

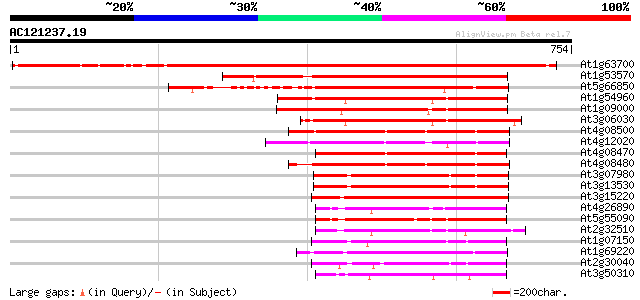
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121237.19 + phase: 0 /pseudo
(754 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g63700 putative protein kinase 860 0.0
At1g53570 MEK kinase MAP3Ka, putative 413 e-115
At5g66850 MAP protein kinase like protein 375 e-104
At1g54960 NPK1-related protein kinase 2 (ANP2) 279 5e-75
At1g09000 NPK1-related protein kinase 1S (ANP1) 277 1e-74
At3g06030 NPK1-related protein kinase 3 262 5e-70
At4g08500 MEKK1/MAP kinase kinase kinase 247 2e-65
At4g12020 putative disease resistance protein 233 2e-61
At4g08470 putative mitogen-activated protein kinase 226 4e-59
At4g08480 putative mitogen-activated protein kinase 225 8e-59
At3g07980 putative MAP3K epsilon protein kinase 208 1e-53
At3g13530 MAP3K epsilon protein kinase 206 3e-53
At3g15220 putative MAP kinase 202 4e-52
At4g26890 putative NPK1-related protein kinase 187 2e-47
At5g55090 putative protein 185 9e-47
At2g32510 putative protein kinase 178 9e-45
At1g07150 hypothetical protein 169 5e-42
At1g69220 serine/threonine kinase (SIK1) 169 7e-42
At2g30040 putative protein kinase 161 1e-39
At3g50310 protein kinase -like protein 160 2e-39
>At1g63700 putative protein kinase
Length = 883
Score = 860 bits (2221), Expect = 0.0
Identities = 463/734 (63%), Positives = 541/734 (73%), Gaps = 17/734 (2%)
Query: 4 WWGKSSSKETKKKAGKESIIDTLHRKFKFPSEGKRSTISGGSRRRSNDTISEKGDRSPSE 63
WW KS K+ KKK KESIID +RK F SE + S S SRRR ++ +SE+G S
Sbjct: 3 WWSKS--KDEKKKTNKESIIDAFNRKLGFASEDRSSGRSRKSRRRRDEIVSERGAISRLP 60
Query: 64 SRSPSPS-KVARCQSFAERPHAQPLPLPDLHPSSLGRVDSEISISAKSRLEKPSKPSLFL 122
SRSPSPS +V+RCQSFAER A PLP P + P + DS ++ S + L+ KPS +L
Sbjct: 61 SRSPSPSTRVSRCQSFAERSPAVPLPRPIVRPH-VTSTDSGMNGSQRPGLDANLKPS-WL 118
Query: 123 PLPKPSCIRCGPTPADLDGDMVNASVFSDCSADSDEPADSRNRSPLATDSETGTRTAAGS 182
PLPKP P + D ASV S S D P+DS SPLA+D E G RT
Sbjct: 119 PLPKPHGATSIPDNTGAEPDFATASVSSGSSV-GDIPSDSL-LSPLASDCENGNRT---- 172
Query: 183 PSSLVLKDQSSAVSQPNLREVKKTANILSNHTPSTSPKRKPLRHHVPNLQVPPHG-VFYS 241
P ++ +DQS ++ + K N N S SP+R+PL HV NLQ+P V S
Sbjct: 173 PVNISSRDQSMHSNKNSAEMFKPVPN--KNRILSASPRRRPLGTHVKNLQIPQRDLVLCS 230
Query: 242 GPDSSLSSPSRSPLRAFGTDQVLNSAFWAGKPYPEINFVGSGHCSSPGSGHNSGHNSMGG 301
PDS LSSPSRSP+R+F DQV N KPY +++ +GSG CSSPGSG+NSG+NS+GG
Sbjct: 231 APDSLLSSPSRSPMRSFIPDQVSNHGLLISKPYSDVSLLGSGQCSSPGSGYNSGNNSIGG 290
Query: 302 DMSGPLFWQPSRGSPEYSPIPSPRMTSPGPSSRIQSGAVTPIHPRAGGTPTESQTGRADD 361
DM+ LFW SR SPE SP+PSPRMTSPGPSSRIQSGAVTP+HPRAGG+ T S T R DD
Sbjct: 291 DMATQLFWPQSRCSPECSPVPSPRMTSPGPSSRIQSGAVTPLHPRAGGSTTGSPTRRLDD 350
Query: 362 GKQQSHRLPLPPLTVTNTSPFSHSNSAATSPSMPRSPARADSPMSSGSRWKKGKLLGRGT 421
+QQSHRLPLPPL ++NT PFS + SAATSPS+PRSPARA++ +S GSRWKKG+LLG G+
Sbjct: 351 NRQQSHRLPLPPLLISNTCPFSPTYSAATSPSVPRSPARAEATVSPGSRWKKGRLLGMGS 410
Query: 422 FGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLRHPNIVQYYGSET 481
FGHVY+GFNS+SGEMCAMKEVTL SDD KS ESA+QL QE+ +LSRLRH NIVQYYGSET
Sbjct: 411 FGHVYLGFNSESGEMCAMKEVTLCSDDPKSRESAQQLGQEISVLSRLRHQNIVQYYGSET 470
Query: 482 VDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQQILSGLAYLHAKNTLHRDIKGAN 541
VDDKLYIYLEYVSGGSI+KLLQEYGQFGE AIR+YTQQILSGLAYLHAKNT+HRDIKGAN
Sbjct: 471 VDDKLYIYLEYVSGGSIYKLLQEYGQFGENAIRNYTQQILSGLAYLHAKNTVHRDIKGAN 530
Query: 542 ILVDPNGRVKVADFGMAKHITGQYCPLSFKGSPYWMAPEVIKNSKECSLGVDIWSLGCTV 601
ILVDP+GRVKVADFGMAKHIT Q PLSFKGSPYWMAPEVIKNS +L VDIWSLGCTV
Sbjct: 531 ILVDPHGRVKVADFGMAKHITAQSGPLSFKGSPYWMAPEVIKNSNGSNLAVDIWSLGCTV 590
Query: 602 LEMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHLSNEGKDFVRKCLQRNPRDRPSASEL 661
LEMATTKPPWSQYEGV AMFKIGNSKELP IPDHLS EGKDFVRKCLQRNP +RP+A++L
Sbjct: 591 LEMATTKPPWSQYEGVPAMFKIGNSKELPDIPDHLSEEGKDFVRKCLQRNPANRPTAAQL 650
Query: 662 LDHPFVKGAAPLERPIMVPEASDPITGITHGTKALGIGQGRNLSALDSDKLLVHSSRVLK 721
LDH FV+ P+ERPI+ E ++ + + ++L IG R+L LDS+ + + LK
Sbjct: 651 LDHAFVRNVMPMERPIVSGEPAEAMNVASSTMRSLDIGHARSLPCLDSEDATNYQQKGLK 710
Query: 722 NNPHESLKSFHQFP 735
H S S Q P
Sbjct: 711 ---HGSGFSISQSP 721
>At1g53570 MEK kinase MAP3Ka, putative
Length = 609
Score = 413 bits (1062), Expect = e-115
Identities = 217/390 (55%), Positives = 271/390 (68%), Gaps = 18/390 (4%)
Query: 286 SSPGSGHNSGHNSMGG-DMSGPLFWQPSRGSPEYSPIPSPRM---TSPGPSSRIQSGAVT 341
S+ GS S +S G D L RG +++ +PR SP ++ I + +
Sbjct: 93 STSGSTSVSSVSSSGSADDQSQLVASRGRGDVKFNVAAAPRSPERVSP-KAATITTRPTS 151
Query: 342 PIHPRAGGTPT-ESQTGRADDGKQQS--HRLPLPPLTVTNTSPFSHSNSAATSPSMPRSP 398
P H R G + ES TGR DDG+ S H LP PP + T+ S S + P
Sbjct: 152 PRHQRLSGVVSLESSTGRNDDGRSSSECHPLPRPPTSPTSPSAVHGSRIGGGYETSP--- 208
Query: 399 ARADSPMSSGSRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQL 458
S S WKKGK LG GTFG VY+GFNS+ G+MCA+KEV + SDD S E KQL
Sbjct: 209 -------SGFSTWKKGKFLGSGTFGQVYLGFNSEKGKMCAIKEVKVISDDQTSKECLKQL 261
Query: 459 MQEVHLLSRLRHPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQ 518
QE++LL++L HPNIVQYYGSE ++ L +YLEYVSGGSIHKLL++YG F E I++YT+
Sbjct: 262 NQEINLLNQLCHPNIVQYYGSELSEETLSVYLEYVSGGSIHKLLKDYGSFTEPVIQNYTR 321
Query: 519 QILSGLAYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHITGQYCPLSFKGSPYWMA 578
QIL+GLAYLH +NT+HRDIKGANILVDPNG +K+ADFGMAKH+T LSFKGSPYWMA
Sbjct: 322 QILAGLAYLHGRNTVHRDIKGANILVDPNGEIKLADFGMAKHVTAFSTMLSFKGSPYWMA 381
Query: 579 PEVIKNSKECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHLSN 638
PEV+ + + VDIWSLGCT+LEMAT+KPPWSQ+EGVAA+FKIGNSK+ P IPDHLSN
Sbjct: 382 PEVVMSQNGYTHAVDIWSLGCTILEMATSKPPWSQFEGVAAIFKIGNSKDTPEIPDHLSN 441
Query: 639 EGKDFVRKCLQRNPRDRPSASELLDHPFVK 668
+ K+F+R CLQRNP RP+AS+LL+HPF++
Sbjct: 442 DAKNFIRLCLQRNPTVRPTASQLLEHPFLR 471
>At5g66850 MAP protein kinase like protein
Length = 533
Score = 375 bits (962), Expect = e-104
Identities = 224/471 (47%), Positives = 295/471 (62%), Gaps = 67/471 (14%)
Query: 214 TPSTSPKRKPLRHHVPNLQVPPHG-VFYSGPD-----SSLSSPSRSPLRAFGTDQVLNSA 267
+P SP+RK H +P +PP +S PD S L P+ + AF TD NS
Sbjct: 10 SPVPSPQRKSTGHDLPFFYLPPKSNQAWSAPDMPLDTSGLPPPAFYDITAFSTD---NSP 66
Query: 268 FWAGKPYPEINFVGSGHCSSPGSGHNSGHNSMGGDMSGPLFWQPSRGSPEYSPIPSPRMT 327
+ +P S + P QPSR S SP+ S +
Sbjct: 67 IHSPQP-----------------------RSPRKQIRSP---QPSRPS---SPLHSVDSS 97
Query: 328 SPGPSSRIQSGAVTPIHPRAGGTPTESQTGRADDGKQQSHRLPLPPLTVTNTSPFSHSNS 387
+P P + S P+HPR T+ GR D H LPLPP ++S S
Sbjct: 98 AP-PRDSVSS----PLHPRLS---TDVTNGRRDCCNV--HPLPLPPGATCSSS------S 141
Query: 388 AATSPSMPRSPARADS-PMSSGSRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFS 446
AA+ PS P++P + DS PM+S +WKKGKL+GRGTFG VY+ NS++G +CAMKEV LF
Sbjct: 142 AASVPS-PQAPLKLDSFPMNS--QWKKGKLIGRGTFGSVYVASNSETGALCAMKEVELFP 198
Query: 447 DDAKSLESAKQLMQEVHLLSRLRHPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLLQEY- 505
DD KS E KQL QE+ LLS L+HPNIVQY+GSETV+D+ +IYLEYV GSI+K ++++
Sbjct: 199 DDPKSAECIKQLEQEIKLLSNLQHPNIVQYFGSETVEDRFFIYLEYVHPGSINKYIRDHC 258
Query: 506 GQFGELAIRSYTQQILSGLAYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHITGQY 565
G E +R++T+ ILSGLAYLH K T+HRDIKGAN+LVD +G VK+ADFGMAKH+TGQ
Sbjct: 259 GTMTESVVRNFTRHILSGLAYLHNKKTVHRDIKGANLLVDASGVVKLADFGMAKHLTGQR 318
Query: 566 CPLSFKGSPYWMAPEVIK------NSKECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAA 619
LS KGSPYWMAPE+++ ++ + + VDIWSLGCT++EM T KPPWS++EG AA
Sbjct: 319 ADLSLKGSPYWMAPELMQAVMQKDSNPDLAFAVDIWSLGCTIIEMFTGKPPWSEFEGAAA 378
Query: 620 MFKIGNSKELPTIPDHLSNEGKDFVRKCLQRNPRDRPSASELLDHPFVKGA 670
MFK+ ++ P IP+ +S EGKDF+R C QRNP +RP+AS LL+H F+K +
Sbjct: 379 MFKV--MRDSPPIPESMSPEGKDFLRLCFQRNPAERPTASMLLEHRFLKNS 427
>At1g54960 NPK1-related protein kinase 2 (ANP2)
Length = 651
Score = 279 bits (713), Expect = 5e-75
Identities = 155/318 (48%), Positives = 201/318 (62%), Gaps = 12/318 (3%)
Query: 360 DDGKQQSHRLPLPPLTVTN-TSPFSHSNSAATSPSMPRSPARADSPMSSGSRWKKGKLLG 418
DD Q++H P P L TS S A S S P + P RW+KG+L+G
Sbjct: 19 DDENQENHPPPFPSLLADKITSCIRKSMVFAKSQSPPNNSTVQIKPPI---RWRKGQLIG 75
Query: 419 RGTFGHVYIGFNSQSGEMCAMKEVTLFSDDA---KSLESAKQLMQEVHLLSRLRHPNIVQ 475
RG FG VY+G N SGE+ A+K+V + S+ A K+ ++L +EV LL L HPNIV+
Sbjct: 76 RGAFGTVYMGMNLDSGELLAVKQVLITSNCASKEKTQAHIQELEEEVKLLKNLSHPNIVR 135
Query: 476 YYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQQILSGLAYLHAKNTLHR 535
Y G+ D+ L I LE+V GGSI LL+++G F E +R+YT Q+L GL YLH +HR
Sbjct: 136 YLGTVREDETLNILLEFVPGGSISSLLEKFGAFPESVVRTYTNQLLLGLEYLHNHAIMHR 195
Query: 536 DIKGANILVDPNGRVKVADFGMAKHITGQYC---PLSFKGSPYWMAPEVIKNSKECSLGV 592
DIKGANILVD G +K+ADFG +K + S KG+PYWMAPEVI + S
Sbjct: 196 DIKGANILVDNQGCIKLADFGASKQVAELATISGAKSMKGTPYWMAPEVILQTGH-SFSA 254
Query: 593 DIWSLGCTVLEMATTKPPWS-QYEGVAAMFKIGNSKELPTIPDHLSNEGKDFVRKCLQRN 651
DIWS+GCTV+EM T K PWS QY+ +AA+F IG +K P IPD++S++ DF+ KCLQ+
Sbjct: 255 DIWSVGCTVIEMVTGKAPWSQQYKEIAAIFHIGTTKSHPPIPDNISSDANDFLLKCLQQE 314
Query: 652 PRDRPSASELLDHPFVKG 669
P RP+ASELL HPFV G
Sbjct: 315 PNLRPTASELLKHPFVTG 332
>At1g09000 NPK1-related protein kinase 1S (ANP1)
Length = 666
Score = 277 bits (709), Expect = 1e-74
Identities = 152/321 (47%), Positives = 198/321 (61%), Gaps = 14/321 (4%)
Query: 359 ADDGKQQSHRLPLPPLTVTNTSPFSHSNSAATSPSMPRSPARADSPMSSGSRWKKGKLLG 418
+ D Q ++ P P + + + PS P M+ W+KG+L+G
Sbjct: 17 SSDDDNQENQPPFPGVLADKITSCIRKSKIFIKPSFSPPPPANTVDMAPPISWRKGQLIG 76
Query: 419 RGTFGHVYIGFNSQSGEMCAMKEVTL---FSDDAKSLESAKQLMQEVHLLSRLRHPNIVQ 475
RG FG VY+G N SGE+ A+K+V + F+ K+ ++L +EV LL L HPNIV+
Sbjct: 77 RGAFGTVYMGMNLDSGELLAVKQVLIAANFASKEKTQAHIQELEEEVKLLKNLSHPNIVR 136
Query: 476 YYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQQILSGLAYLHAKNTLHR 535
Y G+ DD L I LE+V GGSI LL+++G F E +R+YT+Q+L GL YLH +HR
Sbjct: 137 YLGTVREDDTLNILLEFVPGGSISSLLEKFGPFPESVVRTYTRQLLLGLEYLHNHAIMHR 196
Query: 536 DIKGANILVDPNGRVKVADFGMAKHI------TGQYCPLSFKGSPYWMAPEVIKNSKECS 589
DIKGANILVD G +K+ADFG +K + TG S KG+PYWMAPEVI + S
Sbjct: 197 DIKGANILVDNKGCIKLADFGASKQVAELATMTG---AKSMKGTPYWMAPEVILQTGH-S 252
Query: 590 LGVDIWSLGCTVLEMATTKPPWS-QYEGVAAMFKIGNSKELPTIPDHLSNEGKDFVRKCL 648
DIWS+GCTV+EM T K PWS QY+ VAA+F IG +K P IPD LS++ KDF+ KCL
Sbjct: 253 FSADIWSVGCTVIEMVTGKAPWSQQYKEVAAIFFIGTTKSHPPIPDTLSSDAKDFLLKCL 312
Query: 649 QRNPRDRPSASELLDHPFVKG 669
Q P RP+ASELL HPFV G
Sbjct: 313 QEVPNLRPTASELLKHPFVMG 333
>At3g06030 NPK1-related protein kinase 3
Length = 651
Score = 262 bits (670), Expect = 5e-70
Identities = 146/306 (47%), Positives = 191/306 (61%), Gaps = 15/306 (4%)
Query: 392 PSMPRSPARADSPMSSGSRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDA-- 449
P +P +P + ++P RW+KG+L+G G FG VY+G N SGE+ A+K+V + A
Sbjct: 53 PGLP-APRKEEAP---SIRWRKGELIGCGAFGRVYMGMNLDSGELLAIKQVLIAPSSASK 108
Query: 450 -KSLESAKQLMQEVHLLSRLRHPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQF 508
K+ ++L +EV LL L HPNIV+Y G+ D L I +E+V GGSI LL+++G F
Sbjct: 109 EKTQGHIRELEEEVQLLKNLSHPNIVRYLGTVRESDSLNILMEFVPGGSISSLLEKFGSF 168
Query: 509 GELAIRSYTQQILSGLAYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHITGQYC-- 566
E I YT+Q+L GL YLH +HRDIKGANILVD G +++ADFG +K +
Sbjct: 169 PEPVIIMYTKQLLLGLEYLHNNGIMHRDIKGANILVDNKGCIRLADFGASKKVVELATVN 228
Query: 567 -PLSFKGSPYWMAPEVIKNSKECSLGVDIWSLGCTVLEMATTKPPWS-QYEGVAAMFKIG 624
S KG+PYWMAPEVI + S DIWS+GCTV+EMAT KPPWS QY+ AA+ IG
Sbjct: 229 GAKSMKGTPYWMAPEVILQTGH-SFSADIWSVGCTVIEMATGKPPWSEQYQQFAAVLHIG 287
Query: 625 NSKELPTIPDHLSNEGKDFVRKCLQRNPRDRPSASELLDHPFVKGAAPLERPI---MVPE 681
+K P IP+ LS E KDF+ KCL + P R SA+ELL HPFV G P + E
Sbjct: 288 RTKAHPPIPEDLSPEAKDFLMKCLHKEPSLRLSATELLQHPFVTGKRQEPYPAYRNSLTE 347
Query: 682 ASDPIT 687
+PIT
Sbjct: 348 CGNPIT 353
>At4g08500 MEKK1/MAP kinase kinase kinase
Length = 608
Score = 247 bits (630), Expect = 2e-65
Identities = 144/300 (48%), Positives = 191/300 (63%), Gaps = 7/300 (2%)
Query: 375 TVTNTSPFSHSNSAATSPSMPRSPARADSPMSSGSRWKKGKLLGRGTFGHVYIGFNSQSG 434
T T F+ + ++S SP D S W+KG+LLGRG+FG VY G S G
Sbjct: 298 TADETCSFTTNEGDSSSTVSNTSPIYPDGGAIITS-WQKGQLLGRGSFGSVYEGI-SGDG 355
Query: 435 EMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLRHPNIVQYYGSETVDDKLYIYLEYVS 494
+ A+KEV+L +++ E +QL E+ LLS+L+H NIV+Y G+ LYI+LE V+
Sbjct: 356 DFFAVKEVSLLDQGSQAQECIQQLEGEIKLLSQLQHQNIVRYRGTAKDGSNLYIFLELVT 415
Query: 495 GGSIHKLLQEYGQFGELAIRSYTQQILSGLAYLHAKNTLHRDIKGANILVDPNGRVKVAD 554
GS+ KL Q Y Q + + YT+QIL GL YLH K +HRDIK ANILVD NG VK+AD
Sbjct: 416 QGSLLKLYQRY-QLRDSVVSLYTRQILDGLKYLHDKGFIHRDIKCANILVDANGAVKLAD 474
Query: 555 FGMAKHITGQYCPLSFKGSPYWMAPEVI--KNSKECSLGVDIWSLGCTVLEMATTKPPWS 612
FG+AK ++ S KG+P+WMAPEVI K+S DIWSLGCTVLEM T + P+S
Sbjct: 475 FGLAK-VSKFNDIKSCKGTPFWMAPEVINRKDSDGYGSPADIWSLGCTVLEMCTGQIPYS 533
Query: 613 QYEGVAAMFKIGNSKELPTIPDHLSNEGKDFVRKCLQRNPRDRPSASELLDHPFVKGAAP 672
E V A+F+IG LP +PD LS + + F+ KCL+ NP +RP+A+ELL+HPFV+ P
Sbjct: 534 DLEPVQALFRIGRG-TLPEVPDTLSLDARLFILKCLKVNPEERPTAAELLNHPFVRRPLP 592
>At4g12020 putative disease resistance protein
Length = 1895
Score = 233 bits (595), Expect = 2e-61
Identities = 143/333 (42%), Positives = 195/333 (57%), Gaps = 16/333 (4%)
Query: 345 PRAGGTPTESQTGRADDGKQQSHRLPLPPLTVTNTSPFSHSNSAATSPSMPRSPARADSP 404
P P+ S R ++ + + PL T F+ + ++ SP A S
Sbjct: 1561 PETVAPPSSSSEAREEEVETEETGAMFIPLGDKETCSFTVNKGDSSRTISNTSPIYA-SE 1619
Query: 405 MSSGSRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHL 464
S + W+KG+LLGRG+ G VY G S G+ A KEV+L +++ E +Q+ + L
Sbjct: 1620 GSFITCWQKGQLLGRGSLGSVYEGI-SADGDFFAFKEVSLLDQGSQAHEWIQQVEGGIAL 1678
Query: 465 LSRLRHPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQQILSGL 524
LS+L+H NIV+Y G+ + LYI+LE V+ GS+ KL Q Q G+ + YT+QIL GL
Sbjct: 1679 LSQLQHQNIVRYRGTTKDESNLYIFLELVTQGSLRKLYQR-NQLGDSVVSLYTRQILDGL 1737
Query: 525 AYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHITGQYCPLSFKGSPY--WMAPEVI 582
YLH K +HR+IK AN+LVD NG VK+ADFG+AK +S +PY WMAPEVI
Sbjct: 1738 KYLHDKGFIHRNIKCANVLVDANGTVKLADFGLAK-------VMSLWRTPYWNWMAPEVI 1790
Query: 583 KNSKE---CSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHLSNE 639
N K+ DIWSLGCTVLEM T + P+S E A++ IG K LP IPD LS +
Sbjct: 1791 LNPKDYDGYGTPADIWSLGCTVLEMLTGQIPYSDLEIGTALYNIGTGK-LPKIPDILSLD 1849
Query: 640 GKDFVRKCLQRNPRDRPSASELLDHPFVKGAAP 672
+DF+ CL+ NP +RP+A+ELL+HPFV P
Sbjct: 1850 ARDFILTCLKVNPEERPTAAELLNHPFVNMPLP 1882
>At4g08470 putative mitogen-activated protein kinase
Length = 560
Score = 226 bits (576), Expect = 4e-59
Identities = 127/259 (49%), Positives = 170/259 (65%), Gaps = 6/259 (2%)
Query: 411 WKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLRH 470
W KG+LLGRG++ VY S+ G+ A+KEV+L ++ E +QL E+ LLS+L+H
Sbjct: 303 WLKGQLLGRGSYASVYEAI-SEDGDFFAVKEVSLLDKGIQAQECIQQLEGEIALLSQLQH 361
Query: 471 PNIVQYYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQQILSGLAYLHAK 530
NIV+Y G+ KLYI+LE V+ GS+ KL + Y Q + YT+QIL+GL YLH K
Sbjct: 362 QNIVRYRGTAKDVSKLYIFLELVTQGSVQKLYERY-QLSYTVVSLYTRQILAGLNYLHDK 420
Query: 531 NTLHRDIKGANILVDPNGRVKVADFGMAKHITGQYCPLSFKGSPYWMAPEVI--KNSKEC 588
+HRDIK AN+LVD NG VK+ADFG+A+ +S KG+ +WMAPEVI K+S
Sbjct: 421 GFVHRDIKCANMLVDANGTVKLADFGLAEASKFNDI-MSCKGTLFWMAPEVINRKDSDGN 479
Query: 589 SLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHLSNEGKDFVRKCL 648
DIWSLGCTVLEM T + P+S + + A FKIG LP +PD LS + + F+ CL
Sbjct: 480 GSPADIWSLGCTVLEMCTGQIPYSDLKPIQAAFKIGRG-TLPDVPDTLSLDARHFILTCL 538
Query: 649 QRNPRDRPSASELLDHPFV 667
+ NP +RP+A+ELL HPFV
Sbjct: 539 KVNPEERPTAAELLHHPFV 557
>At4g08480 putative mitogen-activated protein kinase
Length = 773
Score = 225 bits (573), Expect = 8e-59
Identities = 134/300 (44%), Positives = 184/300 (60%), Gaps = 24/300 (8%)
Query: 375 TVTNTSPFSHSNSAATSPSMPRSPARADSPMSSGSRWKKGKLLGRGTFGHVYIGFNSQSG 434
TV+NTSP S S + W+KG+LL +G+FG VY S+ G
Sbjct: 483 TVSNTSPICVSGG------------------SINTSWQKGQLLRQGSFGSVYEAI-SEDG 523
Query: 435 EMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLRHPNIVQYYGSETVDDKLYIYLEYVS 494
+ A+KEV+L +++ E +QL E+ LLS+L H NI++Y G++ LYI+LE V+
Sbjct: 524 DFFAVKEVSLLDQGSQAQECIQQLEGEIALLSQLEHQNILRYRGTDKDGSNLYIFLELVT 583
Query: 495 GGSIHKLLQEYGQFGELAIRSYTQQILSGLAYLHAKNTLHRDIKGANILVDPNGRVKVAD 554
GS+ +L + Y Q + I YT+QIL GL YLH K +HRDIK A ILVD NG VK+AD
Sbjct: 584 QGSLLELYRRY-QIRDSLISLYTKQILDGLKYLHHKGFIHRDIKCATILVDANGTVKLAD 642
Query: 555 FGMAKHITGQYCPLSFKGSPYWMAPEVI--KNSKECSLGVDIWSLGCTVLEMATTKPPWS 612
FG+AK ++ S K + +WMAPEVI K++ DIWSLGCTVLEM T + P+S
Sbjct: 643 FGLAK-VSKLNDIKSRKETLFWMAPEVINRKDNDGYRSPADIWSLGCTVLEMCTGQIPYS 701
Query: 613 QYEGVAAMFKIGNSKELPTIPDHLSNEGKDFVRKCLQRNPRDRPSASELLDHPFVKGAAP 672
E V A+F+I LP +PD LS + + F+ KCL+ NP +RP+A+ELL+HPFV+ P
Sbjct: 702 DLEPVEALFRIRRG-TLPEVPDTLSLDARHFILKCLKLNPEERPTATELLNHPFVRRPLP 760
>At3g07980 putative MAP3K epsilon protein kinase
Length = 1367
Score = 208 bits (529), Expect = 1e-53
Identities = 108/265 (40%), Positives = 165/265 (61%), Gaps = 8/265 (3%)
Query: 409 SRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRL 468
+++ G +G+G +G VYIG + ++G+ A+K+V+L + + L + +MQE+ LL L
Sbjct: 18 NKYMLGDEIGKGAYGRVYIGLDLENGDFVAIKQVSLENIGQEDLNT---IMQEIDLLKNL 74
Query: 469 RHPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLLQ--EYGQFGELAIRSYTQQILSGLAY 526
H NIV+Y GS L+I LEYV GS+ +++ ++G F E + Y Q+L GL Y
Sbjct: 75 NHKNIVKYLGSLKTKTHLHIILEYVENGSLANIIKPNKFGPFPESLVTVYIAQVLEGLVY 134
Query: 527 LHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHIT-GQYCPLSFKGSPYWMAPEVIKNS 585
LH + +HRDIKGANIL G VK+ADFG+A + + S G+PYWMAPEVI+ S
Sbjct: 135 LHEQGVIHRDIKGANILTTKEGLVKLADFGVATKLNEADFNTHSVVGTPYWMAPEVIELS 194
Query: 586 KECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHLSNEGKDFVR 645
C+ DIWS+GCT++E+ T PP+ + + A+++I + P IPD LS + DF+R
Sbjct: 195 GVCAAS-DIWSVGCTIIELLTCVPPYYDLQPMPALYRIVQD-DTPPIPDSLSPDITDFLR 252
Query: 646 KCLQRNPRDRPSASELLDHPFVKGA 670
C +++ R RP A LL HP+++ +
Sbjct: 253 LCFKKDSRQRPDAKTLLSHPWIRNS 277
>At3g13530 MAP3K epsilon protein kinase
Length = 1368
Score = 206 bits (525), Expect = 3e-53
Identities = 109/265 (41%), Positives = 164/265 (61%), Gaps = 8/265 (3%)
Query: 409 SRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRL 468
+++ G +G+G +G VY G + ++G+ A+K+V+L + + L + +MQE+ LL L
Sbjct: 18 NKYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIVQEDLNT---IMQEIDLLKNL 74
Query: 469 RHPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLLQ--EYGQFGELAIRSYTQQILSGLAY 526
H NIV+Y GS L+I LEYV GS+ +++ ++G F E + Y Q+L GL Y
Sbjct: 75 NHKNIVKYLGSSKTKTHLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGLVY 134
Query: 527 LHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHIT-GQYCPLSFKGSPYWMAPEVIKNS 585
LH + +HRDIKGANIL G VK+ADFG+A + S G+PYWMAPEVI+ S
Sbjct: 135 LHEQGVIHRDIKGANILTTKEGLVKLADFGVATKLNEADVNTHSVVGTPYWMAPEVIEMS 194
Query: 586 KECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHLSNEGKDFVR 645
C+ DIWS+GCTV+E+ T PP+ + + A+F+I + P IPD LS + DF+R
Sbjct: 195 GVCAAS-DIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQD-DNPPIPDSLSPDITDFLR 252
Query: 646 KCLQRNPRDRPSASELLDHPFVKGA 670
+C +++ R RP A LL HP+++ +
Sbjct: 253 QCFKKDSRQRPDAKTLLSHPWIRNS 277
>At3g15220 putative MAP kinase
Length = 690
Score = 202 bits (515), Expect = 4e-52
Identities = 108/266 (40%), Positives = 164/266 (61%), Gaps = 6/266 (2%)
Query: 406 SSGSRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLL 465
++G+R+ + +L+GRG+FG VY F+ + A+K + L +S + + + +E+ +L
Sbjct: 10 AAGARFSQIELIGRGSFGDVYKAFDKDLNKEVAIKVIDL----EESEDEIEDIQKEISVL 65
Query: 466 SRLRHPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQQILSGLA 525
S+ R P I +YYGS KL+I +EY++GGS+ LLQ E +I T+ +L +
Sbjct: 66 SQCRCPYITEYYGSYLHQTKLWIIMEYMAGGSVADLLQSNNPLDETSIACITRDLLHAVE 125
Query: 526 YLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHITGQYC-PLSFKGSPYWMAPEVIKN 584
YLH + +HRDIK ANIL+ NG VKVADFG++ +T +F G+P+WMAPEVI+N
Sbjct: 126 YLHNEGKIHRDIKAANILLSENGDVKVADFGVSAQLTRTISRRKTFVGTPFWMAPEVIQN 185
Query: 585 SKECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHLSNEGKDFV 644
S+ + DIWSLG TV+EMA +PP + + +F I P + +H S + K+FV
Sbjct: 186 SEGYNEKADIWSLGITVIEMAKGEPPLADLHPMRVLFIIPRETP-PQLDEHFSRQVKEFV 244
Query: 645 RKCLQRNPRDRPSASELLDHPFVKGA 670
CL++ P +RPSA EL+ H F+K A
Sbjct: 245 SLCLKKAPAERPSAKELIKHRFIKNA 270
>At4g26890 putative NPK1-related protein kinase
Length = 444
Score = 187 bits (475), Expect = 2e-47
Identities = 107/263 (40%), Positives = 159/263 (59%), Gaps = 20/263 (7%)
Query: 411 WKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLRH 470
W +G ++GRG+ V I +S SGE+ A+K L S+ L +E +LS L
Sbjct: 5 WTRGPIIGRGSTATVSIAISS-SGELFAVKSA--------DLSSSSLLQKEQSILSTLSS 55
Query: 471 PNIVQYYGSETVDDK----LYIYLEYVSGGSIHKLLQEYG-QFGELAIRSYTQQILSGLA 525
P++V+Y G+ + I +EYVSGG++H L++ G + E IRSYT+QIL+GL
Sbjct: 56 PHMVKYIGTGLTRESNGLVYNILMEYVSGGNLHDLIKNSGGKLPEPEIRSYTRQILNGLV 115
Query: 526 YLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHITGQYCPLSFKGSPYWMAPEVIKNS 585
YLH + +H D+K N+LV+ NG +K+AD G AK + F G+P +MAPEV +
Sbjct: 116 YLHERGIVHCDLKSHNVLVEENGVLKIADMGCAKSVDKS----EFSGTPAFMAPEVAR-G 170
Query: 586 KECSLGVDIWSLGCTVLEMATTKPPWSQY-EGVAAMFKIGNSKELPTIPDHLSNEGKDFV 644
+E D+W+LGCT++EM T PW + + VAAM+KIG S E P IP +S++ KDF+
Sbjct: 171 EEQRFPADVWALGCTMIEMMTGSSPWPELNDVVAAMYKIGFSGESPAIPAWISDKAKDFL 230
Query: 645 RKCLQRNPRDRPSASELLDHPFV 667
+ CL+ + + R + ELL HPF+
Sbjct: 231 KNCLKEDQKQRWTVEELLKHPFL 253
>At5g55090 putative protein
Length = 448
Score = 185 bits (469), Expect = 9e-47
Identities = 109/264 (41%), Positives = 161/264 (60%), Gaps = 20/264 (7%)
Query: 411 WKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLRH 470
W +G ++GRG+ V +G + SG+ A+K S+ L +E +LS+L
Sbjct: 6 WIRGPIIGRGSTATVSLGITN-SGDFFAVKSA--------EFSSSAFLQREQSILSKLSS 56
Query: 471 PNIVQYYGSETV--DDKLY--IYLEYVSGGSIHKLLQEYG-QFGELAIRSYTQQILSGLA 525
P IV+Y GS +DKL + +EYVSGGS+H L++ G + E IRSYT+QIL GL
Sbjct: 57 PYIVKYIGSNVTKENDKLMYNLLMEYVSGGSLHDLIKNSGGKLPEPLIRSYTRQILKGLM 116
Query: 526 YLHAKNTLHRDIKGANILVDPNGRV-KVADFGMAKHITGQYCPLSFKGSPYWMAPEVIKN 584
YLH + +H D+K N+++ G + K+ D G AK + L F G+P +M+PEV +
Sbjct: 117 YLHDQGIVHCDVKSQNVMI--GGEIAKIVDLGCAKTVEENE-NLEFSGTPAFMSPEVARG 173
Query: 585 SKECSLGVDIWSLGCTVLEMATTKPPWSQYEGV-AAMFKIGNSKELPTIPDHLSNEGKDF 643
++ S D+W+LGCTV+EMAT PW + V AA++KIG + E P IP LS +G+DF
Sbjct: 174 EEQ-SFPADVWALGCTVIEMATGSSPWPELNDVVAAIYKIGFTGESPVIPVWLSEKGQDF 232
Query: 644 VRKCLQRNPRDRPSASELLDHPFV 667
+RKCL+++P+ R + ELL HPF+
Sbjct: 233 LRKCLRKDPKQRWTVEELLQHPFL 256
>At2g32510 putative protein kinase
Length = 372
Score = 178 bits (452), Expect = 9e-45
Identities = 102/294 (34%), Positives = 168/294 (56%), Gaps = 22/294 (7%)
Query: 411 WKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLRH 470
W +G++LGRG+ VY S E+ A+K + + ++ L +E +LS L
Sbjct: 3 WTRGRILGRGSTATVYAAAGHNSDEILAVK--------SSEVHRSEFLQREAKILSSLSS 54
Query: 471 PNIVQYYGSETVDDK----LY-IYLEYVSGGSI-HKLLQEYGQFGELAIRSYTQQILSGL 524
P ++ Y GSET + +Y + +EY G++ ++ G+ E + YT+ IL GL
Sbjct: 55 PYVIGYRGSETKRESNGVVMYNLLMEYAPYGTLTDAAAKDGGRVDETRVVKYTRDILKGL 114
Query: 525 AYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHITGQYCPLSFKGSPYWMAPEVIKN 584
Y+H+K +H D+KG+N+++ G K+ADFG AK + + G+P +MAPEV +
Sbjct: 115 EYIHSKGIVHCDVKGSNVVISEKGEAKIADFGCAKRVDPVF-ESPVMGTPAFMAPEVARG 173
Query: 585 SKECSLGVDIWSLGCTVLEMATTKPPW----SQYEGVAAMFKIGNSKELPTIPDHLSNEG 640
K+ DIW++GCT++EM T PPW S+ + V+ ++++G S E P +P L+ E
Sbjct: 174 EKQ-GKESDIWAVGCTMIEMVTGSPPWTKADSREDPVSVLYRVGYSSETPELPCLLAEEA 232
Query: 641 KDFVRKCLQRNPRDRPSASELLDHPFVKGAAPLERPIMVPE-ASDPITGITHGT 693
KDF+ KCL+R +R +A++LL+HPF+ +E P++VP S+ T +T T
Sbjct: 233 KDFLEKCLKREANERWTATQLLNHPFLTTKPDIE-PVLVPGLISNSPTSVTDQT 285
>At1g07150 hypothetical protein
Length = 499
Score = 169 bits (428), Expect = 5e-42
Identities = 100/267 (37%), Positives = 146/267 (54%), Gaps = 10/267 (3%)
Query: 406 SSGSRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLL 465
S S W +G +GRG FG V + +GE+ A+K V L + ES L E+ +
Sbjct: 18 SPSSFWVRGACIGRGCFGAVSTAISKTNGEVFAVKSVDLATSLPTQSES---LENEISVF 74
Query: 466 SRLR-HPNIVQYYGS----ETVDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQQI 520
L+ HP IV++ G E +YLEY+ G + E ++ YT +
Sbjct: 75 RSLKPHPYIVKFLGDGVSKEGTTTFRNLYLEYLPNGDVASHRAGGKIEDETLLQRYTACL 134
Query: 521 LSGLAYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHITGQYCPLSFKGSPYWMAPE 580
+S L ++H++ +H D+K NILV + VK+ADFG A I ++ +GSP WMAPE
Sbjct: 135 VSALRHVHSQGFVHCDVKARNILVSQSSMVKLADFGSAFRIHTPRALITPRGSPLWMAPE 194
Query: 581 VIKNSKECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHLSNEG 640
VI+ + D+WSLGCT++EM T KP W + G+ ++ +I S ELP P LS G
Sbjct: 195 VIRREYQ-GPESDVWSLGCTIIEMFTGKPAWEDH-GIDSLSRISFSDELPVFPSKLSEIG 252
Query: 641 KDFVRKCLQRNPRDRPSASELLDHPFV 667
+DF+ KCL+R+P R S +LL HPF+
Sbjct: 253 RDFLEKCLKRDPNQRWSCDQLLQHPFL 279
>At1g69220 serine/threonine kinase (SIK1)
Length = 836
Score = 169 bits (427), Expect = 7e-42
Identities = 104/287 (36%), Positives = 164/287 (56%), Gaps = 14/287 (4%)
Query: 386 NSAATSPSMPRSPARADSPMSSGSRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLF 445
NS ++ S+P S R D ++++ LG+G++G VY + ++ E+ A+K ++L
Sbjct: 228 NSKMSTTSLPDSITREDPT----TKYEFLNELGKGSYGSVYKARDLKTSEIVAVKVISLT 283
Query: 446 SDDAKSLESAKQLMQEVHLLSRLRHPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLLQEY 505
+ E +++ E+ +L + HPN+V+Y GS +D L+I +EY GGS+ L+
Sbjct: 284 EGE----EGYEEIRGEIEMLQQCNHPNVVRYLGSYQGEDYLWIVMEYCGGGSVADLMNVT 339
Query: 506 GQ-FGELAIRSYTQQILSGLAYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHITGQ 564
+ E I ++ L GLAYLH+ +HRDIKG NIL+ G VK+ DFG+A +T
Sbjct: 340 EEALEEYQIAYICREALKGLAYLHSIYKVHRDIKGGNILLTEQGEVKLGDFGVAAQLTRT 399
Query: 565 YCPL-SFKGSPYWMAPEVIKNSKECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKI 623
+F G+P+WMAPEVI+ ++ VD+W+LG + +EMA PP S + +F I
Sbjct: 400 MSKRNTFIGTPHWMAPEVIQENRYDG-KVDVWALGVSAIEMAEGLPPRSSVHPMRVLFMI 458
Query: 624 GNSKELPTIPD--HLSNEGKDFVRKCLQRNPRDRPSASELLDHPFVK 668
+ + P + D S DFV KCL + PR RP+A+E+L H FV+
Sbjct: 459 -SIEPAPMLEDKEKWSLVFHDFVAKCLTKEPRLRPTAAEMLKHKFVE 504
>At2g30040 putative protein kinase
Length = 463
Score = 161 bits (408), Expect = 1e-39
Identities = 101/270 (37%), Positives = 146/270 (53%), Gaps = 19/270 (7%)
Query: 406 SSGSRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEV---TLFSDDAKSLESAKQLMQEV 462
SS S W +G +GRG FG V + G + A+K + T A+SLE+ + V
Sbjct: 12 SSSSSWIRGSCVGRGCFGTVSKALSKIDGGLFAVKSIDLATCLPSQAESLEN-----EIV 66
Query: 463 HLLSRLRHPNIVQYYGSETVDDKLY----IYLEYVSGGSIHKLLQEYGQFGELAIRSYTQ 518
L S HPNIV++ G + + ++LEY G + G E +R Y
Sbjct: 67 ILRSMKSHPNIVRFLGDDVSKEGTASFRNLHLEYSPEGDV----ANGGIVNETLLRRYVW 122
Query: 519 QILSGLAYLHAKNTLHRDIKGANILVDPNGR-VKVADFGMAKHITGQYCPLSFKGSPYWM 577
++S L+++H+ +H D+K N+LV G VK+ADFG A +S +GSP WM
Sbjct: 123 CLVSALSHVHSNGIVHCDVKSKNVLVFNGGSSVKLADFGSAVEFEKSTIHVSPRGSPLWM 182
Query: 578 APEVIKNSKECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHLS 637
APEV++ + D+WSLGCTV+EM T KP W + G ++ +IG S +LP IP LS
Sbjct: 183 APEVVRREYQGPES-DVWSLGCTVIEMLTGKPAWEDH-GFDSLSRIGFSNDLPFIPVGLS 240
Query: 638 NEGKDFVRKCLQRNPRDRPSASELLDHPFV 667
G+DF+ KCL+R+ R S +LL HPF+
Sbjct: 241 ELGRDFLEKCLKRDRSQRWSCDQLLQHPFL 270
>At3g50310 protein kinase -like protein
Length = 342
Score = 160 bits (405), Expect = 2e-39
Identities = 103/272 (37%), Positives = 151/272 (54%), Gaps = 21/272 (7%)
Query: 411 WKKGKLLGRGTFGHVYIGFNSQ-SGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLR 469
W +G+ +G GTF V S+ SG+ A+ + + S DA A L E +L L
Sbjct: 3 WVRGETIGFGTFSTVSTATKSRNSGDFPAL--IAVKSTDAYG---AASLSNEKSVLDSLG 57
Query: 470 H-PNIVQYYGSETV----DDKLYIYLEYVSGGSIHKLLQEYGQFG--ELAIRSYTQQILS 522
P I++ YG ++ ++ + LEY S GS+ +++ G G E +R +T +L
Sbjct: 58 DCPEIIRCYGEDSTVENGEEMHNLLLEYASRGSLASYMKKLGGEGLPESTVRRHTGSVLR 117
Query: 523 GLAYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHITGQYCPL----SFKGSPYWMA 578
GL ++HAK H DIK ANIL+ +G VK+ADFG+A + G L +G+P +MA
Sbjct: 118 GLRHIHAKGFAHCDIKLANILLFNDGSVKIADFGLAMRVDGDLTALRKSVEIRGTPLYMA 177
Query: 579 PEVIKNSKECSLGVDIWSLGCTVLEMATTKPPWSQYEG---VAAMFKIGNSKELPTIPDH 635
PE + N E D+W+LGC V+EM + K WS EG ++ + +IG ELP IP+
Sbjct: 178 PECV-NDNEYGSAADVWALGCAVVEMFSGKTAWSVKEGSHFMSLLIRIGVGDELPKIPEM 236
Query: 636 LSNEGKDFVRKCLQRNPRDRPSASELLDHPFV 667
LS EGKDF+ KC ++P R +A LL+H FV
Sbjct: 237 LSEEGKDFLSKCFVKDPAKRWTAEMLLNHSFV 268
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.131 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,890,549
Number of Sequences: 26719
Number of extensions: 948641
Number of successful extensions: 5918
Number of sequences better than 10.0: 1091
Number of HSP's better than 10.0 without gapping: 894
Number of HSP's successfully gapped in prelim test: 198
Number of HSP's that attempted gapping in prelim test: 2760
Number of HSP's gapped (non-prelim): 1513
length of query: 754
length of database: 11,318,596
effective HSP length: 107
effective length of query: 647
effective length of database: 8,459,663
effective search space: 5473401961
effective search space used: 5473401961
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC121237.19


