

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149495.11 - phase: 0
(421 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
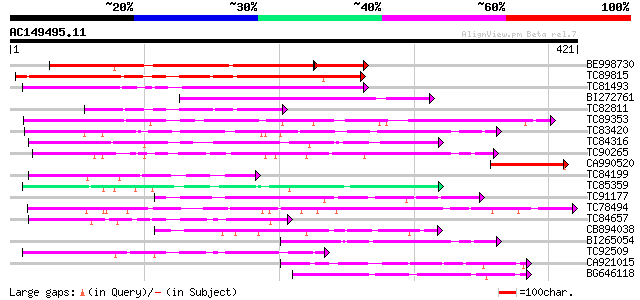
Score E
Sequences producing significant alignments: (bits) Value
BE998730 232 2e-74
TC89815 236 1e-62
TC81493 weakly similar to PIR|F86249|F86249 protein F25C20.23 [i... 174 6e-44
BI272761 155 3e-38
TC82811 116 2e-26
TC89353 weakly similar to GP|9279704|dbj|BAB01261.1 gb|AAF25964.... 97 9e-21
TC83420 weakly similar to GP|5903055|gb|AAD55614.1| May be a pse... 92 5e-19
TC84316 weakly similar to GP|11994612|dbj|BAB02749. gb|AAC24186.... 91 1e-18
TC90265 weakly similar to GP|9294666|dbj|BAB03015.1 gene_id:F14O... 90 1e-18
CA990520 75 5e-14
TC84199 similar to GP|9294656|dbj|BAB03005.1 gb|AAF30317.1~gene_... 74 8e-14
TC85359 weakly similar to PIR|C96592|C96592 hypothetical protein... 74 8e-14
TC91177 similar to GP|9294656|dbj|BAB03005.1 gb|AAF30317.1~gene_... 73 2e-13
TC78494 72 4e-13
TC84657 weakly similar to GP|9294656|dbj|BAB03005.1 gb|AAF30317.... 72 4e-13
CB894038 71 7e-13
BI265054 68 6e-12
TC92509 weakly similar to PIR|T49129|T49129 hypothetical protein... 67 1e-11
CA921015 homologue to SP|Q8Y7G1|DPO3 DNA polymerase III polC-typ... 66 2e-11
BG646118 66 3e-11
>BE998730
Length = 820
Score = 232 bits (592), Expect(2) = 2e-74
Identities = 125/202 (61%), Positives = 141/202 (68%), Gaps = 2/202 (0%)
Frame = +3
Query: 30 KSLLRFRSTSKSLRSLIDSHKFINLHLKSSLNFKLILRHKTNLYQLD--FPNLTKSIIPL 87
KSLLRFRSTSKSL+S+IDSH F NLHLK+SLN LI+R ++YQ+D F +LT + PL
Sbjct: 81 KSLLRFRSTSKSLKSIIDSHNFTNLHLKNSLNRSLIIRRNYDIYQIDDDFSDLTTAEFPL 260
Query: 88 NHPFTTNIDPFTLNSIKALIGSCNGLLAIYNGPIAFTHPNDATEITIWNTNTRKHWIIPF 147
NHPF+ N TL IGSCNGLLAI NG IA T PNDA +I IWN NTRKH IIPF
Sbjct: 261 NHPFSGNSTKITL------IGSCNGLLAISNGLIALTQPNDANQIAIWNPNTRKHHIIPF 422
Query: 148 LPLPIPNIVESENIERGGVCIHGFGFDPLTGDYKLLRISWLVSLQNSFYHSHARLFSSKT 207
LPLPI + + +HGFGFD LT DYKLLRISWLV+ N FY SH LFS KT
Sbjct: 423 LPLPITPHPPPDMYCY--LYLHGFGFDLLTADYKLLRISWLVTQHNPFYDSHVTLFSLKT 596
Query: 208 NSWKIIPIMPYTVYYAQAMGVF 229
NSWK IP PY + Y QAMG F
Sbjct: 597 NSWKTIPSTPYALQYVQAMGGF 662
Score = 65.1 bits (157), Expect(2) = 2e-74
Identities = 30/40 (75%), Positives = 34/40 (85%)
Frame = +1
Query: 227 GVFVENSIHWIMEKNLDGSDLCLIVAFNLTLEVFKEVPLP 266
GVFV+NS+HW+M K LDGS LIVAFNLTLE+F EVPLP
Sbjct: 655 GVFVQNSLHWVMAKKLDGSYPWLIVAFNLTLEIFYEVPLP 774
>TC89815
Length = 807
Score = 236 bits (602), Expect = 1e-62
Identities = 136/267 (50%), Positives = 166/267 (61%), Gaps = 7/267 (2%)
Frame = +1
Query: 5 LPSFLPVASDLPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHLKSSLNFKL 64
+P P+A+ +PP+I +I S LP LLRFRSTSKSL+S+IDSH F NLHLK+S NF L
Sbjct: 19 IPLRSPMAN-IPPEIFIDILSLLPPHPLLRFRSTSKSLKSIIDSHTFTNLHLKNSNNFYL 195
Query: 65 ILRHKTNLYQLDFPNLTKSIIPLNHPFTTNIDPFTLNSIKALIGSCNGLLAIYNGPIAFT 124
I+RH NLYQLDFPNLT IPLNHP + + T L GSCNGL+ I
Sbjct: 196 IIRHNANLYQLDFPNLTPP-IPLNHPLMSYSNRIT------LFGSCNGLICI-------- 330
Query: 125 HPNDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVCIHGFGFDPLTGDYKLLR 184
N A +I WN N RKH IIP+LP P + V HGFG+D GDYKL+R
Sbjct: 331 -SNIADDIAFWNPNIRKHRIIPYLP-TTPRSESDTTLFAARV--HGFGYDSFAGDYKLVR 498
Query: 185 ISWLVSLQNSFYHSHARLFSSKTNSWKIIPIMPYTVYYAQAMGVFVE-------NSIHWI 237
IS+ V LQN + S R+FS K NSWK +P M Y + A+ MGVFVE NS+HW+
Sbjct: 499 ISYFVDLQNRSFDSQVRVFSLKMNSWKELPSMNYALCCARTMGVFVEDSNNLNSNSLHWV 678
Query: 238 MEKNLDGSDLCLIVAFNLTLEVFKEVP 264
+ + L+ LIVAFNLTLE+F EVP
Sbjct: 679 VTRKLEPFQPDLIVAFNLTLEIFNEVP 759
>TC81493 weakly similar to PIR|F86249|F86249 protein F25C20.23 [imported] -
Arabidopsis thaliana, partial (9%)
Length = 728
Score = 174 bits (441), Expect = 6e-44
Identities = 107/258 (41%), Positives = 144/258 (55%), Gaps = 1/258 (0%)
Frame = +1
Query: 10 PVASDLPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHLKSSLNFKLILRHK 69
P +DLP ++ EI S +P K LLR RST K R+LIDS FI LHL S + +ILR
Sbjct: 1 PTMADLPTEVTTEILSRVPAKPLLRLRSTCKWWRNLIDSTDFIFLHLSKSRDSVIILRQH 180
Query: 70 TNLYQLDFPNLTKSIIPLNHPFTTNIDPFTLNSIKALIGSCNGLLAIYNGPIAFTHPNDA 129
+ LY+LD ++ + + L+HP N IK L GSCNGLL I N A
Sbjct: 181 SRLYELDLNSMDR-VKELDHPLMCYS-----NRIKVL-GSCNGLLCICN---------IA 312
Query: 130 TEITIWNTNTRKHWIIPFLPLPIPNIVESENIERG-GVCIHGFGFDPLTGDYKLLRISWL 188
+I WN RKH IIP PL E+ I ++GFG+D T DYKL+ IS
Sbjct: 313 DDIAFWNPTIRKHRIIPSEPLIRKETNENNTITTLLAAHVYGFGYDSATDDYKLVSISNF 492
Query: 189 VSLQNSFYHSHARLFSSKTNSWKIIPIMPYTVYYAQAMGVFVENSIHWIMEKNLDGSDLC 248
V L N Y SH +++ ++ W +P +PY + A+ MGVFV ++HW++ + L+
Sbjct: 493 VDLHNRSYDSHVTIYTMGSDVWMPLPGVPYALCCARPMGVFVSGALHWVVPRALEPDSRD 672
Query: 249 LIVAFNLTLEVFKEVPLP 266
LIVAF+L EVF+EV LP
Sbjct: 673 LIVAFDLRFEVFREVALP 726
>BI272761
Length = 600
Score = 155 bits (392), Expect = 3e-38
Identities = 81/190 (42%), Positives = 114/190 (59%), Gaps = 1/190 (0%)
Frame = +2
Query: 127 NDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERG-GVCIHGFGFDPLTGDYKLLRI 185
N A +I WN RKH IIP PL E+ I ++GFG+D T DYKL+ I
Sbjct: 5 NIADDIAFWNPTIRKHRIIPSEPLIRKETNENNTITTLLAAHVYGFGYDSATDDYKLVSI 184
Query: 186 SWLVSLQNSFYHSHARLFSSKTNSWKIIPIMPYTVYYAQAMGVFVENSIHWIMEKNLDGS 245
S+ V L N + SH ++++ +T+ WK +P MPY + A+ MGVFV ++HW++ ++L+
Sbjct: 185 SYFVDLHNRSFDSHVKIYTMRTDVWKTLPSMPYALCCARTMGVFVSGALHWVVTRDLEPE 364
Query: 246 DLCLIVAFNLTLEVFKEVPLPAVIRGEEVNNNQSFDLEVAVLGGCLSMIVNYQTTKIDVW 305
LIVAF+L EVF+EV LP + G+ FD++VA+L G L +I N + DVW
Sbjct: 365 SRDLIVAFDLRFEVFREVALPGTVDGK-------FDMDVALLXGMLCIIENRGSDGFDVW 523
Query: 306 VMKEYGSRDS 315
VM EYGS DS
Sbjct: 524 VMXEYGSHDS 553
>TC82811
Length = 961
Score = 116 bits (290), Expect = 2e-26
Identities = 71/151 (47%), Positives = 84/151 (55%)
Frame = +1
Query: 56 LKSSLNFKLILRHKTNLYQLDFPNLTKSIIPLNHPFTTNIDPFTLNSIKALIGSCNGLLA 115
LK+S NF LI+RH NLYQLDFPNLT I PLNHP + + TL GSCNGL+
Sbjct: 1 LKNSNNFYLIIRHNANLYQLDFPNLTPPI-PLNHPLMSYSNRITL------FGSCNGLIC 159
Query: 116 IYNGPIAFTHPNDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVCIHGFGFDP 175
I N A +I WN N RKH IIP+LP P + V HGFG+D
Sbjct: 160 I---------SNIADDIAFWNPNIRKHRIIPYLPTT-PRSESDTTLFAARV--HGFGYDS 303
Query: 176 LTGDYKLLRISWLVSLQNSFYHSHARLFSSK 206
GDYKL+RIS+ V LQN + S L + K
Sbjct: 304 FAGDYKLVRISYFVDLQNRSFDSQG*LLAPK 396
>TC89353 weakly similar to GP|9279704|dbj|BAB01261.1
gb|AAF25964.1~gene_id:MYA6.2~similar to unknown protein
{Arabidopsis thaliana}, partial (10%)
Length = 1482
Score = 97.4 bits (241), Expect = 9e-21
Identities = 101/416 (24%), Positives = 180/416 (42%), Gaps = 21/416 (5%)
Frame = +3
Query: 11 VASDLPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHLKSSLNFKLILRHKT 70
+ +D+P +I+ EI LPV+SLL+FR K ++LI +F H+ S + ++
Sbjct: 93 ITADMPEEIIVEILLRLPVRSLLQFRCVCKLWKTLISDPQFAKKHVSISTAYPQLVSVFV 272
Query: 71 NLYQLDFPNLTKSIIPLNHPFTTNIDPFTLNSI----KALIGSCNGLLAIYNGPIAFTHP 126
++ + + + + L++P ++P I +IGSCNGLL +
Sbjct: 273 SIAKCNLVSYPLKPL-LDNPSAHRVEPADFEMIHTTSMTIIGSCNGLLCL---------- 419
Query: 127 NDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVCIHGFGFDPLTGDYKLLRIS 186
+D + T+WN + I P P I+ ++ + GFG+D + YK+L +
Sbjct: 420 SDFYQFTLWNPS------IKLKSKPSPTIIAFDSFDSKRFLYRGFGYDQVNDRYKVLAV- 578
Query: 187 WLVSLQNSFYHSHAR--LFSSKTNSWKIIPIMPYTVYYAQ----AMGVFVENSIHWIMEK 240
+QN + + +++ W I P +G FV +++WI+ K
Sbjct: 579 ----VQNCYNLDETKTLIYTFGGKDWTTIQKFPCDPSRCDLGRLGVGKFVSGNLNWIVSK 746
Query: 241 NLDGSDLCLIVAFNLTLEVFKEVPLPAVIRGEE----VNNNQ---SFDLEVAVLGGCLSM 293
+IV F++ E + E+ LP + V++N+ SFD
Sbjct: 747 K-------VIVFFDIEKETYGEMSLPQDYGDKNTVLYVSSNRIYVSFD------------ 869
Query: 294 IVNYQTTKIDVWVMKEYGSRDSWCKIFTLVKSRFTL-RLKSLRPLGYSSDGNKVLLQIDC 352
+ T VW+MKEYG +SW K+ + + + T L + S+ +L++
Sbjct: 870 --HSNKTHWVVWMMKEYGVVESWTKLMIIPQDKLTSPGPYCLSDALFISEHGVLLMRPQH 1043
Query: 353 KKLVWYDLKSEQVI-YVEGIPNLDEAMICV--ESLVPPSFPVDNSSKKENRLSKSK 405
KL Y+L ++ + Y I + + ESLV P + + +S KK R K K
Sbjct: 1044SKLSVYNLNNDGGLDYCTTISGQFARYLHIYNESLVSPHW-LKHSQKKNKRKRKQK 1208
>TC83420 weakly similar to GP|5903055|gb|AAD55614.1| May be a pseudogene of
F6D8.29. {Arabidopsis thaliana}, partial (10%)
Length = 1210
Score = 91.7 bits (226), Expect = 5e-19
Identities = 100/410 (24%), Positives = 170/410 (41%), Gaps = 56/410 (13%)
Frame = +1
Query: 12 ASDLPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINL---------HLKSSLNF 62
+S +P D+ I S LP+KSL RF KS L + F+N+ H + N
Sbjct: 79 SSHIPDDLTFSILSKLPLKSLTRFTCAQKSWSLLFQNSIFMNMFRTNFLLSKHDEDDENT 258
Query: 63 KLILR-------HKTNLYQLDFPNLTKSI-IPLNHPFTTNIDPFTLNSIKALIGS-CNGL 113
++L+ + T+LY L S+ + L P N D F I+ L + NG+
Sbjct: 259 CVLLKQMEGRRPYHTSLYTLSGEKFENSVRLDLPTPLLEN-DSF----IRILCSAGINGV 423
Query: 114 LAIYNGPIAFTHPNDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVCIHGFGF 173
L +YN + N I +WN T + ++P P NI + FG+
Sbjct: 424 LCLYNDSL-----NANKTIVLWNPATEEFKVVPHSHQPYENIEFNPR-------PFAFGY 567
Query: 174 DPLTGDYKLLRI---------SWL---------------VSLQNSFYHSH---------- 199
DP+T DYK++R+ +W+ + Q FY SH
Sbjct: 568 DPVTNDYKVIRVANYHIYFEDNWIYLPRKESLLWRKDDPLWKQYMFYDSHFWEGHELQVY 747
Query: 200 ---ARLFSSKTNSWKIIPIMPYTVYYAQAMGVFVENSIHWIMEKNLDGSDLCLIVAFNLT 256
++S ++NSW+ + M +V+ + + HW + + +F+ T
Sbjct: 748 EPFLEIYSLRSNSWRKLD-MDLSVFLHGRCMMNLNELCHWQVNDQM--------ASFDFT 900
Query: 257 LEVFKEVPLPAVIRGEEVNNNQSFDLEVAVLGGCLSMIVN-YQTTKIDVWVMKEYGSRDS 315
E+F E LP+ ++ + S ++AVL G ++ I N T + +W++ E +S
Sbjct: 901 NEIFFETTLPSF----DLKDGMSTKKDLAVLHGSVAFIHNVVNTGYLHIWILGELAVNES 1068
Query: 316 WCKIFTLVKSRFTLRLKSLRPLGYSSDGNKVLLQIDCKKLVWYDLKSEQV 365
W K+F L RP+G L+ D ++L W DL ++++
Sbjct: 1069WTKLFVLGPVPCI-----GRPIGVRIKSVMFYLKED-EELAWIDLSTQRI 1200
>TC84316 weakly similar to GP|11994612|dbj|BAB02749.
gb|AAC24186.1~gene_id:MGL6.4~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (10%)
Length = 892
Score = 90.5 bits (223), Expect = 1e-18
Identities = 76/313 (24%), Positives = 142/313 (45%), Gaps = 5/313 (1%)
Frame = +3
Query: 15 LPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLH-LKSSLNFKLILRHKTNLY 73
LP +++ I LPV+SLLRF+ KS ++L F N H L S++ +L+ + Y
Sbjct: 42 LPEELIVIILLRLPVRSLLRFKCVCKSWKTLFSDTHFANNHFLISTVYPQLVACESVSAY 221
Query: 74 QLDFPNLTKSIIPLNHPFTTNIDPF--TLNSIKALIGSCNGLLAIYNGPIAFTHPNDATE 131
+ + T I L +T + P T + ++GSCNG L +Y+ N
Sbjct: 222 RT-WEIKTYPIESLLENSSTTVIPVSNTGHQRYTILGSCNGFLCLYD--------NYQRC 374
Query: 132 ITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVCIHGFGFDPLTGDYKLLRISWLVSL 191
+ +WN P N+ + +GFG+D + YKLL + +
Sbjct: 375 VRLWN--------------PSINLKSKSSPTIDRFIYYGFGYDQVNHKYKLLAVKAFSRI 512
Query: 192 QNSFYHSHARLFSSKTNSWKIIPIMPYTVY--YAQAMGVFVENSIHWIMEKNLDGSDLCL 249
+ ++ NS K + + + Y + +G FV +++WI+++ DG
Sbjct: 513 TETMIYTFGE------NSCKNVEVKDFPRYPPNRKHLGKFVSGTLNWIVDER-DGR--AT 665
Query: 250 IVAFNLTLEVFKEVPLPAVIRGEEVNNNQSFDLEVAVLGGCLSMIVNYQTTKIDVWVMKE 309
I++F++ E +++V LP + + + VL C+ + ++ T+ +W+MK+
Sbjct: 666 ILSFDIEKETYRQVLLPQ-------HGYAVYSPGLYVLSNCICVCTSFLDTRWQLWMMKK 824
Query: 310 YGSRDSWCKIFTL 322
YG +SW K+ ++
Sbjct: 825 YGVAESWTKLMSI 863
>TC90265 weakly similar to GP|9294666|dbj|BAB03015.1
gene_id:F14O13.16~pir||T05437~similar to unknown protein
{Arabidopsis thaliana}, partial (6%)
Length = 1373
Score = 90.1 bits (222), Expect = 1e-18
Identities = 103/399 (25%), Positives = 177/399 (43%), Gaps = 53/399 (13%)
Frame = +3
Query: 18 DILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHLKSSLN--------FKLILR-- 67
++ I S LP+KSL RFR KS L ++H F++ K+ ++ L+L+
Sbjct: 93 ELAFSILSKLPLKSLKRFRCVRKSWTLLFENHHFMSTFCKNLISNHHYYYGDVSLLLQIG 272
Query: 68 ----------HKTNLYQLDFPNLTKSIIPLNHPFTTNIDPF--TLNSIKALIGSCNGLLA 115
N+ +LD+PN PF DP+ L+S GS G+L
Sbjct: 273 DRDLLSLSSDRYENMVKLDWPN----------PFQEE-DPWFCILDS-----GSITGILC 404
Query: 116 IYNGPIAFTHPNDATEITI-WNTNTRKHWIIPFLPLPIPNIVESENIERGGVCIHGFGFD 174
+YN N E T+ WN T++ +IP P P+ + + G +HGFG+D
Sbjct: 405 LYN------RNNRNNERTVFWNPATKEFKVIP--PSPLEAVPTYQGF---GTVLHGFGYD 551
Query: 175 PLTGDYKLLRISWL------------VSLQNSFY-----HSHARLFSSKTNSWKIIPIMP 217
DYKL+R + +SLQ+ + S ++S ++NSWK + I
Sbjct: 552 HARDDYKLIRYLYYFLPSSRDFEDLGISLQDVPWGDISNDSFWEIYSLRSNSWKKLDINM 731
Query: 218 YT---------VYYAQAMGVFVENSIHWIMEKNLDGSDLCLIVA-FNLTLEVFKE--VPL 265
Y ++ ++++ HW +D D +A F+L EVF +PL
Sbjct: 732 YLGDIRCSFSGFDCVKSQRLYLDGRCHWW--HLIDHPDAKRALASFDLVNEVFFTTLIPL 905
Query: 266 PAVIRGEEVNNNQSFDLEVAVLGGCLSMIV-NYQTTKIDVWVMKEYGSRDSWCKIFTLVK 324
+ +++ + S L + L G +++I+ ++ T D++V+ E G ++SW K+FT+
Sbjct: 906 DPPLDVDDIFSVFSRPLYLVALSGSIALILWDFGTPTFDIYVLGEVGVKESWTKLFTIGP 1085
Query: 325 SRFTLRLKSLRPLGYSSDGNKVLLQIDCKKLVWYDLKSE 363
RP+G S G V + ++VW++ S+
Sbjct: 1086LACI-----QRPIGVGSKG--VFFIKEDGEIVWFNWNSQ 1181
>CA990520
Length = 592
Score = 75.1 bits (183), Expect = 5e-14
Identities = 39/60 (65%), Positives = 44/60 (73%), Gaps = 2/60 (3%)
Frame = +2
Query: 358 YDLKSEQVIYVEGIPNLDEAMICVESLVPPSFPVDNSSKKENRLSKSKRRYFF--IDYLS 415
YD +S QV YVEGIP+ + M V SLVPPSFPVDNS KKENR SKSKR + F D+LS
Sbjct: 2 YDFRSAQVSYVEGIPDFHDVMFSVGSLVPPSFPVDNSRKKENRTSKSKRSFSFHRDDFLS 181
>TC84199 similar to GP|9294656|dbj|BAB03005.1
gb|AAF30317.1~gene_id:F14O13.6~similar to unknown
protein {Arabidopsis thaliana}, partial (14%)
Length = 673
Score = 74.3 bits (181), Expect = 8e-14
Identities = 53/178 (29%), Positives = 81/178 (44%), Gaps = 6/178 (3%)
Frame = +1
Query: 15 LPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHL----KSSLNFKLILRHKT 70
LP +++ +I LPVKSLLRF+ KS SLI F N H K S +L H
Sbjct: 109 LPLELIIQILLWLPVKSLLRFKCVCKSWFSLISDTHFANSHFQITAKHSRRVLFMLNHVP 288
Query: 71 NLYQLDFPNL--TKSIIPLNHPFTTNIDPFTLNSIKALIGSCNGLLAIYNGPIAFTHPND 128
LDF L ++ + +P ++P +S+ SC G + ++N P
Sbjct: 289 TTLSLDFEALHCDNAVSEIPNPIPNFVEP-PCDSLDTNSSSCRGFIFLHNDP-------- 441
Query: 129 ATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVCIHGFGFDPLTGDYKLLRIS 186
++ IWN +TR + IP P N C++GFG+D L DY ++ ++
Sbjct: 442 --DLFIWNPSTRVYKQIPLSP----------NDSNSFHCLYGFGYDQLRDDYLVVSVT 579
>TC85359 weakly similar to PIR|C96592|C96592 hypothetical protein T7N22.2
[imported] - Arabidopsis thaliana, partial (7%)
Length = 1955
Score = 74.3 bits (181), Expect = 8e-14
Identities = 90/364 (24%), Positives = 140/364 (37%), Gaps = 51/364 (14%)
Frame = +2
Query: 10 PVASDLPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHLKSSLNFKLILRH- 68
P +LP + I LP+KSLL R K +LI F L + + ++R+
Sbjct: 497 PYFDNLPSQLTTHILLKLPIKSLLICRCVCKIWNTLISEPHFAKLQFERA-PVSFVIRNL 673
Query: 69 -----KTNLYQLD-----FPNLTKSIIPLNHPFT-------------------------T 93
NLY L+ F +K+ + L+ F +
Sbjct: 674 DNIGVSRNLYLLECEAEKFEIGSKNHVKLDPIFELPLCKDISSRDKNDAKFYKVIKKKKS 853
Query: 94 NIDPFTLNSIK---ALIGSCNGLLAIYNGPIAFTHPNDATEITIWNTNTRKHWIIPFLPL 150
I FTL S + ++ SCNGLL + I + + I N TR+ I+P L
Sbjct: 854 KIRYFTLTSSRDKFGIVNSCNGLLCLSETSIG-------SPLVICNPVTREFTILPELT- 1009
Query: 151 PIPNIVESENIERGGVCIH-GFGFDPLTGDYKLLRISWLVSLQNSFYHSHARLFSSK--- 206
S+ R + GFGF P T +YK++ I W N + + RL +
Sbjct: 1010-----TTSDWFNRARARVQAGFGFQPKTNEYKVI-IMW-----NKYVRRNNRLVFERVVL 1156
Query: 207 ------TNSWKIIPIMPYTVYYAQAMGVFVENSIHWIM-EKNLDGSDLCLIVAFNLTLEV 259
T SW+ + + P + V ++HWI+ E S LC FN E
Sbjct: 1157EIHTLGTPSWRKVEVDPQISFLKLLNPTCVNGALHWIIFETGQQKSILC----FNFESER 1324
Query: 260 FKEVPLPAVIRGEEVNNNQ-SFDLEVAVLGGCLSMIVNYQTTKIDVWVMKEYGSRDSWCK 318
+ P P + G N S + + L G L + + +WVM EYG +SW
Sbjct: 1325LQSFPSPPHVFGNHDNGFPLSMPIRLGELKGFLYICHISSLENVTMWVMNEYGIGESWTI 1504
Query: 319 IFTL 322
++++
Sbjct: 1505VYSI 1516
>TC91177 similar to GP|9294656|dbj|BAB03005.1
gb|AAF30317.1~gene_id:F14O13.6~similar to unknown
protein {Arabidopsis thaliana}, partial (9%)
Length = 1298
Score = 73.2 bits (178), Expect = 2e-13
Identities = 69/251 (27%), Positives = 111/251 (43%), Gaps = 6/251 (2%)
Frame = +2
Query: 108 GSCNGLLAIYNGPIAFTHPNDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVC 167
GSC G + + N + + IWN +T H IPF + +
Sbjct: 356 GSCRGFILL----------NCYSSLCIWNPSTGFHKRIPFTTI---------DSNPDANY 478
Query: 168 IHGFGFDPLTGDYKLLRISWLVSLQNSFYHSHARLFSSKTNSWKIIPIMPYTVYYAQAMG 227
+GFG+D T DY ++ +S+ S + SH +FS + N W + +Y ++
Sbjct: 479 FYGFGYDESTDDYLVISMSYEPSPSSDGMLSHLGIFSLRANVWTRVEGGNLLLYSQNSLL 658
Query: 228 VFVEN----SIHWIMEKNLDGSDLCLIVAFNLTLEVFKEVPLPAVIRGEEVNNNQSFDLE 283
VE+ +IHW+ +N + +IVAF+L E+ LP E+ N S +
Sbjct: 659 NLVESLSNGAIHWLAFRN--DISMPVIVAFHLMERKLLELRLP-----NEIINGPSRAYD 817
Query: 284 VAVLGGCLSM--IVNYQTTKIDVWVMKEYGSRDSWCKIFTLVKSRFTLRLKSLRPLGYSS 341
+ V GCL++ I+ + T +WVM++Y + SW K TLV S S P Y+
Sbjct: 818 LWVYRGCLALWHILPDRVT-FQIWVMEKYNVQSSWTK--TLVLSFDGNPAHSFWPKYYTK 988
Query: 342 DGNKVLLQIDC 352
G+ V + C
Sbjct: 989 SGDIVGRNMRC 1021
>TC78494
Length = 1450
Score = 72.0 bits (175), Expect = 4e-13
Identities = 100/447 (22%), Positives = 185/447 (41%), Gaps = 39/447 (8%)
Frame = +1
Query: 14 DLPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLH------LKSSLNFKLILR 67
+LP ++++ I S LP + LL+ + K+ +LI F N + L++ L++R
Sbjct: 184 NLPHELVSNILSRLPSRELLKNKLVCKTWYNLITDSHFTNNYYSFHNQLQNQEEHLLVIR 363
Query: 68 H------KT--NLYQLDFPNLTKSIIP--LNHPFTTNIDPFTLNSIKALIGSCNGLLAIY 117
KT +L+ F + K++ LN P N + + I +G CNG+ +
Sbjct: 364 RPFISSLKTTISLHSSTFNDPKKNVCSSLLNPPEQYNSEHKYWSEI---MGPCNGIYLLQ 534
Query: 118 NGPIAFTHPNDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVCIHGFGFDPLT 177
P + + + F LP ++ +S I FGFDP T
Sbjct: 535 GNPNVLMNAS----------------LQQFKALPESHLTDSNGIY-SLTDYASFGFDPKT 663
Query: 178 GDYKLLRIS--WLVSL---QNSFYHSHARLFSSKTNSWKIIPI----MPYTVYYAQAMG- 227
DYK++ + WL Q ++ L+S +NSWK + +P + + +
Sbjct: 664 NDYKVIVLKDLWLKETDERQKGYWTGE--LYSLNSNSWKKLDAETLPLPIEICGSSSSSS 837
Query: 228 ----VFVENSIHWIMEKN----LDGSDLCLIVAFNLTLEVFKEVPLPAVIRGEEVNNNQS 279
+V N HW N G + +++F++ EVF+++ +P + +
Sbjct: 838 SRVYTYVNNCCHWWSFVNNHDESQGMNQDFVLSFDIVNEVFRKIKVPRICESSQETFVTL 1017
Query: 280 FDLEVAVLGGCLSMIVNYQTTKIDVWVMKEYGSRDSWCKIFTLVKSRFTLRLKSLRPLGY 339
E + G + + DVWVM++Y SW K +++ ++ L+ R +G+
Sbjct: 1018APFEESSTIGFIVNPIRGNVKHFDVWVMRDYWDEGSWIKQYSV----GSIELEIDRLMGF 1185
Query: 340 SSDGNKVLLQIDCKKLVW--YDLKSEQVIYVEGIPNLDE---AMICVESLVPPSFPVDNS 394
N+ L + + +LV +D + + I V+ D+ A++ ESLV
Sbjct: 1186IG-SNRFLWKCNDDELVLHEHDSQKRRDIKVKDYGKYDDSFRAVVYKESLVSL-----QR 1347
Query: 395 SKKENRLSKSKRRYFFIDYLSFLLCIS 421
K +K R + I Y+ F+ C +
Sbjct: 1348GK*YYSTNKGPLRVYVIFYIYFVNCFT 1428
>TC84657 weakly similar to GP|9294656|dbj|BAB03005.1
gb|AAF30317.1~gene_id:F14O13.6~similar to unknown
protein {Arabidopsis thaliana}, partial (14%)
Length = 884
Score = 72.0 bits (175), Expect = 4e-13
Identities = 60/208 (28%), Positives = 98/208 (46%), Gaps = 12/208 (5%)
Frame = +1
Query: 15 LPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHLKSS-------LNFKLILR 67
LP +++ +I LPVKSL+RF+ KSL +LI H F H + S + F L
Sbjct: 328 LPHELIIQIMLRLPVKSLIRFKCVCKSLLALISDHNFAKSHFELSTATHTNRIVFMSTLA 507
Query: 68 HKTNLYQLDFP---NLTKSIIPLNHPFTTNIDPFTLNSIKALIGSCNGLLAIYNGPIAFT 124
+T +DF N + LN F + P + +S++ + SC G + +
Sbjct: 508 LETR--SIDFEASLNDDSASTSLNLNF---MPPESYSSLE-IKSSCRGFIVL-------- 645
Query: 125 HPNDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVCIHGFGFDPLTGDYKLLR 184
++ I +WN +T H IPF P N+ + C++GFG+D L DY ++
Sbjct: 646 --TCSSNIYLWNPSTGHHKQIPF---PASNL-----DAKYSCCLYGFGYDHLRDDYLVVS 795
Query: 185 ISWLVSLQ--NSFYHSHARLFSSKTNSW 210
+S+ S+ + SH + FS + N+W
Sbjct: 796 VSYNTSIDPVDDNISSHLKFFSLRANTW 879
>CB894038
Length = 855
Score = 71.2 bits (173), Expect = 7e-13
Identities = 65/245 (26%), Positives = 114/245 (46%), Gaps = 31/245 (12%)
Frame = +1
Query: 108 GSCNGLLAIYNGPIAFTHPNDATEITIWNTNTRKHWIIPFLPLPIPNIV---ESENIERG 164
GS NG+L + G T I++WN T + +IP +P+IV +S+ ++
Sbjct: 88 GSVNGILCLQYGE---------TRISLWNPTTNEFKVIPPAGTRLPHIVHKFKSKLVDPF 240
Query: 165 GV--CIHGFGFDPLTGDYKLL---------------RISWLVSLQNSFYHSHARLFSSKT 207
+ IHGFG+D + DYKL+ R+ + LQ+ ++S +
Sbjct: 241 YIQTTIHGFGYDSVADDYKLICLQSFEPYYFYNDKQRMKQSLLLQHKSLQPFWMIYSLTS 420
Query: 208 NSWKIIPI-MPYT------VYYAQAMGVFVENSIHWIMEKNLDGSDLCLIVAFNLTLEVF 260
NSWK + + MP + YY ++++ HW+ +L S C+ V+F+L E F
Sbjct: 421 NSWKKLYVNMPRSSPTFQLEYYHGNHRLYMDGVCHWL---SLPTSGACM-VSFDLNNETF 588
Query: 261 KEVPLPAVIRGEEVNNNQSFDLEVAVLGGCLSMIV----NYQTTKIDVWVMKEYGSRDSW 316
P+P+ I + + ++ V+ ++++ N QT I +W E G ++SW
Sbjct: 589 FVTPIPSYI----LRVRRRAWQQLMVVNHSIALVSLPYHNTQTYHISIW--GEVGVKESW 750
Query: 317 CKIFT 321
K+FT
Sbjct: 751 IKLFT 765
>BI265054
Length = 679
Score = 68.2 bits (165), Expect = 6e-12
Identities = 45/166 (27%), Positives = 87/166 (52%), Gaps = 2/166 (1%)
Frame = -1
Query: 202 LFSSKTNSWKIIPI-MPYTVYYAQAMGVFVENSIHWIMEKNLDGSDLCLIVAFNLTLEVF 260
++S ++NSW+ + I MP Y + V+++ HW+ E+N CL V+F L EVF
Sbjct: 670 IYSXRSNSWRKLDIDMPSXSYCIEGTQVYMDGVCHWLCEENTPAGP-CL-VSFYLRNEVF 497
Query: 261 KEVPLPAVIRGEEVNNNQSFDLEVAVLGGCLSMIVNY-QTTKIDVWVMKEYGSRDSWCKI 319
P+P+ ++ ++ + + +L G +S+I + +TT D+ ++ E G ++SW K+
Sbjct: 496 CITPIPS--DEDDCFKFKASSINLVLLNGSISLIAFHRETTTFDIAILGELGIKESWTKL 323
Query: 320 FTLVKSRFTLRLKSLRPLGYSSDGNKVLLQIDCKKLVWYDLKSEQV 365
FT+ RP+ + G ++ Q +L W+DL ++ +
Sbjct: 322 FTVGPLSCV-----ERPIRVGTKG-EIFFQRKDNELAWFDLSTQVI 203
>TC92509 weakly similar to PIR|T49129|T49129 hypothetical protein F26G5.80 -
Arabidopsis thaliana, partial (8%)
Length = 858
Score = 67.4 bits (163), Expect = 1e-11
Identities = 66/238 (27%), Positives = 99/238 (40%), Gaps = 10/238 (4%)
Frame = +3
Query: 10 PVASDLPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHLKSSLN-FKLILRH 68
P LP +++AEI LPVK L + R KS SLI KF HL SS LILR
Sbjct: 159 PPLPTLPFELVAEILCRLPVKLLXQLRCLCKSFNSLISDPKFAKKHLHSSTTPHHLILRS 338
Query: 69 KTNLYQLDF------PNLTKSIIPLNHPFTTNIDPFTLNSIKAL---IGSCNGLLAIYNG 119
+ L+ S +P+ P T P L A SC+G++ +
Sbjct: 339 NNGSGRFALIVSPIQSVLSTSTVPV--PQTQLTYPTCLTEEFASPYEWCSCDGIICL--- 503
Query: 120 PIAFTHPNDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVCIHGFGFDPLTGD 179
D + +WN K +P P+ I +++R C+ FG+DP +
Sbjct: 504 ------TTDYSSAVLWNPFINKFKTLP----PLKYI----SLKRSPSCLFTFGYDPFADN 641
Query: 180 YKLLRISWLVSLQNSFYHSHARLFSSKTNSWKIIPIMPYTVYYAQAMGVFVENSIHWI 237
YK+ I++ V H+ T+SW+ I P + + G+FV +HW+
Sbjct: 642 YKVFAITFCVKRTTVEVHTMG------TSSWRRIEDFPSWSFIPDS-GIFVAGYVHWL 794
>CA921015 homologue to SP|Q8Y7G1|DPO3 DNA polymerase III polC-type (EC
2.7.7.7) (PolIII). {Listeria monocytogenes}, partial
(0%)
Length = 824
Score = 66.2 bits (160), Expect = 2e-11
Identities = 58/196 (29%), Positives = 101/196 (50%), Gaps = 10/196 (5%)
Frame = -1
Query: 202 LFSSKTNSWKIIPIMPYTVYYAQAMGVFVENSIHWIMEKNLDGSDLCLIVAFNLTLEVFK 261
+++ T+ WK I +PY ++ GVFV +++W+ SD I++ ++ E ++
Sbjct: 755 VYTLGTDYWKRIEDIPYNIFEE---GVFVSGTVNWL------ASDDSFILSLDVEKESYQ 603
Query: 262 EVPLPAVIRGEEVNNNQSFDLEV-AVLGGCLSMIVNYQTTKIDVWVMKEYGSRDSWCKIF 320
+V LP + DL + VL CL ++ +DVW+M EYG+++SW K++
Sbjct: 602 QVLLP----------DTENDLWILGVLRNCLCILAT-SNLFLDVWIMNEYGNQESWTKLY 456
Query: 321 TLVKSRFTLRLKSLRPLGYSSDGNKVLLQI-----DCKKLVWYDLKSEQVIYVEGIPNLD 375
+ V + L++ L YSS+ +++LL+ D KLV YD K+ + E N D
Sbjct: 455 S-VPNMQDHGLEAYTVL-YSSEDDQLLLEFNEMRSDKVKLVVYDSKTGTLNIPEFQNNYD 282
Query: 376 EAMIC----VESLVPP 387
+ IC +ESL+ P
Sbjct: 281 Q--ICQNFYIESLISP 240
>BG646118
Length = 764
Score = 65.9 bits (159), Expect = 3e-11
Identities = 53/185 (28%), Positives = 86/185 (45%), Gaps = 8/185 (4%)
Frame = -1
Query: 211 KIIPIMPYTVYYAQAMGVFVENSIHWIMEKNLDGSDLCLIVAFNLTLEVFKEVPLPAVIR 270
K P P+ Q +G V +I+W++ + D IV+ +L E KEV LP
Sbjct: 761 KPFPKFPFGCVPLQFLGKSVSGTINWLVARQYDNKFQDCIVSLDLGSESCKEVLLPK--- 591
Query: 271 GEEVNNNQSFDLEVAVLGGCLSMIVNYQTTKIDVWVMKEYGSRDSWCKIFTLVKSR-FTL 329
E + + VL CL ++ + DVW+MKE+G+ DSW K++++ R F
Sbjct: 590 -EVDTSTLRLRWYLGVLRDCLCLVFGH-----DVWIMKEHGNNDSWAKLYSVSFMRDFPS 429
Query: 330 RLKSLRPLGYSSDGNKVLLQIDC-------KKLVWYDLKSEQVIYVEGIPNLDEAMICVE 382
++ L DG L +DC +KLV+++ ++ + E D +CVE
Sbjct: 428 SFATIEVLHIFKDG---CLLLDCIYEGERTRKLVFHNSRNGTFKFSESKIMRD---VCVE 267
Query: 383 SLVPP 387
SL+ P
Sbjct: 266 SLISP 252
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.323 0.140 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,732,217
Number of Sequences: 36976
Number of extensions: 256868
Number of successful extensions: 1637
Number of sequences better than 10.0: 123
Number of HSP's better than 10.0 without gapping: 1566
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1579
length of query: 421
length of database: 9,014,727
effective HSP length: 99
effective length of query: 322
effective length of database: 5,354,103
effective search space: 1724021166
effective search space used: 1724021166
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC149495.11


