ SFR I+G S+F+G S+P YF++Y +
Sbjct: 567 IWALTVSLGLSNLRYTQTASFRNITIVGVSLFLGLSIPAYFQQYQPLSSLILPSYYIPFG 626
Query: 458 ----GPVHTNARWFNDMINVPFSSGAFVAGILALFFDVTLHKSDNQTRKDRGMHWWDRFS 513
GP T + +N S V +LA D T+ S +++RG++ W R
Sbjct: 627 AASSGPFQTGIEQLDFAMNAVLSLNMVVTFLLAFILDNTVPGS----KEERGVYVWTRAE 682
Query: 514 SFKTDTRSEEFYSLPFNLNKFF 535
+ D YSLP + F
Sbjct: 683 DMQMDPEMRADYSLPRKFAQIF 704
>At5g27690 unknown protein (At5g27690)
Length = 352
Score = 34.7 bits (78), Expect = 0.14
Identities = 18/40 (45%), Positives = 21/40 (52%), Gaps = 1/40 (2%)
Query: 1 MAGGGGGGGATPPPKQDEFQPHPVKDQLPNVSYCITSPPP 40
M GGGGGGG PP Q++ P V P + I PPP
Sbjct: 191 MEGGGGGGGGGGPP-QNDGPPETVIYSAPQDHHIIHGPPP 229
>At3g19430 putative late embryogenesis abundant protein
Length = 550
Score = 32.0 bits (71), Expect = 0.89
Identities = 14/39 (35%), Positives = 19/39 (47%)
Query: 4 GGGGGGATPPPKQDEFQPHPVKDQLPNVSYCITSPPPWP 42
GG GG TPP P P +P+ + ++ PPP P
Sbjct: 66 GGDDGGYTPPAPVPPVSPPPPTPSVPSPTPPVSPPPPTP 104
>At1g08920 putative sugar transport protein
Length = 470
Score = 29.6 bits (65), Expect = 4.4
Identities = 24/84 (28%), Positives = 36/84 (42%), Gaps = 8/84 (9%)
Query: 360 GLLALTRVGSRRVVQISAGFMIFFSILGKFGAVFASIPM-----PIVAALYCLLFSQ--- 411
GL+ + ++G R ++ S G M FFS+L F F S M PI + + F
Sbjct: 319 GLILVEKMGRRPLLLASTGGMCFFSLLLSFSFCFRSYGMLDELTPIFTCIGVVGFISSFA 378
Query: 412 VGSAGLSFLQFCNLNSFRTKFIIG 435
VG GL ++ + K G
Sbjct: 379 VGMGGLPWIIMSEIFPMNVKVSAG 402
>At5g03680 GT2 -like protein
Length = 591
Score = 28.9 bits (63), Expect = 7.5
Identities = 18/44 (40%), Positives = 22/44 (49%)
Query: 325 PPSVLSRGIGWQGVGILLSGIFGTGNGSSVSIENAGLLALTRVG 368
PP L + Q +G L GI G G GSS + N+ L A T G
Sbjct: 50 PPHRLHQFTTDQDMGFLPRGIHGLGGGSSTAGNNSNLNASTSGG 93
>At4g04050 putative transposon protein
Length = 681
Score = 28.9 bits (63), Expect = 7.5
Identities = 20/65 (30%), Positives = 28/65 (42%), Gaps = 3/65 (4%)
Query: 11 TPPPKQDEFQPHPVKDQLPNVSYCITSPPPWPEAIMLGFQHYLVML---GTTVLIPTALV 67
TPPP + E + P + + IT+PPP P + F L+ T L P +
Sbjct: 422 TPPPDEQEEEQTPKQKKRERTPEGITTPPPAPHKKNMRFPLRLLKFCRSATEHLSPKRID 481
Query: 68 SQMGG 72
MGG
Sbjct: 482 FIMGG 486
>At3g06770 unknown protein
Length = 377
Score = 28.9 bits (63), Expect = 7.5
Identities = 21/92 (22%), Positives = 37/92 (39%), Gaps = 14/92 (15%)
Query: 424 NLNSFRTKFIIGFSIFMGFSVPQYFKEYTAIKQYGP-VHTNARWFNDMINVPFSSGAFVA 482
+L S T F+ ++ + P +++ + YG + + + +IN V
Sbjct: 24 SLTSHLTLFLENGAVIVASQDPSHWEVVDPLPSYGRGIDLPGKRYKSLINGNKLHDVVVT 83
Query: 483 GILALFFDVTLHKSDNQTRKDRGMHWWDRFSS 514
G DN T +G+ WWDRF+S
Sbjct: 84 G-------------DNGTIDGQGLVWWDRFTS 102
>At2g19580 putative senescence-associated protein 5
Length = 270
Score = 28.9 bits (63), Expect = 7.5
Identities = 14/40 (35%), Positives = 24/40 (60%), Gaps = 5/40 (12%)
Query: 231 VIFSVIIVWVYAIILTGCGAYKNAEHETQDTCRTDRAGLI 270
+I V+++WVY I C A++NA +T+D R + G +
Sbjct: 236 IITVVVLIWVYVI---ACSAFRNA--QTEDLFRKYKQGWV 270
>At1g62500 putative proline-rich cell wall protein (pir|IS52985);
similar to ESTs gb|AI239404, gb|R89984, and emb|Z17709
Length = 297
Score = 28.9 bits (63), Expect = 7.5
Identities = 10/12 (83%), Positives = 11/12 (91%)
Query: 3 GGGGGGGATPPP 14
GGGGGGG+ PPP
Sbjct: 44 GGGGGGGSKPPP 55
Score = 28.5 bits (62), Expect = 9.8
Identities = 19/43 (44%), Positives = 23/43 (53%), Gaps = 6/43 (13%)
Query: 3 GGGGGGGATPP---PKQDEFQPHPVKDQLPNVSY--CITSPPP 40
GGGGGGG +PP P +P P+ + P V Y I PPP
Sbjct: 80 GGGGGGGKSPPVVRPPPVVVRPPPI-IRPPPVVYPPPIVRPPP 121
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.327 0.142 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,368,796
Number of Sequences: 26719
Number of extensions: 548058
Number of successful extensions: 2985
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 2915
Number of HSP's gapped (non-prelim): 34
length of query: 538
length of database: 11,318,596
effective HSP length: 104
effective length of query: 434
effective length of database: 8,539,820
effective search space: 3706281880
effective search space used: 3706281880
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC126785.11


