

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149547.9 + phase: 0
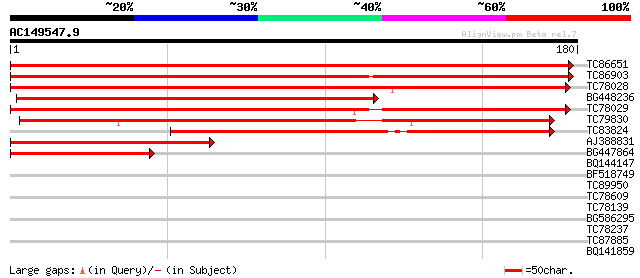
(180 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86651 similar to PIR|F84555|F84555 similar to prolyl 4-hydroxy... 296 3e-81
TC86903 similar to GP|21617881|gb|AAM66931.1 prolyl 4-hydroxylas... 199 5e-52
TC78028 similar to GP|22136524|gb|AAM91340.1 unknown protein {Ar... 184 2e-47
BG448236 similar to GP|21537370|gb putative prolyl 4-hydroxylase... 181 1e-46
TC78029 similar to GP|21593296|gb|AAM65245.1 prolyl 4-hydroxylas... 178 1e-45
TC79830 similar to PIR|G84861|G84861 hypothetical protein At2g43... 165 1e-41
TC83824 similar to GP|17381226|gb|AAL36425.1 unknown protein {Ar... 127 2e-30
AJ388831 weakly similar to GP|10177121|dbj prolyl 4-hydroxylase ... 113 4e-26
BG447864 weakly similar to GP|10177121|db prolyl 4-hydroxylase ... 60 6e-10
BQ144147 39 0.001
BF518749 30 0.40
TC89950 similar to GP|20257495|gb|AAM15917.1 purple acid phospha... 29 0.88
TC78609 similar to GP|21105746|gb|AAM34772.1 nam-like protein 9 ... 28 2.6
TC78139 similar to PIR|S22502|S22502 cysteine proteinase (EC 3.4... 27 4.4
BG586295 27 5.7
TC78237 similar to GP|17933299|gb|AAL48232.1 AT4g17800/dl4935c {... 26 7.5
TC87885 ankyrin-kinase [Medicago truncatula] 26 7.5
BQ141859 26 9.8
>TC86651 similar to PIR|F84555|F84555 similar to prolyl 4-hydroxylase alpha
subunit [imported] - Arabidopsis thaliana, partial (85%)
Length = 1329
Score = 296 bits (758), Expect = 3e-81
Identities = 137/179 (76%), Positives = 153/179 (84%)
Frame = +2
Query: 1 MHKSAVIDEETGNGVDSRERTSSGAFLKRGSDRIVKNIERRIADFTFIPVEHGENFNVLH 60
MHKS V+D ETG DSR RTSSG FL RG D+IV+NIE++IADFTFIPVEHGE VLH
Sbjct: 446 MHKSTVVDSETGKSKDSRVRTSSGTFLARGRDKIVRNIEKKIADFTFIPVEHGEGLQVLH 625
Query: 61 YEVGQKYEPHYDYFMDTFSTTYAGQRIATMLMYLSDVEEGGETVFPNAKGNFSSVPWWNE 120
YEVGQKYEPHYDYF+D F+T GQRIAT+LMYL+DVEEGGETVFP AKGNFS+VPW+NE
Sbjct: 626 YEVGQKYEPHYDYFLDEFNTKNGGQRIATVLMYLTDVEEGGETVFPAAKGNFSNVPWYNE 805
Query: 121 LSDCGKGGLSIKPKMGNAILFWSMKPDATLDPSSLHGACPVIKGDKWLCAKWMHVGEFK 179
LSDCGK GLSIKPK G+A+LFWSMKPDATLD SSLHG CPVIKG+KW KW+ V E+K
Sbjct: 806 LSDCGKKGLSIKPKRGDALLFWSMKPDATLDASSLHGGCPVIKGNKWSSTKWIRVNEYK 982
>TC86903 similar to GP|21617881|gb|AAM66931.1 prolyl 4-hydroxylase putative
{Arabidopsis thaliana}, partial (68%)
Length = 1310
Score = 199 bits (506), Expect = 5e-52
Identities = 93/179 (51%), Positives = 126/179 (69%)
Frame = +3
Query: 1 MHKSAVIDEETGNGVDSRERTSSGAFLKRGSDRIVKNIERRIADFTFIPVEHGENFNVLH 60
+ KS V D E+G + S RTSSG FL + D IV IE RIA +TF+PVE+GE+ VLH
Sbjct: 402 LEKSMVADNESGKSIQSEVRTSSGMFLNKQQDEIVSGIEARIAAWTFLPVENGESMQVLH 581
Query: 61 YEVGQKYEPHYDYFMDTFSTTYAGQRIATMLMYLSDVEEGGETVFPNAKGNFSSVPWWNE 120
Y G+KYEPH+D+F D + G R+AT+LMYLS+VE+GGET+FP+A+G S P
Sbjct: 582 YMNGEKYEPHFDFFHDKANQRLGGHRVATVLMYLSNVEKGGETIFPHAEGKLSQ-PKDES 758
Query: 121 LSDCGKGGLSIKPKMGNAILFWSMKPDATLDPSSLHGACPVIKGDKWLCAKWMHVGEFK 179
S+C G ++KP+ G+A+LF+S+ DAT D SLHG+CPVI+G+KW KW+HV +F+
Sbjct: 759 WSECAHKGYAVKPRKGDALLFFSLHLDATTDSKSLHGSCPVIEGEKWSATKWIHVADFE 935
>TC78028 similar to GP|22136524|gb|AAM91340.1 unknown protein {Arabidopsis
thaliana}, partial (86%)
Length = 1217
Score = 184 bits (467), Expect = 2e-47
Identities = 90/182 (49%), Positives = 123/182 (67%), Gaps = 4/182 (2%)
Frame = +1
Query: 1 MHKSAVIDEETGNGVDSRERTSSGAFLKRGSDRIVKNIERRIADFTFIPVEHGENFNVLH 60
+ +SAV D +G S RTSSG F+ + D IV IE +I+ +TF+P E+GE+ VL
Sbjct: 223 LKRSAVADNLSGESKLSEVRTSSGMFISKNKDAIVSGIEDKISSWTFLPKENGEDIQVLR 402
Query: 61 YEVGQKYEPHYDYFMDTFSTTYAGQRIATMLMYLSDVEEGGETVFPNAKGNFSSVPWWNE 120
YE GQKY+PHYDYF D + G R+AT+LMYL++V +GGETVFPNA+ S +E
Sbjct: 403 YEHGQKYDPHYDYFADKVNIARGGHRVATVLMYLTNVTKGGETVFPNAELQESPRHKLSE 582
Query: 121 ----LSDCGKGGLSIKPKMGNAILFWSMKPDATLDPSSLHGACPVIKGDKWLCAKWMHVG 176
LS+CGK G+++KP+ G+A+LF+S+ P+A D SLH CPVI+G+KW KW+HV
Sbjct: 583 TDEDLSECGKKGVAVKPRRGDALLFFSLHPNAIPDTLSLHAGCPVIEGEKWSATKWIHVD 762
Query: 177 EF 178
F
Sbjct: 763 SF 768
>BG448236 similar to GP|21537370|gb putative prolyl 4-hydroxylase alpha
subunit {Arabidopsis thaliana}, partial (52%)
Length = 682
Score = 181 bits (459), Expect = 1e-46
Identities = 84/115 (73%), Positives = 97/115 (84%)
Frame = +3
Query: 3 KSAVIDEETGNGVDSRERTSSGAFLKRGSDRIVKNIERRIADFTFIPVEHGENFNVLHYE 62
KS+V+D +TG +SR RTSSG FLKRG D+I++NIERRIADFTFIPVE+GE VLHY
Sbjct: 303 KSSVVDSKTGKSTESRVRTSSGMFLKRGKDKIIQNIERRIADFTFIPVENGEGLQVLHYG 482
Query: 63 VGQKYEPHYDYFMDTFSTTYAGQRIATMLMYLSDVEEGGETVFPNAKGNFSSVPW 117
VG+KYEPHYDYF+D F+T GQR+AT+LMYLSDVEEGGETVFP A NFSSVPW
Sbjct: 483 VGEKYEPHYDYFLDEFNTKNGGQRVATVLMYLSDVEEGGETVFPAAXANFSSVPW 647
>TC78029 similar to GP|21593296|gb|AAM65245.1 prolyl 4-hydroxylase alpha
subunit-like protein {Arabidopsis thaliana}, partial
(89%)
Length = 1126
Score = 178 bits (451), Expect = 1e-45
Identities = 87/184 (47%), Positives = 124/184 (67%), Gaps = 6/184 (3%)
Frame = +3
Query: 1 MHKSAVIDEETGNGVDSRERTSSGAFLKRGSDRIVKNIERRIADFTFIPVEHGENFNVLH 60
+ +SAV D +G+ S RTSSG F+ + D IV IE RI+ +TF+P E+GE+ VL
Sbjct: 315 LKRSAVADNLSGDSQLSDVRTSSGMFISKNKDPIVSGIEDRISAWTFLPKENGEDIQVLR 494
Query: 61 YEVGQKYEPHYDYFMDTFSTTYAGQRIATMLMYLSDVEEGGETVFPNA------KGNFSS 114
YE GQKY+PHYDYF D + G R+AT+LMYL++V +GGETVFP A +G+ S
Sbjct: 495 YEHGQKYDPHYDYFADKVNIVQGGHRLATVLMYLTNVTKGGETVFPEAEEPPRRRGSKKS 674
Query: 115 VPWWNELSDCGKGGLSIKPKMGNAILFWSMKPDATLDPSSLHGACPVIKGDKWLCAKWMH 174
++LS+C K G+++KP+ G+A+LF+S+ +A D +SLH CPV++G+KW KW+H
Sbjct: 675 ----SDLSECAKKGIAVKPRRGDALLFFSLDTNAIPDTNSLHAGCPVLEGEKWSATKWIH 842
Query: 175 VGEF 178
V F
Sbjct: 843 VDSF 854
>TC79830 similar to PIR|G84861|G84861 hypothetical protein At2g43080
[imported] - Arabidopsis thaliana, partial (87%)
Length = 1130
Score = 165 bits (417), Expect = 1e-41
Identities = 88/174 (50%), Positives = 108/174 (61%), Gaps = 4/174 (2%)
Frame = +1
Query: 4 SAVIDEETGNGVDSRERTSSGAFLKRGSDR--IVKNIERRIADFTFIPVEHGENFNVLHY 61
S V+D TG G+ S RTSSG FL + ++ IE+RI+ ++ IP+E+GE VL Y
Sbjct: 421 STVVDANTGKGIKSDVRTSSGMFLSHEERKYPMIHAIEKRISVYSQIPIENGELMQVLRY 600
Query: 62 EVGQKYEPHYDYFMDTFSTTYAGQRIATMLMYLSDVEEGGETVFPNAKGNFSSVPWWNEL 121
E Q Y PH+DYF DTF+ GQRIATMLMYL D EGGET FP+A +E
Sbjct: 601 EKNQYYRPHHDYFSDTFNLKRGGQRIATMLMYLGDNVEGGETHFPSAGS--------DEC 756
Query: 122 SDCGK--GGLSIKPKMGNAILFWSMKPDATLDPSSLHGACPVIKGDKWLCAKWM 173
S GK GL +KP GNA+LFWSM D DP S+HG CPV+ G+KW KWM
Sbjct: 757 SCGGKLTKGLCVKPVKGNAVLFWSMGLDGQSDPDSVHGGCPVLAGEKWSATKWM 918
>TC83824 similar to GP|17381226|gb|AAL36425.1 unknown protein {Arabidopsis
thaliana}, partial (43%)
Length = 680
Score = 127 bits (320), Expect = 2e-30
Identities = 61/122 (50%), Positives = 82/122 (67%)
Frame = +3
Query: 52 HGENFNVLHYEVGQKYEPHYDYFMDTFSTTYAGQRIATMLMYLSDVEEGGETVFPNAKGN 111
HGE FN+L YEVGQ+Y HYD F QR+A+ L+YL+DVEEGGET+FP G
Sbjct: 3 HGEAFNILRYEVGQRYNSHYDAFNPDEYGPQKSQRVASFLLYLTDVEEGGETMFPFENGL 182
Query: 112 FSSVPWWNELSDCGKGGLSIKPKMGNAILFWSMKPDATLDPSSLHGACPVIKGDKWLCAK 171
+ E DC GL +KP+ G+ +LF+S+ P+ T+D +SLHG+CPVIKG+KW+ K
Sbjct: 183 NMDGTYGYE--DC--VGLRVKPRQGDGLLFYSLLPNGTIDQTSLHGSCPVIKGEKWVATK 350
Query: 172 WM 173
W+
Sbjct: 351 WI 356
>AJ388831 weakly similar to GP|10177121|dbj prolyl 4-hydroxylase alpha
subunit-like protein {Arabidopsis thaliana}, partial
(33%)
Length = 505
Score = 113 bits (283), Expect = 4e-26
Identities = 56/65 (86%), Positives = 59/65 (90%)
Frame = +3
Query: 1 MHKSAVIDEETGNGVDSRERTSSGAFLKRGSDRIVKNIERRIADFTFIPVEHGENFNVLH 60
MHKSAVIDEETGNGVDS ERTSSGAFLKRGSDRIVKNIERRIADFTFIP EHGENFN L
Sbjct: 291 MHKSAVIDEETGNGVDSSERTSSGAFLKRGSDRIVKNIERRIADFTFIPXEHGENFNGLX 470
Query: 61 YEVGQ 65
++ +
Sbjct: 471 MKLAK 485
>BG447864 weakly similar to GP|10177121|db prolyl 4-hydroxylase alpha
subunit-like protein {Arabidopsis thaliana}, partial
(50%)
Length = 639
Score = 59.7 bits (143), Expect = 6e-10
Identities = 28/46 (60%), Positives = 32/46 (68%)
Frame = +3
Query: 1 MHKSAVIDEETGNGVDSRERTSSGAFLKRGSDRIVKNIERRIADFT 46
MHKS V+D ETG DS RT SG FL RG D I +NI ++IADFT
Sbjct: 501 MHKSTVVDSETGKSKDSXVRTXSGTFLARGRDXIXRNIXKKIADFT 638
>BQ144147
Length = 729
Score = 38.9 bits (89), Expect = 0.001
Identities = 19/70 (27%), Positives = 37/70 (52%)
Frame = +2
Query: 46 TFIPVEHGENFNVLHYEVGQKYEPHYDYFMDTFSTTYAGQRIATMLMYLSDVEEGGETVF 105
T +P+E+G+N ++ Y+ G + P+ +Y + G R T+LM +++ +G F
Sbjct: 92 TSLPIENGDNLHIWRYKHGHNHNPNDNYSTHKINIVQGGHRPPTVLMLITNETKGNRN*F 271
Query: 106 PNAKGNFSSV 115
+ G SS+
Sbjct: 272 -SQSGRTSSM 298
>BF518749
Length = 416
Score = 30.4 bits (67), Expect = 0.40
Identities = 17/53 (32%), Positives = 30/53 (56%)
Frame = +1
Query: 8 DEETGNGVDSRERTSSGAFLKRGSDRIVKNIERRIADFTFIPVEHGENFNVLH 60
++ +GNG S++ +S D IVK IE R++ +TF+ E+ + V+H
Sbjct: 268 EKSSGNGGYSKKEETSLDM----DDDIVKRIEERLSVWTFLSKENSKPLQVMH 414
>TC89950 similar to GP|20257495|gb|AAM15917.1 purple acid phosphatase
{Arabidopsis thaliana}, partial (59%)
Length = 1014
Score = 29.3 bits (64), Expect = 0.88
Identities = 16/50 (32%), Positives = 23/50 (46%)
Frame = +1
Query: 114 SVPWWNELSDCGKGGLSIKPKMGNAILFWSMKPDATLDPSSLHGACPVIK 163
SV WWN G+G L PK + + DP+ L G+CP ++
Sbjct: 127 SVVWWNPTQARGRGWL---PKATMRLRVSLLSNHMPSDPTMLVGSCPTMR 267
>TC78609 similar to GP|21105746|gb|AAM34772.1 nam-like protein 9 {Petunia x
hybrida}, partial (39%)
Length = 1931
Score = 27.7 bits (60), Expect = 2.6
Identities = 11/38 (28%), Positives = 20/38 (51%)
Frame = +1
Query: 5 AVIDEETGNGVDSRERTSSGAFLKRGSDRIVKNIERRI 42
+ +D++ G G + T G + G DR++KN E +
Sbjct: 304 SALDKKYGTGCRTNRATDRGYWKTTGKDRVIKNGEETV 417
>TC78139 similar to PIR|S22502|S22502 cysteine proteinase (EC 3.4.22.-) -
kidney bean, complete
Length = 1720
Score = 26.9 bits (58), Expect = 4.4
Identities = 18/68 (26%), Positives = 25/68 (36%)
Frame = +3
Query: 50 VEHGENFNVLHYEVGQKYEPHYDYFMDTFSTTYAGQRIATMLMYLSDVEEGGETVFPNAK 109
V H N N + K D F TTYAG ++ M+ G ++ N
Sbjct: 630 VMHVHNTNKMDKPYKLKLNKFADMTNHEFKTTYAGSKVNHHRMFRGTPRVSGTFMYENFT 809
Query: 110 GNFSSVPW 117
+SV W
Sbjct: 810 KAPASVDW 833
>BG586295
Length = 807
Score = 26.6 bits (57), Expect = 5.7
Identities = 27/117 (23%), Positives = 48/117 (40%)
Frame = -3
Query: 4 SAVIDEETGNGVDSRERTSSGAFLKRGSDRIVKNIERRIADFTFIPVEHGENFNVLHYEV 63
SA ID T +DS+ RTS RG R+V ++ ++D HGE + Y
Sbjct: 370 SASIDRPTNISIDSQRRTSIDEATNRG--RLVPKMKSDMSD----THNHGEEISTDTYAT 209
Query: 64 GQKYEPHYDYFMDTFSTTYAGQRIATMLMYLSDVEEGGETVFPNAKGNFSSVPWWNE 120
+++ + + D Q+I ++D+ G+ + G W+N+
Sbjct: 208 LMRHQFNLESLGDRL------QKIENTTATMNDIWCRGDEAIRHFTGT-----WFNK 71
>TC78237 similar to GP|17933299|gb|AAL48232.1 AT4g17800/dl4935c {Arabidopsis
thaliana}, partial (59%)
Length = 1166
Score = 26.2 bits (56), Expect = 7.5
Identities = 8/15 (53%), Positives = 9/15 (59%)
Frame = +2
Query: 112 FSSVPWWNELSDCGK 126
F VPWW S CG+
Sbjct: 581 FECVPWWRPRSSCGR 625
>TC87885 ankyrin-kinase [Medicago truncatula]
Length = 1605
Score = 26.2 bits (56), Expect = 7.5
Identities = 10/21 (47%), Positives = 16/21 (75%)
Frame = +1
Query: 145 KPDATLDPSSLHGACPVIKGD 165
+P+A +DPSSL+ A + +GD
Sbjct: 1228 QPEANIDPSSLYVAPEIYRGD 1290
>BQ141859
Length = 862
Score = 25.8 bits (55), Expect = 9.8
Identities = 12/33 (36%), Positives = 17/33 (51%)
Frame = -1
Query: 107 NAKGNFSSVPWWNELSDCGKGGLSIKPKMGNAI 139
+AKGN +PW E ++ KG I+ G I
Sbjct: 271 SAKGNVFDIPWQYEENEIPKGVFGIRGGTGGHI 173
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.137 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,852,311
Number of Sequences: 36976
Number of extensions: 78133
Number of successful extensions: 345
Number of sequences better than 10.0: 37
Number of HSP's better than 10.0 without gapping: 339
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 339
length of query: 180
length of database: 9,014,727
effective HSP length: 90
effective length of query: 90
effective length of database: 5,686,887
effective search space: 511819830
effective search space used: 511819830
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 55 (25.8 bits)
Medicago: description of AC149547.9


