

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148718.4 + phase: 0
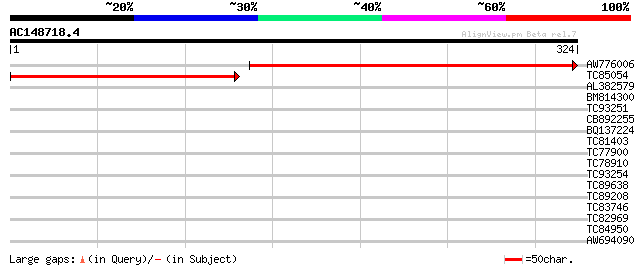
(324 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AW776006 similar to PIR|F84456|F84 hypothetical protein At2g0436... 386 e-108
TC85054 262 2e-70
AL382579 32 0.43
BM814300 similar to PIR|T47833|T478 hypothetical protein T2O9.70... 31 0.57
TC93251 similar to PIR|T39903|T39903 serine-rich protein - fissi... 31 0.57
CB892255 30 0.97
BQ137224 similar to GP|1934808|emb| unknown {Bacillus subtilis},... 30 0.97
TC81403 similar to PIR|T06752|T06752 carboxy-terminal proteinase... 28 4.8
TC77900 similar to GP|9759182|dbj|BAB09797.1 ATP-dependent Clp p... 28 4.8
TC78910 similar to GP|20260424|gb|AAM13110.1 putative protein {A... 28 4.8
TC93254 weakly similar to GP|10176972|dbj|BAB10190. contains sim... 28 6.3
TC89638 similar to GP|9294416|dbj|BAB02497.1 gene_id:MOE17.21~re... 28 6.3
TC89208 homologue to GP|15912211|gb|AAL08239.1 At2g31160/T16B12.... 27 8.2
TC83746 homologue to GP|9955582|emb|CAC05509.1 rec-like protein ... 27 8.2
TC82969 similar to SP|Q43793|G6PC_TOBAC Glucose-6-phosphate 1-de... 27 8.2
TC84950 27 8.2
AW694090 weakly similar to GP|11994279|dbj gb|AAD43616.1~gene_id... 27 8.2
>AW776006 similar to PIR|F84456|F84 hypothetical protein At2g04360 [imported]
- Arabidopsis thaliana, partial (44%)
Length = 697
Score = 386 bits (991), Expect = e-108
Identities = 187/187 (100%), Positives = 187/187 (100%)
Frame = +3
Query: 138 MNEVLVSEWYIMGFWPLVYSMLLLPTGRSSKSSVPVWPFLSASFFGGIYALLPYFVLWKP 197
MNEVLVSEWYIMGFWPLVYSMLLLPTGRSSKSSVPVWPFLSASFFGGIYALLPYFVLWKP
Sbjct: 3 MNEVLVSEWYIMGFWPLVYSMLLLPTGRSSKSSVPVWPFLSASFFGGIYALLPYFVLWKP 182
Query: 198 PPPPVEEAELKTWPLNFLESKITAMILLASGIGIIIYAGLAGEDVWKEFFQYCRESKFIH 257
PPPPVEEAELKTWPLNFLESKITAMILLASGIGIIIYAGLAGEDVWKEFFQYCRESKFIH
Sbjct: 183 PPPPVEEAELKTWPLNFLESKITAMILLASGIGIIIYAGLAGEDVWKEFFQYCRESKFIH 362
Query: 258 ITSIDFTLLSTLAPFWVYNDMTARKWFDKGSWLLPVSLIPLLGPALYILLRPSLSTSAVA 317
ITSIDFTLLSTLAPFWVYNDMTARKWFDKGSWLLPVSLIPLLGPALYILLRPSLSTSAVA
Sbjct: 363 ITSIDFTLLSTLAPFWVYNDMTARKWFDKGSWLLPVSLIPLLGPALYILLRPSLSTSAVA 542
Query: 318 QSPAESE 324
QSPAESE
Sbjct: 543 QSPAESE 563
>TC85054
Length = 437
Score = 262 bits (669), Expect = 2e-70
Identities = 131/131 (100%), Positives = 131/131 (100%)
Frame = +3
Query: 1 MVLVVAKPNLISCNFPSLKTKKSIQPHITVVASGCCCRLSSFKPLVAMKTPFVAATTSFR 60
MVLVVAKPNLISCNFPSLKTKKSIQPHITVVASGCCCRLSSFKPLVAMKTPFVAATTSFR
Sbjct: 45 MVLVVAKPNLISCNFPSLKTKKSIQPHITVVASGCCCRLSSFKPLVAMKTPFVAATTSFR 224
Query: 61 RKSSSSSIVICNDSNKQNSSVEEKEEVKRDWTTSILLFLLWAALIYYVSFLSPNQTPSRD 120
RKSSSSSIVICNDSNKQNSSVEEKEEVKRDWTTSILLFLLWAALIYYVSFLSPNQTPSRD
Sbjct: 225 RKSSSSSIVICNDSNKQNSSVEEKEEVKRDWTTSILLFLLWAALIYYVSFLSPNQTPSRD 404
Query: 121 MYFLKKLLNLK 131
MYFLKKLLNLK
Sbjct: 405 MYFLKKLLNLK 437
>AL382579
Length = 408
Score = 31.6 bits (70), Expect = 0.43
Identities = 17/54 (31%), Positives = 29/54 (53%)
Frame = +2
Query: 161 LPTGRSSKSSVPVWPFLSASFFGGIYALLPYFVLWKPPPPPVEEAELKTWPLNF 214
+P+ +SS+ P P+ + SFF + P + KPP PP++E ++ NF
Sbjct: 242 IPSRKSSRQRPPPKPYTTPSFFKDTESR-PNRNVQKPPTPPLKEKQISCPACNF 400
>BM814300 similar to PIR|T47833|T478 hypothetical protein T2O9.70 -
Arabidopsis thaliana, partial (27%)
Length = 626
Score = 31.2 bits (69), Expect = 0.57
Identities = 24/66 (36%), Positives = 32/66 (48%), Gaps = 6/66 (9%)
Frame = -2
Query: 256 IHITSIDFTLLSTLAPFWVYNDMTARKWFDKGSWLLPV----SLIPLL--GPALYILLRP 309
+HI+S F+ TL PFW RK F ++ S+ PLL G L +LL P
Sbjct: 472 LHISSSSFSFFFTLIPFW-----RLRKSFSSFCMIIAFSRFKSIEPLLGGGAVLDLLLAP 308
Query: 310 SLSTSA 315
LS S+
Sbjct: 307 PLSCSS 290
>TC93251 similar to PIR|T39903|T39903 serine-rich protein - fission yeast
(Schizosaccharomyces pombe), partial (4%)
Length = 781
Score = 31.2 bits (69), Expect = 0.57
Identities = 30/99 (30%), Positives = 43/99 (43%), Gaps = 12/99 (12%)
Frame = -3
Query: 16 PSLKTKKSIQPHITVVASGCCCRLSSFKPLVAMKTPFVAATTSFRRKS----------SS 65
PS + S+QP + + CRLSS KPL P F K+ SS
Sbjct: 671 PSSQPGMSLQP--SSPSPSPSCRLSSAKPLPIHSWPLSPLDGRFGPKAAGRATKDVLLSS 498
Query: 66 SSIVICNDSNKQNSSVEEKEEVKRDWTTSILL--FLLWA 102
++ + +V+EK+ VKRD T+ L L+WA
Sbjct: 497 VKVMAAFGVKMEKGAVKEKKVVKRDRGTAAWLTTALVWA 381
>CB892255
Length = 853
Score = 30.4 bits (67), Expect = 0.97
Identities = 13/27 (48%), Positives = 18/27 (66%)
Frame = -2
Query: 176 FLSASFFGGIYALLPYFVLWKPPPPPV 202
FL+AS F G +L P+FV+W P P +
Sbjct: 318 FLTASVFTGHTSLPPFFVIWHPFAPRI 238
>BQ137224 similar to GP|1934808|emb| unknown {Bacillus subtilis}, partial
(17%)
Length = 1046
Score = 30.4 bits (67), Expect = 0.97
Identities = 20/53 (37%), Positives = 25/53 (46%), Gaps = 7/53 (13%)
Frame = -2
Query: 164 GRSSKSSVPVWPFLSASFF-------GGIYALLPYFVLWKPPPPPVEEAELKT 209
GRS S P PFLS FF G + LLP+ L PPP+ L++
Sbjct: 280 GRSGGRSSPSCPFLSPLFFFPCTFATGLFFFLLPFSSLAGSIPPPLRACMLRS 122
>TC81403 similar to PIR|T06752|T06752 carboxy-terminal proteinase homolog
F15B8.130 - Arabidopsis thaliana, partial (28%)
Length = 674
Score = 28.1 bits (61), Expect = 4.8
Identities = 18/63 (28%), Positives = 32/63 (50%)
Frame = -2
Query: 5 VAKPNLISCNFPSLKTKKSIQPHITVVASGCCCRLSSFKPLVAMKTPFVAATTSFRRKSS 64
+A+P +SCN P+ + P +T+ SG C + + ++ M F + + F R SS
Sbjct: 211 IARP--LSCNAPAKISLALALPSLTITTSGSC--VIACPSIILMGDMFPSLSIVFTRVSS 44
Query: 65 SSS 67
S+
Sbjct: 43 PSN 35
>TC77900 similar to GP|9759182|dbj|BAB09797.1 ATP-dependent Clp protease
regulatory subunit CLPX {Arabidopsis thaliana}, partial
(61%)
Length = 2144
Score = 28.1 bits (61), Expect = 4.8
Identities = 15/41 (36%), Positives = 24/41 (57%)
Frame = +1
Query: 169 SSVPVWPFLSASFFGGIYALLPYFVLWKPPPPPVEEAELKT 209
+S+P PFLSA+ I ++ +L P PPP+ + +KT
Sbjct: 334 TSLPSNPFLSAASISSIRVVI-IILLLPPLPPPLSKTLIKT 453
>TC78910 similar to GP|20260424|gb|AAM13110.1 putative protein {Arabidopsis
thaliana}, partial (40%)
Length = 1216
Score = 28.1 bits (61), Expect = 4.8
Identities = 24/67 (35%), Positives = 33/67 (48%), Gaps = 3/67 (4%)
Frame = -1
Query: 160 LLPTGRSSKSSVPVWPFLSASFFGGIYALL---PYFVLWKPPPPPVEEAELKTWPLNFLE 216
LLP+ R KSS P LS+S LL PY L PPPPP+ + + L LE
Sbjct: 331 LLPSRR--KSSTLSLPPLSSSPPPVSQPLLQPAPYSPLTSPPPPPLSSQQREETKLRRLE 158
Query: 217 SKITAMI 223
+ + ++
Sbjct: 157 TAPSTLL 137
>TC93254 weakly similar to GP|10176972|dbj|BAB10190. contains similarity to
DNA repair protein~gene_id:MTG13.7 {Arabidopsis
thaliana}, partial (8%)
Length = 647
Score = 27.7 bits (60), Expect = 6.3
Identities = 12/28 (42%), Positives = 15/28 (52%)
Frame = -3
Query: 25 QPHITVVASGCCCRLSSFKPLVAMKTPF 52
+PH T+ S CC R S F + K PF
Sbjct: 354 RPHFTLGTSNCCNRKSHFHLHLKFKIPF 271
>TC89638 similar to GP|9294416|dbj|BAB02497.1
gene_id:MOE17.21~ref|NP_002083.1~similar to unknown
protein {Arabidopsis thaliana}, partial (76%)
Length = 1253
Score = 27.7 bits (60), Expect = 6.3
Identities = 14/46 (30%), Positives = 23/46 (49%), Gaps = 5/46 (10%)
Frame = +3
Query: 179 ASFFGG-----IYALLPYFVLWKPPPPPVEEAELKTWPLNFLESKI 219
+SF+G +Y PY + +PPPPP L+ P + E+ +
Sbjct: 156 SSFYGSPGSSYMYNPSPYGYVSQPPPPPFPVVRLRGLPFDCTETDV 293
>TC89208 homologue to GP|15912211|gb|AAL08239.1 At2g31160/T16B12.3
{Arabidopsis thaliana}, partial (17%)
Length = 678
Score = 27.3 bits (59), Expect = 8.2
Identities = 14/38 (36%), Positives = 18/38 (46%)
Frame = +1
Query: 56 TTSFRRKSSSSSIVICNDSNKQNSSVEEKEEVKRDWTT 93
TT+ SSSS N NSS + + +RDW T
Sbjct: 487 TTTTTASGSSSSSAASTIINSPNSSSRYENQKRRDWNT 600
>TC83746 homologue to GP|9955582|emb|CAC05509.1 rec-like protein
{Arabidopsis thaliana}, partial (15%)
Length = 402
Score = 27.3 bits (59), Expect = 8.2
Identities = 23/81 (28%), Positives = 40/81 (48%), Gaps = 4/81 (4%)
Frame = +2
Query: 155 VYSMLLLPTGRSSKSSVPVWPFLSASFFGG----IYALLPYFVLWKPPPPPVEEAELKTW 210
VY++LL SS S + PF S SFF + LLP+ ++ PP + + +
Sbjct: 98 VYALLLAH*IPSSSLSSLLSPF-SLSFFAFSLILVMFLLPFILMMLNPPQMPKIMQNRRN 274
Query: 211 PLNFLESKITAMILLASGIGI 231
N L++ + +I++ S G+
Sbjct: 275 VTNALDTNLQGLIIVESAEGV 337
>TC82969 similar to SP|Q43793|G6PC_TOBAC Glucose-6-phosphate 1-dehydrogenase
chloroplast precursor (EC 1.1.1.49) (G6PD). [Common
tobacco], partial (18%)
Length = 610
Score = 27.3 bits (59), Expect = 8.2
Identities = 15/52 (28%), Positives = 25/52 (47%), Gaps = 3/52 (5%)
Frame = -2
Query: 81 VEEKEEVKRDWTTSILLFLLWAALIYYVSF---LSPNQTPSRDMYFLKKLLN 129
V +E V +WTT+ + +LW +SF + +Q S +K LL+
Sbjct: 306 VSGREIVSTNWTTTTIRIVLWGYFFSSISFNTGVKSDQAASSSSLRIKSLLS 151
>TC84950
Length = 826
Score = 27.3 bits (59), Expect = 8.2
Identities = 11/35 (31%), Positives = 21/35 (59%)
Frame = +2
Query: 91 WTTSILLFLLWAALIYYVSFLSPNQTPSRDMYFLK 125
W LL+L+W + +Y V LS + + ++YF++
Sbjct: 629 WREVTLLWLVWFSYVYIVYSLSLFRQHTSELYFIR 733
>AW694090 weakly similar to GP|11994279|dbj
gb|AAD43616.1~gene_id:F16J14.3~strong similarity to
unknown protein {Arabidopsis thaliana}, partial (4%)
Length = 659
Score = 27.3 bits (59), Expect = 8.2
Identities = 13/42 (30%), Positives = 21/42 (49%)
Frame = +3
Query: 90 DWTTSILLFLLWAALIYYVSFLSPNQTPSRDMYFLKKLLNLK 131
+W TS+LLFL W V+ P + F+ K+ ++K
Sbjct: 342 NWVTSLLLFLFWQRFSKTVTSQIP*PSLHSSRVFVSKVTSIK 467
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.137 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,322,525
Number of Sequences: 36976
Number of extensions: 227521
Number of successful extensions: 1632
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 1570
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1629
length of query: 324
length of database: 9,014,727
effective HSP length: 96
effective length of query: 228
effective length of database: 5,465,031
effective search space: 1246027068
effective search space used: 1246027068
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC148718.4


