

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125481.10 + phase: 0 /pseudo
(116 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
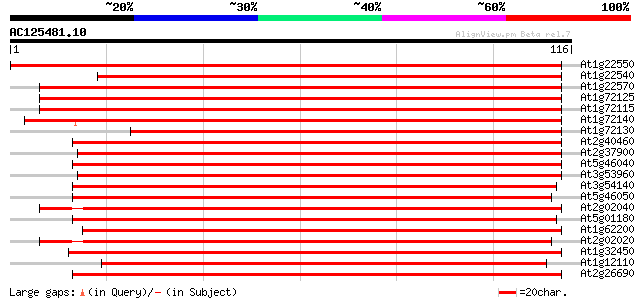
At1g22550 Similar to LeOPT1 [Lycopersicon esculentum 156 2e-39
At1g22540 Similar to peptide transporter 155 3e-39
At1g22570 Similar to LeOPT1 [Lycopersicon esculentum 152 3e-38
At1g72125 oligopeptide like transporter 149 2e-37
At1g72115 oligopeptide like transporter 140 1e-34
At1g72140 peptide transporter like protein (PTR2-B) 128 4e-31
At1g72130 peptide transporter PTR2-B -like protein 127 1e-30
At2g40460 putative PTR2 family peptide transporter 114 1e-26
At2g37900 putative peptide/amino acid transporter 111 7e-26
At5g46040 peptide transporter 108 4e-25
At3g53960 transporter like protein 107 7e-25
At3g54140 peptide transport - like protein 106 2e-24
At5g46050 peptide transporter 105 5e-24
At2g02040 histidine transport protein (PTR2-B) 104 6e-24
At5g01180 oligopeptide transporter - like protein 99 4e-22
At1g62200 unknown protein (At1g62200) 98 8e-22
At2g02020 putative peptide/amino acid transporter 97 1e-21
At1g32450 peptide transporter PTR2-B, putative 94 1e-20
At1g12110 nitrate/chlorate transporter CHL1 93 2e-20
At2g26690 putative nitrate transporter 92 5e-20
>At1g22550 Similar to LeOPT1 [Lycopersicon esculentum
Length = 564
Score = 156 bits (394), Expect = 2e-39
Identities = 76/114 (66%), Positives = 94/114 (81%)
Query: 1 MAGGDCETRLLLETVDGVVDFKGRPVLRSSSGGWRAAAFIISVEVAERFAYYGINSNLIN 60
MA + E L+ ++V VD +G P +SS+GGWR+A +II VEV ERFAY+GI SNLI
Sbjct: 1 MAIAEEEAALIEDSVSDSVDHRGLPAGKSSTGGWRSAWYIIGVEVGERFAYFGIGSNLIT 60
Query: 61 YLTGPLGQSTATAAENVNIWSGTASLLPLLGAFLADSFLGRYRTIIIASLVYIL 114
YLTGPLGQSTATAA NVN WSGTAS+LP+LGAF+AD++LGRYRTI++ASL+YIL
Sbjct: 61 YLTGPLGQSTATAAVNVNTWSGTASILPVLGAFIADAYLGRYRTIVVASLIYIL 114
>At1g22540 Similar to peptide transporter
Length = 557
Score = 155 bits (392), Expect = 3e-39
Identities = 74/96 (77%), Positives = 86/96 (89%)
Query: 19 VDFKGRPVLRSSSGGWRAAAFIISVEVAERFAYYGINSNLINYLTGPLGQSTATAAENVN 78
VD++ +P ++SSSGGWR+A FII VEVAERFAYYGI+SNLI YLTGPLGQSTA AA NVN
Sbjct: 19 VDYRNKPAVKSSSGGWRSAGFIIGVEVAERFAYYGISSNLITYLTGPLGQSTAAAAANVN 78
Query: 79 IWSGTASLLPLLGAFLADSFLGRYRTIIIASLVYIL 114
WSGTASLLPLLGAF+ADSFLGR+RTI+ AS +YI+
Sbjct: 79 AWSGTASLLPLLGAFVADSFLGRFRTILAASALYIV 114
>At1g22570 Similar to LeOPT1 [Lycopersicon esculentum
Length = 565
Score = 152 bits (383), Expect = 3e-38
Identities = 75/108 (69%), Positives = 89/108 (81%)
Query: 7 ETRLLLETVDGVVDFKGRPVLRSSSGGWRAAAFIISVEVAERFAYYGINSNLINYLTGPL 66
E LL + V VD +G P +SS+GGWR+A FII VEVAERFAY+GI NLI YLTGPL
Sbjct: 7 EVALLEDYVSDSVDHRGFPAGKSSTGGWRSAWFIIGVEVAERFAYFGIACNLITYLTGPL 66
Query: 67 GQSTATAAENVNIWSGTASLLPLLGAFLADSFLGRYRTIIIASLVYIL 114
GQSTA AA NVN WSGTAS+LP+LGAF+AD++LGRYRTI++ASL+YIL
Sbjct: 67 GQSTAKAAVNVNTWSGTASILPILGAFVADAYLGRYRTIVVASLIYIL 114
>At1g72125 oligopeptide like transporter
Length = 557
Score = 149 bits (376), Expect = 2e-37
Identities = 75/108 (69%), Positives = 87/108 (80%)
Query: 7 ETRLLLETVDGVVDFKGRPVLRSSSGGWRAAAFIISVEVAERFAYYGINSNLINYLTGPL 66
E L E V VD +G RS++G WRAA FII VEVAERFAYYGI SNLI+YLTGPL
Sbjct: 6 EISLQEEYVTDAVDHRGLAARRSNTGRWRAALFIIGVEVAERFAYYGIGSNLISYLTGPL 65
Query: 67 GQSTATAAENVNIWSGTASLLPLLGAFLADSFLGRYRTIIIASLVYIL 114
G+STA AA NVN WSG A+LLP+LGAF+AD+FLGRYRTIII+SL+Y+L
Sbjct: 66 GESTAVAAANVNAWSGIATLLPVLGAFVADAFLGRYRTIIISSLIYVL 113
>At1g72115 oligopeptide like transporter
Length = 561
Score = 140 bits (353), Expect = 1e-34
Identities = 73/108 (67%), Positives = 84/108 (77%)
Query: 7 ETRLLLETVDGVVDFKGRPVLRSSSGGWRAAAFIISVEVAERFAYYGINSNLINYLTGPL 66
+T L E V VD +G RS +G WRAA FII VEVAERFA YGI SNLI+YLTGPL
Sbjct: 6 KTSLQEEYVIDAVDHRGFSARRSITGRWRAAWFIIGVEVAERFANYGIGSNLISYLTGPL 65
Query: 67 GQSTATAAENVNIWSGTASLLPLLGAFLADSFLGRYRTIIIASLVYIL 114
GQSTA AA NVN WSG +++LPLLGAF+AD+FLGRY TIIIAS +Y+L
Sbjct: 66 GQSTAVAAANVNAWSGISTILPLLGAFVADAFLGRYITIIIASFIYVL 113
>At1g72140 peptide transporter like protein (PTR2-B)
Length = 555
Score = 128 bits (322), Expect = 4e-31
Identities = 63/113 (55%), Positives = 83/113 (72%), Gaps = 2/113 (1%)
Query: 4 GDCETRLLL--ETVDGVVDFKGRPVLRSSSGGWRAAAFIISVEVAERFAYYGINSNLINY 61
GD ET ++ E V+ VDF+G P +RSSSG W+++ F + EVAE+FAY+GI SNLI Y
Sbjct: 6 GDNETGAIVSNENVEFSVDFRGNPSIRSSSGAWKSSGFTMCAEVAEKFAYFGIASNLITY 65
Query: 62 LTGPLGQSTATAAENVNIWSGTASLLPLLGAFLADSFLGRYRTIIIASLVYIL 114
T LG+STA AA NVN+W GTA+ LPL+ +ADSFLGR+RTI++ S YI+
Sbjct: 66 FTEALGESTAVAASNVNLWLGTAAFLPLIWGSIADSFLGRFRTILLTSSFYIM 118
>At1g72130 peptide transporter PTR2-B -like protein
Length = 538
Score = 127 bits (318), Expect = 1e-30
Identities = 60/89 (67%), Positives = 75/89 (83%)
Query: 26 VLRSSSGGWRAAAFIISVEVAERFAYYGINSNLINYLTGPLGQSTATAAENVNIWSGTAS 85
+ ++SGGW++A II V++AERFAY+GI SNLI YLTGPLG+STA AA NVN W+GT +
Sbjct: 21 IRENTSGGWKSARLIIVVQMAERFAYFGIASNLIMYLTGPLGESTAAAAANVNAWTGTVA 80
Query: 86 LLPLLGAFLADSFLGRYRTIIIASLVYIL 114
LPLLG FLADS+LGR+RTIII+S +YIL
Sbjct: 81 FLPLLGGFLADSYLGRFRTIIISSSLYIL 109
>At2g40460 putative PTR2 family peptide transporter
Length = 583
Score = 114 bits (284), Expect = 1e-26
Identities = 51/101 (50%), Positives = 71/101 (69%)
Query: 14 TVDGVVDFKGRPVLRSSSGGWRAAAFIISVEVAERFAYYGINSNLINYLTGPLGQSTATA 73
T DG VD +GRPVL S +G WRA +F++ E ER A+YGI SNL+NYLT L + T ++
Sbjct: 8 TQDGTVDLQGRPVLASKTGRWRACSFLLGYEAFERMAFYGIASNLVNYLTKRLHEDTISS 67
Query: 74 AENVNIWSGTASLLPLLGAFLADSFLGRYRTIIIASLVYIL 114
NVN WSG + P+ GA++ADS++GR+ T +SL+Y+L
Sbjct: 68 VRNVNNWSGAVWITPIAGAYIADSYIGRFWTFTASSLIYVL 108
>At2g37900 putative peptide/amino acid transporter
Length = 575
Score = 111 bits (277), Expect = 7e-26
Identities = 48/100 (48%), Positives = 73/100 (73%)
Query: 15 VDGVVDFKGRPVLRSSSGGWRAAAFIISVEVAERFAYYGINSNLINYLTGPLGQSTATAA 74
+D +D +GR LR+ +G WRAA FII++E +ER +Y+G+ +NL+ YLT L Q A
Sbjct: 23 LDSSLDSRGRVPLRARTGAWRAALFIIAIEFSERLSYFGLATNLVVYLTTILNQDLKMAI 82
Query: 75 ENVNIWSGTASLLPLLGAFLADSFLGRYRTIIIASLVYIL 114
NVN WSG +L+PLLG F+AD++LGRY T+++A+ +Y++
Sbjct: 83 RNVNYWSGVTTLMPLLGGFIADAYLGRYATVLVATTIYLM 122
>At5g46040 peptide transporter
Length = 586
Score = 108 bits (270), Expect = 4e-25
Identities = 50/101 (49%), Positives = 70/101 (68%)
Query: 14 TVDGVVDFKGRPVLRSSSGGWRAAAFIISVEVAERFAYYGINSNLINYLTGPLGQSTATA 73
T DG VD +G V RS +G W+A +F++ EV ER AYYGI+SNL+ Y+T L Q T +
Sbjct: 11 TKDGTVDLRGNRVRRSQTGRWKACSFVVVYEVFERMAYYGISSNLVIYMTTKLHQGTVKS 70
Query: 74 AENVNIWSGTASLLPLLGAFLADSFLGRYRTIIIASLVYIL 114
+ NV W GT+ L P+LGA++AD+ GRY T +I+S +Y+L
Sbjct: 71 SNNVTNWVGTSWLTPILGAYVADAHFGRYITFVISSAIYLL 111
>At3g53960 transporter like protein
Length = 620
Score = 107 bits (268), Expect = 7e-25
Identities = 47/100 (47%), Positives = 71/100 (71%)
Query: 15 VDGVVDFKGRPVLRSSSGGWRAAAFIISVEVAERFAYYGINSNLINYLTGPLGQSTATAA 74
+D D +G LR+ +G WRAA FII +E +ER +Y+GI++NL+ YLT L Q A
Sbjct: 22 LDSSTDSRGEIPLRAQTGAWRAALFIIGIEFSERLSYFGISTNLVVYLTTILHQDLKMAV 81
Query: 75 ENVNIWSGTASLLPLLGAFLADSFLGRYRTIIIASLVYIL 114
+N N WSG +L+PLLG F+AD++LGRY T+++A+ +Y++
Sbjct: 82 KNTNYWSGVTTLMPLLGGFVADAYLGRYGTVLLATTIYLM 121
>At3g54140 peptide transport - like protein
Length = 570
Score = 106 bits (265), Expect = 2e-24
Identities = 49/100 (49%), Positives = 63/100 (63%)
Query: 14 TVDGVVDFKGRPVLRSSSGGWRAAAFIISVEVAERFAYYGINSNLINYLTGPLGQSTATA 73
T DG VD P + +G W+A FI+ E ER AYYG+ +NL+NYL L Q ATA
Sbjct: 8 TQDGTVDIHKNPANKEKTGNWKACRFILGNECCERLAYYGMGTNLVNYLESRLNQGNATA 67
Query: 74 AENVNIWSGTASLLPLLGAFLADSFLGRYRTIIIASLVYI 113
A NV WSGT + PL+GAF+AD++LGRY TI +Y+
Sbjct: 68 ANNVTNWSGTCYITPLIGAFIADAYLGRYWTIATFVFIYV 107
>At5g46050 peptide transporter
Length = 582
Score = 105 bits (261), Expect = 5e-24
Identities = 49/99 (49%), Positives = 66/99 (66%)
Query: 14 TVDGVVDFKGRPVLRSSSGGWRAAAFIISVEVAERFAYYGINSNLINYLTGPLGQSTATA 73
T DG VD +G PV RS G W+A +F++ EV ER AYYGI+SNL Y+T L Q T +
Sbjct: 11 TKDGTVDLQGNPVRRSIRGRWKACSFVVVYEVFERMAYYGISSNLFIYMTTKLHQGTVKS 70
Query: 74 AENVNIWSGTASLLPLLGAFLADSFLGRYRTIIIASLVY 112
+ NV W GT+ L P+LGA++ D+ LGRY T +I+ +Y
Sbjct: 71 SNNVTNWVGTSWLTPILGAYVGDALLGRYITFVISCAIY 109
>At2g02040 histidine transport protein (PTR2-B)
Length = 585
Score = 104 bits (260), Expect = 6e-24
Identities = 54/108 (50%), Positives = 65/108 (60%), Gaps = 2/108 (1%)
Query: 7 ETRLLLETVDGVVDFKGRPVLRSSSGGWRAAAFIISVEVAERFAYYGINSNLINYLTGPL 66
E +L E DG VDF G P L+ +G W+A FI+ E ER AYYGI NLI YLT L
Sbjct: 20 EVKLYAE--DGSVDFNGNPPLKEKTGNWKACPFILGNECCERLAYYGIAGNLITYLTTKL 77
Query: 67 GQSTATAAENVNIWSGTASLLPLLGAFLADSFLGRYRTIIIASLVYIL 114
Q +AA NV W GT L PL+GA LAD++ GRY TI S +Y +
Sbjct: 78 HQGNVSAATNVTTWQGTCYLTPLIGAVLADAYWGRYWTIACFSGIYFI 125
>At5g01180 oligopeptide transporter - like protein
Length = 570
Score = 98.6 bits (244), Expect = 4e-22
Identities = 44/100 (44%), Positives = 66/100 (66%)
Query: 14 TVDGVVDFKGRPVLRSSSGGWRAAAFIISVEVAERFAYYGINSNLINYLTGPLGQSTATA 73
T DG +D +P ++ +G W+A FI+ E ER AYYG+++NLINYL + +A
Sbjct: 9 TKDGTLDIHKKPANKNKTGTWKACRFILGTECCERLAYYGMSTNLINYLEKQMNMENVSA 68
Query: 74 AENVNIWSGTASLLPLLGAFLADSFLGRYRTIIIASLVYI 113
+++V+ WSGT PL+GAF+AD++LGRY TI ++YI
Sbjct: 69 SKSVSNWSGTCYATPLIGAFIADAYLGRYWTIASFVVIYI 108
>At1g62200 unknown protein (At1g62200)
Length = 590
Score = 97.8 bits (242), Expect = 8e-22
Identities = 46/99 (46%), Positives = 60/99 (60%)
Query: 16 DGVVDFKGRPVLRSSSGGWRAAAFIISVEVAERFAYYGINSNLINYLTGPLGQSTATAAE 75
DG +D G P + +G W+A FI+ E ER AYYGI NLI Y T L +S +AA
Sbjct: 38 DGSIDIYGNPPSKKKTGNWKACPFILGNECCERLAYYGIAKNLITYYTSELHESNVSAAS 97
Query: 76 NVNIWSGTASLLPLLGAFLADSFLGRYRTIIIASLVYIL 114
+V IW GT + PL+GA +ADS+ GRY TI S +Y +
Sbjct: 98 DVMIWQGTCYITPLIGAVIADSYWGRYWTIASFSAIYFI 136
>At2g02020 putative peptide/amino acid transporter
Length = 545
Score = 97.1 bits (240), Expect = 1e-21
Identities = 47/106 (44%), Positives = 64/106 (60%), Gaps = 2/106 (1%)
Query: 7 ETRLLLETVDGVVDFKGRPVLRSSSGGWRAAAFIISVEVAERFAYYGINSNLINYLTGPL 66
E +L E DG +D G P L+ ++G W+A FI + E ER AYYGI NLI Y T L
Sbjct: 21 EVKLYAE--DGSIDIHGNPPLKQTTGNWKACPFIFANECCERLAYYGIAKNLITYFTNEL 78
Query: 67 GQSTATAAENVNIWSGTASLLPLLGAFLADSFLGRYRTIIIASLVY 112
++ +AA +V W GT + PL+GA +AD++ GRY TI S +Y
Sbjct: 79 HETNVSAARHVMTWQGTCYITPLIGALIADAYWGRYWTIACFSAIY 124
>At1g32450 peptide transporter PTR2-B, putative
Length = 606
Score = 93.6 bits (231), Expect = 1e-20
Identities = 44/102 (43%), Positives = 67/102 (65%)
Query: 13 ETVDGVVDFKGRPVLRSSSGGWRAAAFIISVEVAERFAYYGINSNLINYLTGPLGQSTAT 72
ET DG VD+ GRP +RS+SG W A I+ + A++G+ NL+ +LT L Q+ A
Sbjct: 12 ETRDGTVDYYGRPSIRSNSGQWVAGIVILLNQGLATLAFFGVGVNLVLFLTRVLQQNNAD 71
Query: 73 AAENVNIWSGTASLLPLLGAFLADSFLGRYRTIIIASLVYIL 114
AA NV+ W+GT + L+GAFL+DS+ GRY+T I +++++
Sbjct: 72 AANNVSKWTGTVYIFSLVGAFLSDSYWGRYKTCAIFQVIFVI 113
>At1g12110 nitrate/chlorate transporter CHL1
Length = 590
Score = 93.2 bits (230), Expect = 2e-20
Identities = 46/92 (50%), Positives = 60/92 (65%)
Query: 20 DFKGRPVLRSSSGGWRAAAFIISVEVAERFAYYGINSNLINYLTGPLGQSTATAAENVNI 79
DF+GRP RS +GGW +AA I+ +E ER GI NL+ YLTG + ATAA V
Sbjct: 17 DFQGRPADRSKTGGWASAAMILCIEAVERLTTLGIGVNLVTYLTGTMHLGNATAANTVTN 76
Query: 80 WSGTASLLPLLGAFLADSFLGRYRTIIIASLV 111
+ GT+ +L LLG F+AD+FLGRY TI I + +
Sbjct: 77 FLGTSFMLCLLGGFIADTFLGRYLTIAIFAAI 108
>At2g26690 putative nitrate transporter
Length = 586
Score = 91.7 bits (226), Expect = 5e-20
Identities = 49/101 (48%), Positives = 65/101 (63%)
Query: 14 TVDGVVDFKGRPVLRSSSGGWRAAAFIISVEVAERFAYYGINSNLINYLTGPLGQSTATA 73
TV VD+KGRP +S +GGW AA I+ +EV ER + GI NL+ YL + ++T+
Sbjct: 17 TVADAVDYKGRPADKSKTGGWITAALILGIEVVERLSTMGIAVNLVTYLMETMHLPSSTS 76
Query: 74 AENVNIWSGTASLLPLLGAFLADSFLGRYRTIIIASLVYIL 114
A V + GT+ LL LLG FLADSFLGR++TI I S + L
Sbjct: 77 ANIVTDFMGTSFLLCLLGGFLADSFLGRFKTIGIFSTIQAL 117
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.139 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,247,207
Number of Sequences: 26719
Number of extensions: 76151
Number of successful extensions: 226
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 56
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 168
Number of HSP's gapped (non-prelim): 62
length of query: 116
length of database: 11,318,596
effective HSP length: 92
effective length of query: 24
effective length of database: 8,860,448
effective search space: 212650752
effective search space used: 212650752
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 52 (24.6 bits)
Medicago: description of AC125481.10


