

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC119419.1 + phase: 0 /partial
(107 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
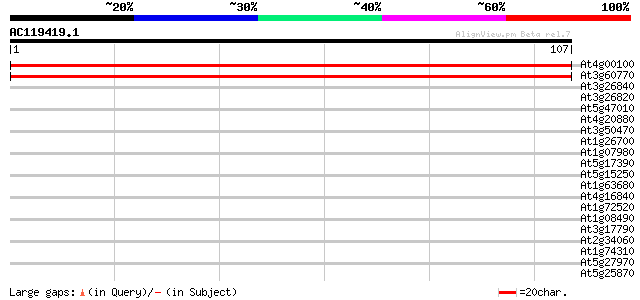
Score E
Sequences producing significant alignments: (bits) Value
At4g00100 putative ribosomal protein S13 207 5e-55
At3g60770 ribosomal protein S13 207 5e-55
At3g26840 unknown protein 27 1.3
At3g26820 unknown protein 27 1.3
At5g47010 UPF1 27 1.7
At4g20880 ethylene-regulated transcript 2 (ERT2) 27 1.7
At3g50470 hypothetical protein 27 1.7
At1g26700 membrane protein Mlo14 27 1.7
At1g07980 unknown protein 27 1.7
At5g17390 putative protein 27 2.2
At5g15250 FtsH-like protein Pftf precursor - like 27 2.2
At1g63680 putative UDP-N-acetylmuramoylalanyl-D-glutamate--2,6-d... 26 2.9
At4g16840 hypothetical protein 26 3.8
At1g72520 putative lipoxygenase 26 3.8
At1g08490 NIFS like protein CpNifsp precursor (NIFS) 26 3.8
At3g17790 acid phosphatase type 5 25 4.9
At2g34060 putative peroxidase 25 4.9
At1g74310 heat shock protein 101 25 4.9
At5g27970 unknown protein 25 6.4
At5g25870 putative protein 25 6.4
>At4g00100 putative ribosomal protein S13
Length = 151
Score = 207 bits (528), Expect = 5e-55
Identities = 102/107 (95%), Positives = 106/107 (98%)
Query: 1 VDETICKFAKKGLTPSQIGVILRDSHGIAQVKAVTGNKILRILKAHGLAPEIPEDLYHLI 60
VDE+ICKFAKKGLTPSQIGVILRDSHGI QVK+VTG+KILRILKAHGLAPEIPEDLYHLI
Sbjct: 33 VDESICKFAKKGLTPSQIGVILRDSHGIPQVKSVTGSKILRILKAHGLAPEIPEDLYHLI 92
Query: 61 KKAVSIRKHLERNRKDKDSKFRLILVESRIHRLARYYKKTKKLPPVW 107
KKAV+IRKHLERNRKDKDSKFRLILVESRIHRLARYYKKTKKLPPVW
Sbjct: 93 KKAVAIRKHLERNRKDKDSKFRLILVESRIHRLARYYKKTKKLPPVW 139
>At3g60770 ribosomal protein S13
Length = 151
Score = 207 bits (528), Expect = 5e-55
Identities = 102/107 (95%), Positives = 106/107 (98%)
Query: 1 VDETICKFAKKGLTPSQIGVILRDSHGIAQVKAVTGNKILRILKAHGLAPEIPEDLYHLI 60
VDE+ICKFAKKGLTPSQIGVILRDSHGI QVK+VTG+KILRILKAHGLAPEIPEDLYHLI
Sbjct: 33 VDESICKFAKKGLTPSQIGVILRDSHGIPQVKSVTGSKILRILKAHGLAPEIPEDLYHLI 92
Query: 61 KKAVSIRKHLERNRKDKDSKFRLILVESRIHRLARYYKKTKKLPPVW 107
KKAV+IRKHLERNRKDKDSKFRLILVESRIHRLARYYKKTKKLPPVW
Sbjct: 93 KKAVAIRKHLERNRKDKDSKFRLILVESRIHRLARYYKKTKKLPPVW 139
>At3g26840 unknown protein
Length = 701
Score = 27.3 bits (59), Expect = 1.3
Identities = 18/60 (30%), Positives = 28/60 (46%), Gaps = 2/60 (3%)
Query: 47 GLAPEIPEDLYHLIKKAVSIRKHLERNRKDKDSKFRLIL-VESRIHRLARYYKKTKKLPP 105
GL P+IP Y+ K + E+ KDK+ L L V+S + + Y K ++ P
Sbjct: 618 GLVPKIPGRFYYYFGKPIETAGK-EKELKDKEKAQELYLQVKSEVEQCIDYLKVKRESDP 676
>At3g26820 unknown protein
Length = 634
Score = 27.3 bits (59), Expect = 1.3
Identities = 19/75 (25%), Positives = 32/75 (42%), Gaps = 2/75 (2%)
Query: 32 KAVTGNKILRILKAHGLAPEIPEDLYHLIKKAVSIRKHLERNRKDKDSKFRLIL-VESRI 90
K N +I GL P+IP Y+ K + + E+ KDK+ + L +S +
Sbjct: 536 KETKANWETKIAIIPGLVPKIPGRFYYYFGKPIDLAGK-EKELKDKEKAQEVYLQAKSEV 594
Query: 91 HRLARYYKKTKKLPP 105
+ Y K ++ P
Sbjct: 595 EQCIAYLKMKRECDP 609
>At5g47010 UPF1
Length = 1254
Score = 26.9 bits (58), Expect = 1.7
Identities = 11/21 (52%), Positives = 15/21 (71%)
Query: 1 VDETICKFAKKGLTPSQIGVI 21
V++ + F K G+ PSQIGVI
Sbjct: 793 VEKLVTAFLKSGVVPSQIGVI 813
>At4g20880 ethylene-regulated transcript 2 (ERT2)
Length = 405
Score = 26.9 bits (58), Expect = 1.7
Identities = 13/43 (30%), Positives = 24/43 (55%)
Query: 61 KKAVSIRKHLERNRKDKDSKFRLILVESRIHRLARYYKKTKKL 103
K+ +IR + + K K +K + +V+ + + R YKK KK+
Sbjct: 253 KEMQTIRDVMFKTEKSKSAKLKSKIVKKIVPKKLRSYKKKKKM 295
>At3g50470 hypothetical protein
Length = 213
Score = 26.9 bits (58), Expect = 1.7
Identities = 14/33 (42%), Positives = 22/33 (66%), Gaps = 1/33 (3%)
Query: 51 EIPEDLYHLIKKAVS-IRKHLERNRKDKDSKFR 82
++ EDL HL++KAVS + + E R++ KFR
Sbjct: 67 KVIEDLKHLLEKAVSLVEAYAELRRRNLLKKFR 99
>At1g26700 membrane protein Mlo14
Length = 554
Score = 26.9 bits (58), Expect = 1.7
Identities = 11/24 (45%), Positives = 17/24 (70%)
Query: 81 FRLILVESRIHRLARYYKKTKKLP 104
F ++VE IHRL+ + +KTK+ P
Sbjct: 29 FVSLIVERSIHRLSNWLQKTKRKP 52
>At1g07980 unknown protein
Length = 206
Score = 26.9 bits (58), Expect = 1.7
Identities = 11/30 (36%), Positives = 19/30 (62%)
Query: 37 NKILRILKAHGLAPEIPEDLYHLIKKAVSI 66
N+I RI+++ AP+I +D L+ KA +
Sbjct: 113 NRIRRIMRSDNSAPQIMQDAVFLVNKATEM 142
>At5g17390 putative protein
Length = 285
Score = 26.6 bits (57), Expect = 2.2
Identities = 18/62 (29%), Positives = 29/62 (46%), Gaps = 11/62 (17%)
Query: 54 EDLYHLIKKAVSIRK--------HLERNRKDKDSKFRLILVESRIHRLARYYKKTKKLPP 105
++L H +KK ++ LE KDK K I+ ES+ +++ +K PP
Sbjct: 171 DELVHTLKKLCQTKRPGIEVEIRRLEGKDKDKGQK---IVEESKKQQVSLLVVGQEKKPP 227
Query: 106 VW 107
VW
Sbjct: 228 VW 229
>At5g15250 FtsH-like protein Pftf precursor - like
Length = 687
Score = 26.6 bits (57), Expect = 2.2
Identities = 13/52 (25%), Positives = 28/52 (53%), Gaps = 5/52 (9%)
Query: 39 ILRILKAHGLAPEIPEDLYHLIKKAVS-----IRKHLERNRKDKDSKFRLIL 85
+LR+L + ++ ++ ED+ +KK + +KH+ NR+ D ++L
Sbjct: 598 VLRMLARNSMSEKLAEDIDSCVKKIIGDAYEVAKKHVRNNREAIDKLVDVLL 649
>At1g63680 putative
UDP-N-acetylmuramoylalanyl-D-glutamate--2,
6-diaminopimelate ligase
Length = 767
Score = 26.2 bits (56), Expect = 2.9
Identities = 12/32 (37%), Positives = 17/32 (52%)
Query: 50 PEIPEDLYHLIKKAVSIRKHLERNRKDKDSKF 81
PE PED H + K I++ R RK ++ F
Sbjct: 59 PEAPEDSMHGVSKFQQIQRQAARARKLEEEDF 90
>At4g16840 hypothetical protein
Length = 661
Score = 25.8 bits (55), Expect = 3.8
Identities = 15/41 (36%), Positives = 21/41 (50%), Gaps = 8/41 (19%)
Query: 22 LRDSHGIAQVKAVTGNKIL--------RILKAHGLAPEIPE 54
LR HG+ +T N +L R+++AH L EIPE
Sbjct: 81 LRVFHGMRAKNTITWNSLLIGISKDPSRMMEAHQLFDEIPE 121
>At1g72520 putative lipoxygenase
Length = 926
Score = 25.8 bits (55), Expect = 3.8
Identities = 14/61 (22%), Positives = 31/61 (49%)
Query: 40 LRILKAHGLAPEIPEDLYHLIKKAVSIRKHLERNRKDKDSKFRLILVESRIHRLARYYKK 99
+R + + G+ I EDL ++ + RK + ++K KF++ V + ++ +K+
Sbjct: 49 MRAVNSSGVVAAISEDLVKTLRISTVGRKQEKEEEEEKSVKFKVRAVATVRNKNKEDFKE 108
Query: 100 T 100
T
Sbjct: 109 T 109
>At1g08490 NIFS like protein CpNifsp precursor (NIFS)
Length = 463
Score = 25.8 bits (55), Expect = 3.8
Identities = 9/25 (36%), Positives = 14/25 (56%)
Query: 5 ICKFAKKGLTPSQIGVILRDSHGIA 29
+C F +GL P+ + L HG+A
Sbjct: 387 LCSFNVEGLHPTDLATFLDQQHGVA 411
>At3g17790 acid phosphatase type 5
Length = 338
Score = 25.4 bits (54), Expect = 4.9
Identities = 19/79 (24%), Positives = 37/79 (46%), Gaps = 10/79 (12%)
Query: 16 SQIGVILRD-----SHGIAQVKAVTGNKILRILKAHGLAPEIPEDLYHLIKK-----AVS 65
S + +LRD A+ K V G+ +R + HG E+ E+L ++K+ ++
Sbjct: 192 SYVKALLRDLEVSLKSSKARWKIVVGHHAMRSIGHHGDTKELNEELLPILKENGVDLYMN 251
Query: 66 IRKHLERNRKDKDSKFRLI 84
H ++ D+DS + +
Sbjct: 252 GHDHCLQHMSDEDSPIQFL 270
>At2g34060 putative peroxidase
Length = 346
Score = 25.4 bits (54), Expect = 4.9
Identities = 17/38 (44%), Positives = 21/38 (54%), Gaps = 3/38 (7%)
Query: 1 VDETICKFAKKGLTPSQIGVILRDSH--GIAQVKAVTG 36
VD+ I FA KGLT ++ V+L SH G A K G
Sbjct: 188 VDQLIKLFASKGLTVEEL-VVLSGSHTIGFAHCKNFLG 224
>At1g74310 heat shock protein 101
Length = 911
Score = 25.4 bits (54), Expect = 4.9
Identities = 19/59 (32%), Positives = 28/59 (47%), Gaps = 9/59 (15%)
Query: 53 PEDLYHLIKK--AVSIRKHLERNRKDKDSKFRLILV-------ESRIHRLARYYKKTKK 102
PE++ +L +K + I H KDK SK RLI V ++ L Y+K K+
Sbjct: 410 PEEIDNLERKRMQLEIELHALEREKDKASKARLIEVRKELDDLRDKLQPLTMKYRKEKE 468
>At5g27970 unknown protein
Length = 1623
Score = 25.0 bits (53), Expect = 6.4
Identities = 17/47 (36%), Positives = 21/47 (44%), Gaps = 1/47 (2%)
Query: 62 KAVSIRKHLERNRKDKDSKFRLILVESRIHRLAR-YYKKTKKLPPVW 107
KAV + H RN K +LV I RL R Y+ + LP W
Sbjct: 1056 KAVHMLIHHSRNSAQKQWDETFVLVLGGIARLFRSYFPLLESLPNFW 1102
>At5g25870 putative protein
Length = 173
Score = 25.0 bits (53), Expect = 6.4
Identities = 10/25 (40%), Positives = 17/25 (68%)
Query: 54 EDLYHLIKKAVSIRKHLERNRKDKD 78
ED+ LIK + +RK +E R++K+
Sbjct: 2 EDMKSLIKTSKELRKRIETRRENKE 26
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.139 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,349,533
Number of Sequences: 26719
Number of extensions: 83277
Number of successful extensions: 260
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 13
Number of HSP's successfully gapped in prelim test: 17
Number of HSP's that attempted gapping in prelim test: 246
Number of HSP's gapped (non-prelim): 30
length of query: 107
length of database: 11,318,596
effective HSP length: 83
effective length of query: 24
effective length of database: 9,100,919
effective search space: 218422056
effective search space used: 218422056
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 52 (24.6 bits)
Medicago: description of AC119419.1


