

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148718.7 + phase: 0
(541 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
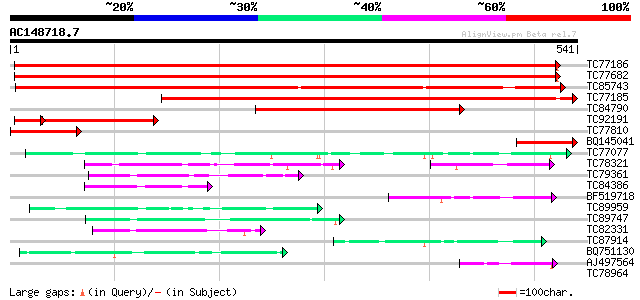
Score E
Sequences producing significant alignments: (bits) Value
TC77186 phosphate transporter 840 0.0
TC77682 phosphate transporter 838 0.0
TC85743 phosphate transporter PT4 [Medicago truncatula] 640 0.0
TC77185 similar to GP|12697486|emb|CAC28219. phosphate transport... 629 0.0
TC84790 similar to GP|13676624|gb|AAK38197.1 phosphate transport... 406 e-114
TC92191 homologue to GP|12697484|emb|CAC28218. phosphate transpo... 178 2e-55
TC77810 similar to GP|13676624|gb|AAK38197.1 phosphate transport... 125 4e-29
BQ145041 similar to GP|13676624|gb| phosphate transporter 2 {Lup... 115 4e-26
TC77077 similar to GP|9293909|dbj|BAB01812.1 sugar transporter p... 70 3e-12
TC78321 similar to GP|21689841|gb|AAM67564.1 unknown protein {Ar... 55 7e-08
TC79361 similar to GP|17380890|gb|AAL36257.1 putative membrane t... 52 6e-07
TC84386 similar to GP|21689841|gb|AAM67564.1 unknown protein {Ar... 52 8e-07
BF519718 weakly similar to PIR|G84564|G84 probable sugar transpo... 52 8e-07
TC89959 similar to GP|15795137|dbj|BAB02515. transporter-like pr... 50 2e-06
TC89747 similar to PIR|F96696|F96696 protein F1N21.12 [imported]... 48 8e-06
TC82331 weakly similar to EGAD|120604|128852 hypothetical protei... 47 1e-05
TC87914 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_14... 44 2e-04
BQ751130 weakly similar to GP|15289796|dbj putative monosacchari... 43 4e-04
AJ497564 weakly similar to PIR|C84593|C845 probable sugar transp... 42 6e-04
TC78964 weakly similar to PIR|T00450|T00450 probable monosacchar... 40 0.002
>TC77186 phosphate transporter
Length = 2314
Score = 840 bits (2169), Expect = 0.0
Identities = 408/524 (77%), Positives = 460/524 (86%), Gaps = 3/524 (0%)
Frame = +1
Query: 5 QLGVLTALDAAKTQWYHFTAIIIAGMGFFTDAYDLFCIPNVTKLLGRIYYTHPGALKPGT 64
+LGVL ALD AKTQ YHFT I+IAGMGFFTDAYDLFCI VTKLLGRIYYT P +PGT
Sbjct: 406 ELGVLNALDVAKTQLYHFTTIVIAGMGFFTDAYDLFCISLVTKLLGRIYYTEPNPTRPGT 585
Query: 65 LPPNVSAAVNGVALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAK 124
LPP+ +AV GVAL GTLAGQLFFGWLGDK+GRKKVYGLTL LMV S+ASGLSFG S K
Sbjct: 586 LPPSAQSAVTGVALVGTLAGQLFFGWLGDKLGRKKVYGLTLILMVVCSVASGLSFGSSPK 765
Query: 125 GTIGTLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVSL 184
+ TLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIA+VFAMQGFGIL GGIV+L
Sbjct: 766 SVMATLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIAAVFAMQGFGILGGGIVAL 945
Query: 185 VVSTAFDHAFKAPPYKVDAAASL-VPEADYVWRIILMFGAVPAGLTYYWRMKMPETARYT 243
+V++ FDH +K P ++ + AASL VP+ DYVWR+ILMFGA+PA LTYYWRMKMPETARYT
Sbjct: 946 IVASIFDHKYKVPTFEENPAASLLVPQFDYVWRLILMFGALPAALTYYWRMKMPETARYT 1125
Query: 244 ALVAKNAKQAAQDMSSVLQVEIEAEQEKVDKIGVQDKNSFGLFSKEFLRRHGLHLLGTTS 303
ALVAKNAKQAA DMS VLQVE+E E+EKV K+ +NS+GLFSK+F RHGL L GT S
Sbjct: 1126ALVAKNAKQAAADMSKVLQVELEVEEEKVQKMTSDKRNSYGLFSKQFAARHGLALFGTCS 1305
Query: 304 TWFLLDIAYYSSNLFQKDIYSSIGWLPPAQDMNAIHEVFKVARATTLIALCGTVPGYWFT 363
TWFLLDIA+YS NLFQKDI+S+IGW+PPA++MNAIHEV+K+ARA TLIALC TVPGYWFT
Sbjct: 1306TWFLLDIAFYSQNLFQKDIFSAIGWIPPAKEMNAIHEVYKIARAQTLIALCSTVPGYWFT 1485
Query: 364 VAFIDVIGRFAIQLMGFFFMTVFMFALAIPYDHWTKRENRIGFLVMYAMTFFFANFGPNS 423
VAFID +GRFAIQ+MGFFFMTVFMFALAIPYDHW+K ENRIGF+VMY++TFFFANFGPN+
Sbjct: 1486VAFIDHMGRFAIQMMGFFFMTVFMFALAIPYDHWSKEENRIGFVVMYSLTFFFANFGPNA 1665
Query: 424 TTFVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQSKDPKKRDAGYPAGIGMK 483
TTFVVPAEIFPARLRSTCHGIS+AAGKAGAI+GAFGFLYA+QSKDP K D GYP GIG+K
Sbjct: 1666TTFVVPAEIFPARLRSTCHGISAAAGKAGAIVGAFGFLYAAQSKDPTKTDKGYPTGIGIK 1845
Query: 484 NTLILLAVVNCLGIFFTFLVPEANGKSLEEMSGENEEE--EETE 525
N+LI+L V+N +G+ T LVPE+ GKSLEE+SGENE E E TE
Sbjct: 1846NSLIMLGVINFVGMLCTLLVPESKGKSLEELSGENEGEGAEATE 1977
>TC77682 phosphate transporter
Length = 2245
Score = 838 bits (2164), Expect = 0.0
Identities = 407/524 (77%), Positives = 460/524 (87%), Gaps = 3/524 (0%)
Frame = +2
Query: 5 QLGVLTALDAAKTQWYHFTAIIIAGMGFFTDAYDLFCIPNVTKLLGRIYYTHPGALKPGT 64
+LGVL ALD AKTQ YHFT I+IAGMGFFTDAYDLFCI VTKLLGRIYYT P +PGT
Sbjct: 128 ELGVLNALDVAKTQLYHFTTIVIAGMGFFTDAYDLFCISLVTKLLGRIYYTEPNPTRPGT 307
Query: 65 LPPNVSAAVNGVALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAK 124
LPP+ +AV GVAL GTLAGQLFFGWLGDK+GRKKVYGLTL LMV S+ASGLSFG S K
Sbjct: 308 LPPSAQSAVTGVALVGTLAGQLFFGWLGDKLGRKKVYGLTLILMVVCSVASGLSFGSSPK 487
Query: 125 GTIGTLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVSL 184
+ TLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIA+VFAMQGFGIL GGIV+L
Sbjct: 488 SVMATLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIAAVFAMQGFGILGGGIVAL 667
Query: 185 VVSTAFDHAFKAPPYKVDAAASL-VPEADYVWRIILMFGAVPAGLTYYWRMKMPETARYT 243
V++ FDH +K P ++ + AASL VP+ DYVWR+ILMFGA+PA LTYYWRMKMPETARYT
Sbjct: 668 TVASIFDHKYKVPTFEENPAASLLVPQFDYVWRLILMFGALPAALTYYWRMKMPETARYT 847
Query: 244 ALVAKNAKQAAQDMSSVLQVEIEAEQEKVDKIGVQDKNSFGLFSKEFLRRHGLHLLGTTS 303
ALVAKNAKQAA DMS VLQVE+E E+EKV+K+ +NS+GLFSK+F RHGL L GT S
Sbjct: 848 ALVAKNAKQAAADMSKVLQVELEVEEEKVEKMTSDKRNSYGLFSKQFAARHGLALFGTCS 1027
Query: 304 TWFLLDIAYYSSNLFQKDIYSSIGWLPPAQDMNAIHEVFKVARATTLIALCGTVPGYWFT 363
TWFLLDIA+YS NLFQKDI+S+IGW+PPA++MNAIHEV+K+ARA TLIALC TVPGYWFT
Sbjct: 1028TWFLLDIAFYSQNLFQKDIFSAIGWIPPAKEMNAIHEVYKIARAQTLIALCSTVPGYWFT 1207
Query: 364 VAFIDVIGRFAIQLMGFFFMTVFMFALAIPYDHWTKRENRIGFLVMYAMTFFFANFGPNS 423
VAFID +GRFAIQ+MGFFFMTVFMFALAIPYDHW+K ENRIGF+V+Y++TFFFANFGPN+
Sbjct: 1208VAFIDHMGRFAIQMMGFFFMTVFMFALAIPYDHWSKEENRIGFVVIYSLTFFFANFGPNA 1387
Query: 424 TTFVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQSKDPKKRDAGYPAGIGMK 483
TTFVVPAEIFPARLRSTCHGIS+AAGKAGAI+GAFGFLYA+QSKDP K D GYP GIG+K
Sbjct: 1388TTFVVPAEIFPARLRSTCHGISAAAGKAGAIVGAFGFLYAAQSKDPTKTDKGYPTGIGIK 1567
Query: 484 NTLILLAVVNCLGIFFTFLVPEANGKSLEEMSGENEEE--EETE 525
N+LI+L V+N +G+ T LVPE+ GKSLEE+SGENE E E TE
Sbjct: 1568NSLIMLGVINFVGMLCTLLVPESKGKSLEELSGENEGEGAEATE 1699
>TC85743 phosphate transporter PT4 [Medicago truncatula]
Length = 1850
Score = 640 bits (1651), Expect = 0.0
Identities = 318/525 (60%), Positives = 393/525 (74%)
Frame = +2
Query: 6 LGVLTALDAAKTQWYHFTAIIIAGMGFFTDAYDLFCIPNVTKLLGRIYYTHPGALKPGTL 65
L VL ALD+A+TQWYH TAI+IAGMGFFTDAYDLFCI V+KLLGR+YY P KPG L
Sbjct: 35 LEVLEALDSARTQWYHVTAIVIAGMGFFTDAYDLFCISTVSKLLGRLYYFDPSTNKPGKL 214
Query: 66 PPNVSAAVNGVALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAKG 125
PP+V+ V GVAL GTL+GQL FGWLGDK+GRKKVYG+TL +MV ++ SGLSFG SAK
Sbjct: 215 PPSVNNVVTGVALVGTLSGQLVFGWLGDKLGRKKVYGVTLIIMVACAICSGLSFGSSAKS 394
Query: 126 TIGTLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVSLV 185
+ TLCFFRFWLGFGIGGDYPLSATIMSEYANK+TRGAFIA+VFAMQG GI+ G+VS+V
Sbjct: 395 VMITLCFFRFWLGFGIGGDYPLSATIMSEYANKRTRGAFIAAVFAMQGVGIIFAGLVSMV 574
Query: 186 VSTAFDHAFKAPPYKVDAAASLVPEADYVWRIILMFGAVPAGLTYYWRMKMPETARYTAL 245
S F ++AP + D S PE D +WR+ILM GAVPA +TYYWRMKMPET RYTA+
Sbjct: 575 FSGIFKAYYQAPRFNEDPILSTQPEGDLLWRLILMIGAVPAAMTYYWRMKMPETGRYTAI 754
Query: 246 VAKNAKQAAQDMSSVLQVEIEAEQEKVDKIGVQDKNSFGLFSKEFLRRHGLHLLGTTSTW 305
V NAKQAA DM+ VL +EI AEQ+K+ + + N + L+S EF RHG HL+GT S W
Sbjct: 755 VEGNAKQAAADMARVLDIEIIAEQDKLAEF--KAANDYPLWSSEFFNRHGRHLIGTMSCW 928
Query: 306 FLLDIAYYSSNLFQKDIYSSIGWLPPAQDMNAIHEVFKVARATTLIALCGTVPGYWFTVA 365
FLLDIA+YS NL QKDIY ++G + ++MNAI EVF+ +RA ++AL GT PGYWFTV
Sbjct: 929 FLLDIAFYSQNLTQKDIYPAMGLIRQDKEMNAIDEVFQTSRAMFVVALFGTFPGYWFTVF 1108
Query: 366 FIDVIGRFAIQLMGFFFMTVFMFALAIPYDHWTKRENRIGFLVMYAMTFFFANFGPNSTT 425
FI+ +GRF IQL+GFF M+ FMF + + Y+ + K EN+ F ++Y +TFFFANFGPNSTT
Sbjct: 1109FIEKLGRFKIQLVGFFMMSFFMFVIGVKYE-YLKDENKNLFALLYGLTFFFANFGPNSTT 1285
Query: 426 FVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQSKDPKKRDAGYPAGIGMKNT 485
FV+PAE+FP R+RSTCH S+A+GKAGA++GAFG Y + P+K ++
Sbjct: 1286FVLPAELFPTRVRSTCHAFSAASGKAGAMVGAFGIQYYTLDGTPRK----------IRRA 1435
Query: 486 LILLAVVNCLGIFFTFLVPEANGKSLEEMSGENEEEEETEAAVVD 530
+++LA N +G F TFLV E G+SLEE+SGE+ E E A D
Sbjct: 1436MMILAFTNLIGFFCTFLVTETKGRSLEEISGEDGRESELTATPND 1570
>TC77185 similar to GP|12697486|emb|CAC28219. phosphate transporter
{Sesbania rostrata}, partial (71%)
Length = 1467
Score = 629 bits (1623), Expect = 0.0
Identities = 304/397 (76%), Positives = 351/397 (87%), Gaps = 1/397 (0%)
Frame = +2
Query: 146 PLSATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVSLVVSTAFDHAFKAPPYKVDAAA 205
PLSATIMSEYANKKTRGAFIA+VFAMQGFGI+AGG+ +L+V+ AFDH ++ P Y+ +A A
Sbjct: 2 PLSATIMSEYANKKTRGAFIAAVFAMQGFGIMAGGVFALIVAAAFDHKYQVPTYEENAKA 181
Query: 206 SLV-PEADYVWRIILMFGAVPAGLTYYWRMKMPETARYTALVAKNAKQAAQDMSSVLQVE 264
SLV P DYVWRIILMFGAVPA LTYYWRMKMPETARYTALVAKN KQAA DMS VLQVE
Sbjct: 182 SLVLPAFDYVWRIILMFGAVPAALTYYWRMKMPETARYTALVAKNGKQAASDMSKVLQVE 361
Query: 265 IEAEQEKVDKIGVQDKNSFGLFSKEFLRRHGLHLLGTTSTWFLLDIAYYSSNLFQKDIYS 324
IEAE+EKV + FGLF+++F +RHGLHLLGTT+TWFLLDIA+YS NLFQKDI++
Sbjct: 362 IEAEEEKVQNLAENQNQKFGLFTRQFAKRHGLHLLGTTTTWFLLDIAFYSQNLFQKDIFT 541
Query: 325 SIGWLPPAQDMNAIHEVFKVARATTLIALCGTVPGYWFTVAFIDVIGRFAIQLMGFFFMT 384
+IGW+PPA++MNAI+E++++ARA TLIALC TVPGYWFTVAFID +GRFAIQLMGFFFMT
Sbjct: 542 AIGWIPPAKEMNAINELYRIARAQTLIALCSTVPGYWFTVAFIDYMGRFAIQLMGFFFMT 721
Query: 385 VFMFALAIPYDHWTKRENRIGFLVMYAMTFFFANFGPNSTTFVVPAEIFPARLRSTCHGI 444
VFMFALAIPYDHWTK+ENRIGF+VMY++TFFFANFGPN+TTFVVPAEIFPARLRSTCHGI
Sbjct: 722 VFMFALAIPYDHWTKKENRIGFVVMYSLTFFFANFGPNATTFVVPAEIFPARLRSTCHGI 901
Query: 445 SSAAGKAGAIIGAFGFLYASQSKDPKKRDAGYPAGIGMKNTLILLAVVNCLGIFFTFLVP 504
S+AAGKAGAI+GAFGFLYA+QSKDP K D GYP GIG+KN+LI+L VVN G+ FTFLVP
Sbjct: 902 SAAAGKAGAIVGAFGFLYAAQSKDPTKTDHGYPTGIGIKNSLIVLGVVNFFGMVFTFLVP 1081
Query: 505 EANGKSLEEMSGENEEEEETEAAVVDPQASSNRTVPV 541
E NGKSLEEMSGENE++ +A ++ A S RTVPV
Sbjct: 1082EPNGKSLEEMSGENEDD---DAEAIEMAAGSARTVPV 1183
>TC84790 similar to GP|13676624|gb|AAK38197.1 phosphate transporter 2
{Lupinus albus}, partial (37%)
Length = 639
Score = 406 bits (1044), Expect = e-114
Identities = 200/200 (100%), Positives = 200/200 (100%)
Frame = +2
Query: 235 KMPETARYTALVAKNAKQAAQDMSSVLQVEIEAEQEKVDKIGVQDKNSFGLFSKEFLRRH 294
KMPETARYTALVAKNAKQAAQDMSSVLQVEIEAEQEKVDKIGVQDKNSFGLFSKEFLRRH
Sbjct: 2 KMPETARYTALVAKNAKQAAQDMSSVLQVEIEAEQEKVDKIGVQDKNSFGLFSKEFLRRH 181
Query: 295 GLHLLGTTSTWFLLDIAYYSSNLFQKDIYSSIGWLPPAQDMNAIHEVFKVARATTLIALC 354
GLHLLGTTSTWFLLDIAYYSSNLFQKDIYSSIGWLPPAQDMNAIHEVFKVARATTLIALC
Sbjct: 182 GLHLLGTTSTWFLLDIAYYSSNLFQKDIYSSIGWLPPAQDMNAIHEVFKVARATTLIALC 361
Query: 355 GTVPGYWFTVAFIDVIGRFAIQLMGFFFMTVFMFALAIPYDHWTKRENRIGFLVMYAMTF 414
GTVPGYWFTVAFIDVIGRFAIQLMGFFFMTVFMFALAIPYDHWTKRENRIGFLVMYAMTF
Sbjct: 362 GTVPGYWFTVAFIDVIGRFAIQLMGFFFMTVFMFALAIPYDHWTKRENRIGFLVMYAMTF 541
Query: 415 FFANFGPNSTTFVVPAEIFP 434
FFANFGPNSTTFVVPAEIFP
Sbjct: 542 FFANFGPNSTTFVVPAEIFP 601
>TC92191 homologue to GP|12697484|emb|CAC28218. phosphate transporter
{Sesbania rostrata}, partial (26%)
Length = 531
Score = 178 bits (452), Expect(2) = 2e-55
Identities = 87/111 (78%), Positives = 92/111 (82%)
Frame = +2
Query: 32 FFTDAYDLFCIPNVTKLLGRIYYTHPGALKPGTLPPNVSAAVNGVALCGTLAGQLFFGWL 91
F DAYDLFCI VTKLLGRIYYT P KPG LPP V AAV GVAL GTL+GQLFFGWL
Sbjct: 197 FSLDAYDLFCISLVTKLLGRIYYTDPTQPKPGVLPPGVQAAVTGVALVGTLSGQLFFGWL 376
Query: 92 GDKMGRKKVYGLTLALMVFSSLASGLSFGHSAKGTIGTLCFFRFWLGFGIG 142
GDK+GRKKVYG+TL LMV SLASGLSFG+S KG + TLCFFRFWLGFGIG
Sbjct: 377 GDKLGRKKVYGITLMLMVGCSLASGLSFGNSPKGVMATLCFFRFWLGFGIG 529
Score = 56.2 bits (134), Expect(2) = 2e-55
Identities = 26/30 (86%), Positives = 27/30 (89%)
Frame = +1
Query: 5 QLGVLTALDAAKTQWYHFTAIIIAGMGFFT 34
QLGVL ALD AKTQ YHFTAI+IAGMGFFT
Sbjct: 115 QLGVLNALDVAKTQMYHFTAIVIAGMGFFT 204
>TC77810 similar to GP|13676624|gb|AAK38197.1 phosphate transporter 2
{Lupinus albus}, partial (11%)
Length = 1035
Score = 125 bits (314), Expect = 4e-29
Identities = 60/68 (88%), Positives = 62/68 (90%)
Frame = +2
Query: 1 MAREQLGVLTALDAAKTQWYHFTAIIIAGMGFFTDAYDLFCIPNVTKLLGRIYYTHPGAL 60
+A EQLGVLT+L AAKTQWY FTAIIIAGMGF TDAYDLFCIPNVTKLLGRIYYTHP AL
Sbjct: 332 LAHEQLGVLTSLHAAKTQWYLFTAIIIAGMGFCTDAYDLFCIPNVTKLLGRIYYTHPDAL 511
Query: 61 KPGTLPPN 68
KPGTLP N
Sbjct: 512 KPGTLPTN 535
>BQ145041 similar to GP|13676624|gb| phosphate transporter 2 {Lupinus albus},
partial (7%)
Length = 522
Score = 115 bits (288), Expect = 4e-26
Identities = 58/58 (100%), Positives = 58/58 (100%)
Frame = +1
Query: 484 NTLILLAVVNCLGIFFTFLVPEANGKSLEEMSGENEEEEETEAAVVDPQASSNRTVPV 541
NTLILLAVVNCLGIFFTFLVPEANGKSLEEMSGENEEEEETEAAVVDPQASSNRTVPV
Sbjct: 1 NTLILLAVVNCLGIFFTFLVPEANGKSLEEMSGENEEEEETEAAVVDPQASSNRTVPV 174
>TC77077 similar to GP|9293909|dbj|BAB01812.1 sugar transporter protein
{Arabidopsis thaliana}, partial (85%)
Length = 1752
Score = 69.7 bits (169), Expect = 3e-12
Identities = 128/565 (22%), Positives = 205/565 (35%), Gaps = 44/565 (7%)
Frame = +1
Query: 16 KTQWYHFTAIIIAGMGFFTDAYDLFCIPNVTKLLGRIYYTHPGALKPGTLPPNVSAAVNG 75
K Y F ++A M YD+ + + R G ++ + G
Sbjct: 94 KRNKYAFACAMLASMTSILLGYDIGVMSGAAIYIKRDLKVSDGKIE----------VLLG 243
Query: 76 VALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAKGTIGTLCFFRF 135
+ +L G G D +GR+ A+ +L G S ++ L F RF
Sbjct: 244 IINIYSLIGSCLAGRTSDWIGRRYTIVFAGAIFFVGALLMGFSPNYNF------LMFGRF 405
Query: 136 WLGFGIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVSLVVSTAFDHAFK 195
G GIG ++ +E + +RG + GIL G I + +AF
Sbjct: 406 VAGVGIGYALMIAPVYTAEVSPASSRGFLTSFPEVFINSGILLGYISN--------YAFS 561
Query: 196 APPYKVDAAASLVPEADYVWRIILMFGAVPAGLTYYWRMKMPETARYTALVAK------- 248
K+ WR++L GA+P+ + + MPE+ R+ + +
Sbjct: 562 KLSLKLG------------WRMMLGVGAIPSVILAVGVLAMPESPRWLVMRGRLGDAIKV 705
Query: 249 -----NAKQAAQ-DMSSVLQVEIEAEQEKVDKIGVQDKNSF-GLFSKEFLR-----RH-- 294
++K+ AQ ++ + Q E D + V+ KN+ G++ + FL RH
Sbjct: 706 LNKTSDSKEEAQLRLAEIKQAAGIPEDCNDDVVEVKVKNTGEGVWKELFLYPTPAVRHIV 885
Query: 295 ----GLHLLGTTSTWFLLDIAYYSSNLFQKDIYSSIGWLPPAQDMNAIHEVFKVARATTL 350
G+H S + + YS +F+K + D + + V TL
Sbjct: 886 IAALGIHFFQQASG--VDAVVLYSPTIFKKAGING--------DTHLLIATIAVGFVKTL 1035
Query: 351 IALCGTVPGYWFTVAFIDVIGRFAIQLMGFFFMTVFMFALAIPY---DHWTKREN----- 402
L T +D GR + L M V + LA+ DH + N
Sbjct: 1036FILVATF--------MLDRYGRRPLLLTSVGGMVVSLLTLAVSLTIIDHSNTKLNWAIGL 1191
Query: 403 RIGFLVMYAMTFFFANFGPNSTTFVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFGFLY 462
I ++ Y TF + G T+V +EIFP RLR+ G + G +
Sbjct: 1192SIATVLSYVATF---SIGAGPITWVYSSEIFPLRLRA--QGAACGVVVNRVTSGVISMTF 1356
Query: 463 ASQSKDPKKRDAGYPAGIGMKNTLILLAVVNCLG-IFFTFLVPEANGKSLEEM------- 514
S SK GI + L + +G IFF ++PE GK+LEEM
Sbjct: 1357LSLSK-----------GITIGGAFFLFGGIATIGWIFFYIMLPETQGKTLEEMEASFGKI 1503
Query: 515 ---SGENEEEEETEAAVVDPQASSN 536
S +E E+ A V Q +N
Sbjct: 1504WRKSKNTKEAEKDNAQVAQVQLGTN 1578
>TC78321 similar to GP|21689841|gb|AAM67564.1 unknown protein {Arabidopsis
thaliana}, partial (80%)
Length = 2093
Score = 55.1 bits (131), Expect = 7e-08
Identities = 64/259 (24%), Positives = 111/259 (42%), Gaps = 11/259 (4%)
Frame = +2
Query: 72 AVNGVALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAKGTIGTLC 131
A+ A+ G + G GW+ D+ GRK ++++V +L S +A TL
Sbjct: 353 AIVSTAIAGAIIGAAIGGWINDRFGRK------VSIIVADTLFLLGSIILAAAPNPATLI 514
Query: 132 FFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVSLVVSTAFD 191
R ++G G+G S +SE + + RGA + ++ F I G +S +++ AF
Sbjct: 515 VGRVFVGLGVGMASMASPLYISEASPTRVRGALV----SLNSFLITGGQFLSYLINLAFT 682
Query: 192 HAFKAPPYKVDAAASLVPEADYVWRIILMFGAVPAGLTYYWRMKMPETARYTALVAKNAK 251
KAP WR +L A PA + + +PE+ R+ K +
Sbjct: 683 ---KAPG---------------TWRWMLGVAAAPAVIQIVLMLSLPESPRWLYRKGKE-E 805
Query: 252 QAAQDMSSVLQV-----EIEAEQEKVD-KIGVQDKNSFGLFSKEFLRRHGLHLLGTTSTW 305
+A + + +V EI+A +E V+ ++ +K S K R GL+ G +
Sbjct: 806 EAKVILKKIYEVEDYDNEIQALKESVEMELKETEKISIMQLVKTTSVRRGLY-AGVGLAF 982
Query: 306 F-----LLDIAYYSSNLFQ 319
F + + YYS ++ Q
Sbjct: 983 FQQFTGINTVMYYSPSIVQ 1039
Score = 45.8 bits (107), Expect = 4e-05
Identities = 32/122 (26%), Positives = 54/122 (44%), Gaps = 3/122 (2%)
Frame = +2
Query: 402 NRIGFLVMYAMTFFFANFGPNSTT--FVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFG 459
+ G++ + A+ + F P T +VV +EI+P R R C GI+S ++ +
Sbjct: 1475 SNFGWIAILALALYIIFFSPGMGTVPWVVNSEIYPLRYRGICGGIASTTVWVSNLVVSQS 1654
Query: 460 FLYASQSKDPKKRDAGYPAGIGMKNTLILLAVVNCLGIFFTFL-VPEANGKSLEEMSGEN 518
FL IG T ++ A++ + IFF + VPE G +EE+
Sbjct: 1655 FL-------------SLTVAIGPAWTFMIFAIIAIVAIFFVIIFVPETKGVPMEEVESML 1795
Query: 519 EE 520
E+
Sbjct: 1796 EK 1801
>TC79361 similar to GP|17380890|gb|AAL36257.1 putative membrane transporter
protein {Arabidopsis thaliana}, partial (55%)
Length = 949
Score = 52.0 bits (123), Expect = 6e-07
Identities = 48/205 (23%), Positives = 86/205 (41%)
Frame = +1
Query: 76 VALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAKGTIGTLCFFRF 135
+A+ G + G F GWL D GRKK L + + ++ +A L R
Sbjct: 313 MAIAGAIVGAAFGGWLNDAYGRKKATLLADVIFILGAILM------AAAPDPYVLIAGRL 474
Query: 136 WLGFGIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVSLVVSTAFDHAFK 195
+G G+G + ++E A + RG+ +++ M I G VS +V+ F
Sbjct: 475 LVGLGVGIASVTAPVYIAEVAPSEIRGSLVSTNVLM----ITGGQFVSYLVNLVF----- 627
Query: 196 APPYKVDAAASLVPEADYVWRIILMFGAVPAGLTYYWRMKMPETARYTALVAKNAKQAAQ 255
+ WR +L VPA + + + +PE+ R+ L KN K A
Sbjct: 628 -------------TQVPGTWRWMLGVSGVPALIQFICMLFLPESPRW--LFIKNRKNEAV 762
Query: 256 DMSSVLQVEIEAEQEKVDKIGVQDK 280
D+ S + ++ ++++D + Q +
Sbjct: 763 DVISKI-YDLSRLEDEIDFLTAQSE 834
>TC84386 similar to GP|21689841|gb|AAM67564.1 unknown protein {Arabidopsis
thaliana}, partial (30%)
Length = 664
Score = 51.6 bits (122), Expect = 8e-07
Identities = 36/122 (29%), Positives = 56/122 (45%)
Frame = +2
Query: 72 AVNGVALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAKGTIGTLC 131
A+ AL G + G GW+ D+ GRKK L AL S+ + + L
Sbjct: 317 AIVSTALAGAIIGASVGGWINDRFGRKKAIILADALFFIGSVIMAAAINPA------ILI 478
Query: 132 FFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVSLVVSTAFD 191
R ++G G+G S +SE + + RGA + ++ GF I G +S V++ AF
Sbjct: 479 VGRVFVGLGVGMASMASPLYISEASPTRVRGALV----SLNGFLITGGQXLSYVINLAFT 646
Query: 192 HA 193
+A
Sbjct: 647 NA 652
>BF519718 weakly similar to PIR|G84564|G84 probable sugar transporter
[imported] - Arabidopsis thaliana, partial (20%)
Length = 659
Score = 51.6 bits (122), Expect = 8e-07
Identities = 50/168 (29%), Positives = 73/168 (42%), Gaps = 8/168 (4%)
Frame = +3
Query: 362 FTVAFIDVIGRFAIQLMGFFFMTVFMFALAIPYDHWTKRENRIGFLVMY-------AMTF 414
F+ +D GR + L+G M V +F L + + + + + A++F
Sbjct: 39 FSALVLDRFGRRPMLLLGSSGMAVSLFGLGMGCTLLHNSDEKPMWAIALCVVAVCAAVSF 218
Query: 415 FFANFGPNSTTFVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQSKDPKKRDA 474
F GP TT+V +EIFP RLR+ G S A I G + S S++
Sbjct: 219 FSIGLGP--TTWVYSSEIFPMRLRA--QGTSLAISVNRLISGVVSMSFLSISEE------ 368
Query: 475 GYPAGIGMKNTLILLAVVNCLG-IFFTFLVPEANGKSLEEMSGENEEE 521
I +LA V L +FF + +PE GKSLEE+ EEE
Sbjct: 369 -----ITFGGMFFVLAGVMVLATLFFYYFLPETKGKSLEEIEALFEEE 497
>TC89959 similar to GP|15795137|dbj|BAB02515. transporter-like protein
{Arabidopsis thaliana}, partial (87%)
Length = 1681
Score = 50.1 bits (118), Expect = 2e-06
Identities = 59/280 (21%), Positives = 107/280 (38%), Gaps = 1/280 (0%)
Frame = +3
Query: 20 YHFTAIIIAGMGFFTDAYDLFCIPNVTKLLGRIYYTHPGALKPGTLPPNVSAAVNGVALC 79
+ +I AG+G+ ++A ++ + + P L + + + V
Sbjct: 186 FQILVLIYAGIGWISEAMEMMLLS----------FVGPAVQSAWNLSSHQESFITSVVFA 335
Query: 80 GTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAKGTIGTLCFFRFWLGF 139
G L G +G + D+ GR+K + +T + + S LS + + L FR +G
Sbjct: 336 GMLIGAYSWGIVSDQHGRRKGFLITATVTATAGFFSSLSPNYVS------LLVFRSLVGL 497
Query: 140 GIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVSLVVSTAFDHAFKAPPY 199
G+GG LS+ + E+ RG ++ GF V T F+
Sbjct: 498 GLGGGPVLSSWFL-EFVPAPNRGTWMV---VFSGFW---------TVGTIFE-------- 614
Query: 200 KVDAAASLVPEADYVWRIILMFGAVPAGLTYYWRMKMPETARYTALVAKNAKQAAQDMSS 259
A+ + + WR +L ++P + PE+ RY L K +
Sbjct: 615 ---ASLAWIVMPRLGWRWLLALSSLPTSFLLLFYKMTPESPRYLCL-----KGRTTEAID 770
Query: 260 VLQVEIEAEQEKVDK-IGVQDKNSFGLFSKEFLRRHGLHL 298
VL+ +K+ + V DK++ + + RRHG L
Sbjct: 771 VLETISRLNGKKLPSGVLVSDKSN*ATKN*QPFRRHGFTL 890
Score = 35.0 bits (79), Expect = 0.073
Identities = 22/105 (20%), Positives = 44/105 (40%)
Frame = +1
Query: 351 IALCGTVPGYWFTVAFIDVIGRFAIQLMGFFFMTVFMFALAIPYDHWTKRENRIGFLVMY 410
IA +PG + +D +GR + FF +F+ + G +
Sbjct: 1141 IASFAELPGLLLSAVAVDKLGRKLSMSIMFFMCCIFLLPVTFYLPEDLTTGLLFGARICI 1320
Query: 411 AMTFFFANFGPNSTTFVVPAEIFPARLRSTCHGISSAAGKAGAII 455
+TF + ++ EI+P +R+T G++S+ G+ G ++
Sbjct: 1321 TVTF--------TIVYIYAPEIYPTSVRTTGVGVASSVGRIGGML 1431
>TC89747 similar to PIR|F96696|F96696 protein F1N21.12 [imported] -
Arabidopsis thaliana, partial (64%)
Length = 1372
Score = 48.1 bits (113), Expect = 8e-06
Identities = 53/254 (20%), Positives = 96/254 (36%), Gaps = 7/254 (2%)
Frame = +3
Query: 73 VNGVALCGTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAKGTIGTLCF 132
V + L G L G L GW+ D +GR++ + L M+ + S +A + +
Sbjct: 504 VVSICLGGALFGCLLSGWIADAVGRRRAFQLCALPMIIGAAMS------AATNNLFGMLV 665
Query: 133 FRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVSLVVSTAFDH 192
R ++G G+G P++A ++E + RG + A + FGIL + + V
Sbjct: 666 GRLFVGTGLGLGPPVAALYVTEVSPAFVRGTYGALIQIATCFGILGSLFIGIPVK----- 830
Query: 193 AFKAPPYKVDAAASLVPEADYVWRIILMFGAVPAGLTYYWRMKMPETARYTALVAKNAKQ 252
E WR+ +PA + E+ + + A+
Sbjct: 831 -----------------EISGWWRVCFWVSTIPAAILALAMDFCAESPHWLYKQGRTAEA 959
Query: 253 AAQDMSSVLQVEIEAEQEKVDKIGVQDKNSFGLFSKEFLRRHGLHLLGTTSTWFLLD--- 309
A+ + E + ++ K+ + FS E L H ++ ST F L
Sbjct: 960 EAEFERLLGVSEAKFAMSQLSKVDRGEDTDTVKFS-ELLHGHHSKVVFIGSTLFALQQLS 1136
Query: 310 ----IAYYSSNLFQ 319
+ Y+SS +F+
Sbjct: 1137GINAVFYFSSTVFK 1178
>TC82331 weakly similar to EGAD|120604|128852 hypothetical protein M01F1.5
{Caenorhabditis elegans}, partial (10%)
Length = 848
Score = 47.4 bits (111), Expect = 1e-05
Identities = 45/168 (26%), Positives = 69/168 (40%), Gaps = 3/168 (1%)
Frame = +3
Query: 80 GTLAGQLFFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAKGTIGTLCFFRFWLGF 139
GTL G L + D GR+K + + V ++ +A T + RF G
Sbjct: 426 GTLIGALSGAYTADWWGRRKSLSFGVVVFVIGNIVQ-----ITAMDTWVHMMMGRFIAGL 590
Query: 140 GIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVSLVVSTAFDHAFKAPPY 199
G+G SE + K+ RGA +AS M FGIL +++ V
Sbjct: 591 GVGNLSVGVPMFQSECSPKEIRGAVVASYQLMITFGILISNLINFGVRN----------- 737
Query: 200 KVDAAASLVPEADYVWRIILMFG---AVPAGLTYYWRMKMPETARYTA 244
+ E+D WRI++ G A+P GL + +PE+ R+ A
Sbjct: 738 --------IQESDASWRIVIGLGIAFAMPLGLGI---LVVPESPRWLA 848
>TC87914 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_140
{Arabidopsis thaliana}, partial (39%)
Length = 1129
Score = 43.9 bits (102), Expect = 2e-04
Identities = 51/209 (24%), Positives = 83/209 (39%), Gaps = 6/209 (2%)
Frame = +3
Query: 310 IAYYSSNLFQKDIYSSIGWLPPAQDMNAIHEVFKVARATTLIALCGTVPGYWFTVAFIDV 369
+ YY+ + ++ + +G+L +++ F ++ TTL+ L P + +D+
Sbjct: 342 VLYYTPQILEQ---AGVGYLLSNLGLSSTSSSFLISAVTTLLML----PCIAVAMRLMDI 500
Query: 370 IGRFAIQLMGFFFMTVFMFALAIPY-----DHWTKRENRIGFLVMYAMTFFFANFGPNST 424
GR + L + V +F L + D + I +V + F FGP
Sbjct: 501 SGRRTLLLTTIPVLIVSLFILVLGSLVDLGDTANASISTISVVVYFCS--FVMGFGPVPN 674
Query: 425 TFVVPAEIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQSKDPKKRDAGYPAGIGMKN 484
++ AEIFP R+R C I + II + S +G+
Sbjct: 675 --ILCAEIFPTRVRGLCIAICALTFWICDIIVTYSLPVMLNS-------------VGLGG 809
Query: 485 TLILLAVVNCLGIFFTFL-VPEANGKSLE 512
L AVV C+ F FL VPE G LE
Sbjct: 810 VFGLYAVVCCIAWVFVFLKVPETKGMPLE 896
>BQ751130 weakly similar to GP|15289796|dbj putative monosaccharide
transporter 3 {Oryza sativa (japonica cultivar-group)},
partial (7%)
Length = 795
Score = 42.7 bits (99), Expect = 4e-04
Identities = 62/265 (23%), Positives = 103/265 (38%), Gaps = 9/265 (3%)
Frame = +3
Query: 10 TALDAAKTQWYHFTAIIIAGMGFFTDAYDLFCIPNVTKLLGRIYYTH-PGALKPGTLPPN 68
T+++A T W + A G YD I V I + PGA + +
Sbjct: 9 TSIEAPVT-WKAYVICAFASFGGIFFGYDSGYINGVLGANQFIEHVEGPGA---EAISES 176
Query: 69 VSAAVNGVALCGTLAGQLFFGWLGDKMGRK-------KVYGLTLALMVFSSLASGLSFGH 121
+ + + CGT G L G + D +GRK +Y + + + +F+ + + L
Sbjct: 177 HQSLIVSILSCGTFFGALIAGDVSDWIGRKWTVIIGCGIYMIGVLIQMFTGMGAPL---- 344
Query: 122 SAKGTIGTLCFFRFWLGFGIGGDYPLSATIMSEYANKKTRGAFIASVFAMQGFGILAGGI 181
G + R G G+G + + MSE KK RGA +A G+L +
Sbjct: 345 ------GAIVAGRLIAGIGVGFESAIVILYMSEICPKKVRGALVAGYQFCITIGLL---L 497
Query: 182 VSLVVSTAFDHAFKAPPYKVDAAASLVPEA-DYVWRIILMFGAVPAGLTYYWRMKMPETA 240
+ VV D D +P A + W +IL G + + + +K A
Sbjct: 498 AACVVYATKDRG--------DTGVYRIPIAIQFPWALILGGGLLLLPESPRFFVKKGRIA 653
Query: 241 RYTALVAKNAKQAAQDMSSVLQVEI 265
TA +++ Q + S +QVE+
Sbjct: 654 DATAALSRLRGQPKE--SEYIQVEL 722
>AJ497564 weakly similar to PIR|C84593|C845 probable sugar transporter
[imported] - Arabidopsis thaliana, partial (17%)
Length = 610
Score = 42.0 bits (97), Expect = 6e-04
Identities = 29/100 (29%), Positives = 46/100 (46%), Gaps = 7/100 (7%)
Frame = +2
Query: 430 AEIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQSKDPKKRDAGYPAGIGMKNTLILL 489
+EIFP RLR+ + + + + GA + S +K I + T +
Sbjct: 5 SEIFPLRLRAQASALGAVGSRVSS--GAISMSFLSVTK-----------AITVAGTFFVF 145
Query: 490 AVVNCLGIFFT-FLVPEANGKSLEEMS------GENEEEE 522
V++C + F + VPE GKSLEE+ GE++E E
Sbjct: 146 GVISCSAVAFVHYCVPETKGKSLEEIEVLFQNVGESQESE 265
>TC78964 weakly similar to PIR|T00450|T00450 probable monosaccharide
transport protein T14N5.7 - Arabidopsis thaliana,
partial (98%)
Length = 1771
Score = 40.0 bits (92), Expect = 0.002
Identities = 99/447 (22%), Positives = 160/447 (35%), Gaps = 19/447 (4%)
Frame = +1
Query: 87 FFGWLGDKMGRKKVYGLTLALMVFSSLASGLSFGHSAKGTIGTLCFFRFWLGFGIGGDYP 146
F +L GRK T+ + S L + ++A I TL R +LG GIG
Sbjct: 334 FASYLTRNKGRKA----TIIVGALSFLIGAIL--NAAAQNIPTLIIGRVFLGGGIGFGNQ 495
Query: 147 LSATIMSEYANKKTRGAFIASVFAMQGFGILAGGIVSLVVSTAFDHAFKAPPYKVDAAAS 206
+SE A +RGA GIL +V+ H
Sbjct: 496 AVPLYLSEMAPASSRGAVNQLFQFTTCAGILIANLVNYFTDKIHPHG------------- 636
Query: 207 LVPEADYVWRIILMFGAVPAGLTYYWRMKMPETARYTALVAK----NAKQAAQDMSSVLQ 262
WRI L +PA L + ET +LV + A++ + +
Sbjct: 637 --------WRISLGLAGIPAVLMLLGGIFCAETP--NSLVEQGRLDEARKVLEKVRGTKN 786
Query: 263 VEIEAEQEK-VDKIGVQDKNSFG-LFSKEFLRRHGLHLLGTTSTWFLL---DIAYYSSNL 317
V+ E E K ++ K+ F L +++ + + LGT + L I +Y+ +
Sbjct: 787 VDAEFEDLKDASELAQAVKSPFKVLLKRKYRPQLIIGALGTPAFQQLTGNNSILFYAPVI 966
Query: 318 FQKDIYSSIGWLPPAQDMNAIHEVFKVARATTLIALCGTVPGYWFTVAFIDVIGRFAIQL 377
FQ + S NA + L+A ++ +D G L
Sbjct: 967 FQSSGFGS----------NAALFSSFITNGALLVATV-------ISMFLVDKFGTRKFFL 1095
Query: 378 MGFFFMTVFMF----ALAIPYDHWTKRENRI-GFLVMYAMTFFFA---NFGPNSTTFVVP 429
F M M LA+ + H + I FLV+ F A ++GP ++VP
Sbjct: 1096EAGFEMICCMIITAVVLAVEFGHGKELSKGISAFLVIMIFWFVLAYGRSWGP--LGWLVP 1269
Query: 430 AEIFPARLRSTCHGISSAAGKAGAIIGAFGFLYASQSKDPKKRDAGYPAGIGMKNTLILL 489
+E+FP +RS I+ + A FL + +K + LL
Sbjct: 1270SELFPLEIRSAAQSIAVCVNMIFTALVAQLFLLSL---------------CHLKYGIFLL 1404
Query: 490 --AVVNCLGIFFTFLVPEANGKSLEEM 514
++ + +F FL+PE +EE+
Sbjct: 1405FGGLIVVMSVFVFFLLPETKQVPIEEI 1485
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.325 0.139 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,898,562
Number of Sequences: 36976
Number of extensions: 208102
Number of successful extensions: 1454
Number of sequences better than 10.0: 65
Number of HSP's better than 10.0 without gapping: 1409
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1428
length of query: 541
length of database: 9,014,727
effective HSP length: 101
effective length of query: 440
effective length of database: 5,280,151
effective search space: 2323266440
effective search space used: 2323266440
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC148718.7


