

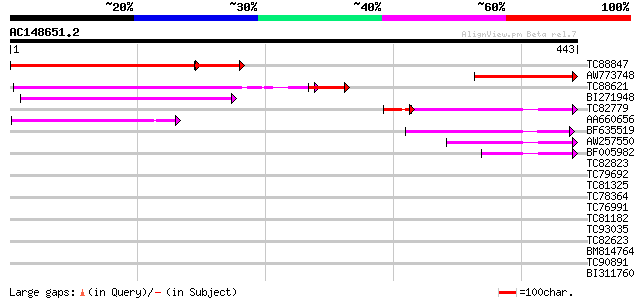
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148651.2 - phase: 0
(443 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC88847 similar to GP|10176892|dbj|BAB10122. Na+-dependent inorg... 298 e-100
AW773748 similar to GP|10176892|dbj Na+-dependent inorganic phos... 157 1e-38
TC88621 similar to PIR|H84698|H84698 hypothetical protein At2g29... 138 2e-36
BI271948 similar to GP|6498428|dbj Similar to Arabidopsis thalia... 121 6e-28
TC82779 similar to GP|6498428|dbj|BAA87831.1 Similar to Arabidop... 93 2e-19
AA660656 weakly similar to GP|6498428|dbj| Similar to Arabidopsi... 88 6e-18
BF635519 similar to GP|17064732|gb putative protein {Arabidopsis... 79 3e-15
AW257550 homologue to GP|17064976|gb putative Na+-dependent inor... 77 1e-14
BF005982 homologue to GP|2252847|gb|A A_IG005I10.nn gene product... 62 6e-10
TC82823 similar to GP|17064732|gb|AAL32520.1 putative protein {A... 40 0.002
TC79692 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_14... 39 0.004
TC81325 similar to GP|17380890|gb|AAL36257.1 putative membrane t... 37 0.015
TC78364 similar to GP|9759266|dbj|BAB09587.1 diacylglycerol kina... 31 0.84
TC76991 similar to SP|Q9SRX2|R19A_ARATH 60S ribosomal protein L1... 30 1.9
TC81182 30 2.4
TC93035 30 2.4
TC82623 weakly similar to GP|17978937|gb|AAL47435.1 AT4g32850/T1... 29 3.2
BM814764 similar to PIR|T10694|T106 hypothetical protein T16I18.... 29 3.2
TC90891 29 4.2
BI311760 similar to GP|21536488|gb unknown {Arabidopsis thaliana... 29 4.2
>TC88847 similar to GP|10176892|dbj|BAB10122. Na+-dependent inorganic
phosphate cotransporter-like protein {Arabidopsis
thaliana}, partial (33%)
Length = 808
Score = 298 bits (762), Expect(2) = e-100
Identities = 147/149 (98%), Positives = 148/149 (98%)
Frame = +2
Query: 1 MQNFPVRYLIVLLTFICTSVCYIERVGFSIAYTVAADAAGINQSTKGTILSTFYYGYACS 60
MQNFPVRYLIVLLTFICTSVCYIERVGFSIAYTVAADAAGINQSTKGTILSTFYYGYACS
Sbjct: 239 MQNFPVRYLIVLLTFICTSVCYIERVGFSIAYTVAADAAGINQSTKGTILSTFYYGYACS 418
Query: 61 QVPGGYFAQKIGGRKILLFSFLLWSLTCALLPLDPNKTLILVTARLLVGVAQGFIFPSIH 120
QVPGGYFAQKIGGRKILLFSFLLWSLTCALLPLDPNKTLILVTARLLVGVAQGFIFPSIH
Sbjct: 419 QVPGGYFAQKIGGRKILLFSFLLWSLTCALLPLDPNKTLILVTARLLVGVAQGFIFPSIH 598
Query: 121 TVLAQWVPPHERSRSVSLTTSGMYLGAAF 149
TVLAQWVPPHERSRSVSLTTSGMYLG +F
Sbjct: 599 TVLAQWVPPHERSRSVSLTTSGMYLGXSF 685
Score = 84.7 bits (208), Expect(2) = e-100
Identities = 38/39 (97%), Positives = 39/39 (99%)
Frame = +3
Query: 145 LGAAFGMLFLPSLVKFKGPQSVFVAESFLGFVWCFIWFR 183
LGAAFGMLFLPSLVKFKGPQSVFVAESFLGFVWCFIWF+
Sbjct: 672 LGAAFGMLFLPSLVKFKGPQSVFVAESFLGFVWCFIWFK 788
>AW773748 similar to GP|10176892|dbj Na+-dependent inorganic phosphate
cotransporter-like protein {Arabidopsis thaliana},
partial (20%)
Length = 596
Score = 157 bits (396), Expect = 1e-38
Identities = 79/80 (98%), Positives = 80/80 (99%)
Frame = -1
Query: 364 HMDVAPRYAGIVMGVSNTAGTLAGIVGVDLTGKLLEAAKASDSDLSTPESWRLVFFIPGL 423
+MDVAPRYAGIVMGVSNTAGTLAGIVGVDLTGKLLEAAKASDSDLSTPESWRLVFFIPGL
Sbjct: 563 NMDVAPRYAGIVMGVSNTAGTLAGIVGVDLTGKLLEAAKASDSDLSTPESWRLVFFIPGL 384
Query: 424 LCVFSSLVFLLFSTGERIFD 443
LCVFSSLVFLLFSTGERIFD
Sbjct: 383 LCVFSSLVFLLFSTGERIFD 324
>TC88621 similar to PIR|H84698|H84698 hypothetical protein At2g29650
[imported] - Arabidopsis thaliana, partial (49%)
Length = 1074
Score = 138 bits (347), Expect(2) = 2e-36
Identities = 85/240 (35%), Positives = 127/240 (52%), Gaps = 1/240 (0%)
Frame = +1
Query: 4 FPVRYLIVLLTFICTSVCYIERVGFSIAYTVAADAAGINQSTKGTILSTFYYGYACSQVP 63
FP R++IV+L F +C ++RV SIA + N +T G + S+F++GY +Q+
Sbjct: 322 FPKRWVIVILCFSAFLLCNMDRVNMSIAILPMSAEYNWNPTTVGLVQSSFFWGYLLTQIA 501
Query: 64 GGYFAQKIGGRKILLFSFLLWSLTCALLPLDPNKTL-ILVTARLLVGVAQGFIFPSIHTV 122
GG +A +GG+++L F + WS+ L P+ L L+ AR +G+ +G P+++ +
Sbjct: 502 GGIWADTVGGKQVLAFGVIWWSVATILTPVAAKLGLPFLLVARAFMGIGEGVAMPAMNNI 681
Query: 123 LAQWVPPHERSRSVSLTTSGMYLGAAFGMLFLPSLVKFKGPQSVFVAESFLGFVWCFIWF 182
L++WVP ERSRS++L SGMYLG+ G+ F P L+ G SVF + LG VW IW
Sbjct: 682 LSKWVPVAERSRSLALVYSGMYLGSVTGLAFSPFLIHQYGWPSVFYSFGSLGTVWFCIWL 861
Query: 183 RYSSDPKSSASGAGESLLPVNKKIDRRVSDLKVGVEKNGGEGKKSGVGIPWVKILTSLPV 242
S SS E + P KK+ G K+ IPW IL+ PV
Sbjct: 862 ---SKAHSSPLDDPE-MRPEEKKL-----------IATNGFSKEPVKEIPWGLILSKPPV 996
Score = 32.7 bits (73), Expect(2) = 2e-36
Identities = 8/32 (25%), Positives = 22/32 (68%)
Frame = +3
Query: 234 VKILTSLPVWAIVVNNFTFHYALYVLMNWLPT 265
+ + + +WA++V++F ++ ++L+ W+PT
Sbjct: 972 INFVETTGIWALIVSHFCHNWGTFILLTWMPT 1067
>BI271948 similar to GP|6498428|dbj Similar to Arabidopsis thaliana
chromosome II BAC T27A16 sequence; hypothetical protein.
(AC005496), partial (33%)
Length = 695
Score = 121 bits (303), Expect = 6e-28
Identities = 62/170 (36%), Positives = 99/170 (57%), Gaps = 1/170 (0%)
Frame = +3
Query: 9 LIVLLTFICTSVCYIERVGFSIAYTVAADAAGINQSTKGTILSTFYYGYACSQVPGGYFA 68
L+ L F +C ++RV SIA + N +T G I S+F++GY +Q+ GG +A
Sbjct: 81 LLYXLCFTAFLLCNMDRVNMSIAILPMSQQFNWNSATVGLIQSSFFWGYLLTQIAGGIWA 260
Query: 69 QKIGGRKILLFSFLLWSLTCALLPLDPNKTL-ILVTARLLVGVAQGFIFPSIHTVLAQWV 127
K+GG+ +L F + WS+ L P+ L L+ R +G+ +G P+++ +L++W+
Sbjct: 261 DKVGGKLVLGFGVVWWSIATVLTPIAAKLGLPYLLVMRAFMGIGEGVAMPAMNNILSKWI 440
Query: 128 PPHERSRSVSLTTSGMYLGAAFGMLFLPSLVKFKGPQSVFVAESFLGFVW 177
P ERSRS++L SGMYLG+ G+ F P L++ G SVF + LG +W
Sbjct: 441 PVSERSRSLALVYSGMYLGSVTGLGFSPFLIQKFGWPSVFYSFGSLGSIW 590
>TC82779 similar to GP|6498428|dbj|BAA87831.1 Similar to Arabidopsis
thaliana chromosome II BAC T27A16 sequence; hypothetical
protein. (AC005496), partial (26%)
Length = 723
Score = 92.8 bits (229), Expect(2) = 2e-19
Identities = 50/129 (38%), Positives = 74/129 (56%)
Frame = +2
Query: 315 RKFLNTIGFLVASFALIVIPSFRTSGGAVFCSSVALGFLALGRAGFAVNHMDVAPRYAGI 374
RK + +IG L +F L + + RT AV C + + G A ++G NH D+ PRYAG+
Sbjct: 65 RKIMQSIGVLGPAFFLTQLSNVRTPAMAVLCMACSQGCDAFSQSGLYSNHQDIGPRYAGV 244
Query: 375 VMGVSNTAGTLAGIVGVDLTGKLLEAAKASDSDLSTPESWRLVFFIPGLLCVFSSLVFLL 434
++G+SNTAG LAG+ G TG +L+ SW VF + +L + +LV+ +
Sbjct: 245 LLGLSNTAGVLAGVFGTAATGYILQRG-----------SWNDVFKVSVVLYLIGTLVWNI 391
Query: 435 FSTGERIFD 443
FSTGE+I D
Sbjct: 392 FSTGEKILD 418
Score = 20.8 bits (42), Expect(2) = 2e-19
Identities = 10/24 (41%), Positives = 16/24 (66%)
Frame = +1
Query: 293 SNIGGVVADYLITRRVMSVTRTRK 316
+NIGG +AD L+++ S+T K
Sbjct: 1 ANIGGWIADTLVSKG-FSITTVAK 69
>AA660656 weakly similar to GP|6498428|dbj| Similar to Arabidopsis thaliana
chromosome II BAC T27A16 sequence; hypothetical protein.
(AC005496), partial (14%)
Length = 634
Score = 88.2 bits (217), Expect = 6e-18
Identities = 47/132 (35%), Positives = 75/132 (56%)
Frame = +3
Query: 2 QNFPVRYLIVLLTFICTSVCYIERVGFSIAYTVAADAAGINQSTKGTILSTFYYGYACSQ 61
+N P+RY ++ T + +C +++V SIA + G N ST G + S+F++GYA SQ
Sbjct: 195 KNLPLRYKLIGTTSLAFVICNMDKVNLSIAIIPMSHQFGWNSSTAGLVQSSFFWGYALSQ 374
Query: 62 VPGGYFAQKIGGRKILLFSFLLWSLTCALLPLDPNKTLILVTARLLVGVAQGFIFPSIHT 121
+PGG+ A+ GG +L+ L+WS+ AL+P L+ R+LVG+ + + PS T
Sbjct: 375 LPGGWLAKIFGGXNVLVVGVLIWSVATALVPFLAGYMPGLILTRILVGIXES-VSPSAAT 551
Query: 122 VLAQWVPPHERS 133
L P ER+
Sbjct: 552 DLIAXSIPLERT 587
>BF635519 similar to GP|17064732|gb putative protein {Arabidopsis thaliana},
partial (25%)
Length = 676
Score = 79.3 bits (194), Expect = 3e-15
Identities = 45/132 (34%), Positives = 73/132 (55%)
Frame = -3
Query: 310 SVTRTRKFLNTIGFLVASFALIVIPSFRTSGGAVFCSSVALGFLALGRAGFAVNHMDVAP 369
SVT T K + +IGF+ +LI + + R A ++A G + G +GF VN ++AP
Sbjct: 641 SVTLTXKIMQSIGFVGPGVSLIGLATARNPSVASAWLTLAFGLKSFGHSGFLVNFQEIAP 462
Query: 370 RYAGIVMGVSNTAGTLAGIVGVDLTGKLLEAAKASDSDLSTPESWRLVFFIPGLLCVFSS 429
+Y+G++ G++NTAGTLA I+G G +E S+R + +L ++
Sbjct: 461 QYSGVIHGMANTAGTLAAIIGTVGAGFFVELV----------GSFRGFLLLTSVLYFLAA 312
Query: 430 LVFLLFSTGERI 441
L + L+STGER+
Sbjct: 311 LFYCLYSTGERV 276
>AW257550 homologue to GP|17064976|gb putative Na+-dependent inorganic
phosphate cotransporter {Arabidopsis thaliana}, partial
(20%)
Length = 723
Score = 77.0 bits (188), Expect = 1e-14
Identities = 42/102 (41%), Positives = 61/102 (59%)
Frame = -2
Query: 342 AVFCSSVALGFLALGRAGFAVNHMDVAPRYAGIVMGVSNTAGTLAGIVGVDLTGKLLEAA 401
AV C + + G A ++G NH D+APRY+GI++G+SNTAG LAG++G TG +L+
Sbjct: 677 AVLCMTCSQGTDAFSQSGLYSNHQDIAPRYSGILLGLSNTAGVLAGVLGTAATGYILQHG 498
Query: 402 KASDSDLSTPESWRLVFFIPGLLCVFSSLVFLLFSTGERIFD 443
SW VF + L + ++V+ LFSTGE+I D
Sbjct: 497 -----------SWDDVFKVSVGLYLVGTVVWNLFSTGEKIID 405
>BF005982 homologue to GP|2252847|gb|A A_IG005I10.nn gene product
{Arabidopsis thaliana}, partial (15%)
Length = 460
Score = 61.6 bits (148), Expect = 6e-10
Identities = 32/75 (42%), Positives = 45/75 (59%)
Frame = +1
Query: 369 PRYAGIVMGVSNTAGTLAGIVGVDLTGKLLEAAKASDSDLSTPESWRLVFFIPGLLCVFS 428
PRYAG+++G+SNTAG LAG+ G TG +L+ SW VF + L +
Sbjct: 4 PRYAGVLLGLSNTAGVLAGVFGTAATGYILQRG-----------SWNDVFKVSVALYIIG 150
Query: 429 SLVFLLFSTGERIFD 443
+LV+ +FSTGE+I D
Sbjct: 151 TLVWNIFSTGEKILD 195
>TC82823 similar to GP|17064732|gb|AAL32520.1 putative protein {Arabidopsis
thaliana}, partial (25%)
Length = 949
Score = 39.7 bits (91), Expect = 0.002
Identities = 19/68 (27%), Positives = 34/68 (49%)
Frame = +1
Query: 7 RYLIVLLTFICTSVCYIERVGFSIAYTVAADAAGINQSTKGTILSTFYYGYACSQVPGGY 66
R +V + + ++C +RV S+A + A G ++ G + S+F +GY S + GG
Sbjct: 307 RVKVVAMLALALALCNADRVVMSVAIVPLSVANGWTRAFSGIVQSSFLWGYLLSPIGGGM 486
Query: 67 FAQKIGGR 74
GG+
Sbjct: 487 LVDNYGGK 510
>TC79692 similar to GP|20453189|gb|AAM19835.1 AT4g35300/F23E12_140
{Arabidopsis thaliana}, partial (49%)
Length = 1353
Score = 38.9 bits (89), Expect = 0.004
Identities = 32/110 (29%), Positives = 51/110 (46%)
Frame = +2
Query: 44 STKGTILSTFYYGYACSQVPGGYFAQKIGGRKILLFSFLLWSLTCALLPLDPNKTLILVT 103
+ +G I++ G G + G R +L+ S LL+ L+ ++ PN IL+
Sbjct: 278 TVEGLIVAMSLIGATVVTTCSGALSDLFGRRPMLIISSLLYFLSSLVMFWSPN-VYILLF 454
Query: 104 ARLLVGVAQGFIFPSIHTVLAQWVPPHERSRSVSLTTSGMYLGAAFGMLF 153
ARLL G+ G + +++ PP R SL T + G+A GM F
Sbjct: 455 ARLLDGLGIGLAVTLVPLYISEIAPPEIRG---SLNTLPQFAGSA-GMFF 592
>TC81325 similar to GP|17380890|gb|AAL36257.1 putative membrane transporter
protein {Arabidopsis thaliana}, partial (32%)
Length = 636
Score = 37.0 bits (84), Expect = 0.015
Identities = 30/113 (26%), Positives = 49/113 (42%), Gaps = 4/113 (3%)
Frame = +3
Query: 48 TILSTFYYGYACSQVPGGYFAQKIGGRKILLFSFLLWSLTCALLPLDPNKTLILVTARLL 107
TI+S G GG+ G +K L + ++++L ++ P+ +L+ RLL
Sbjct: 300 TIVSMALVGAIIGAATGGWINDAFGRKKATLSADVVFTLGSVVMASAPD-AYVLILGRLL 476
Query: 108 VGVAQGFIFPSIHTVLAQWVPPHER----SRSVSLTTSGMYLGAAFGMLFLPS 156
VG+ G + +A+ P R S +V + T G F L LPS
Sbjct: 477 VGIGVGVASVTAPVYIAESSPSEIRGSLVSTNVLMITGGQVFFLTFVNLALPS 635
>TC78364 similar to GP|9759266|dbj|BAB09587.1 diacylglycerol kinase-like
protein {Arabidopsis thaliana}, partial (76%)
Length = 1889
Score = 31.2 bits (69), Expect = 0.84
Identities = 26/109 (23%), Positives = 51/109 (45%), Gaps = 2/109 (1%)
Frame = -3
Query: 121 TVLAQWVPPHERSRSVSLTTSGMYLGAAFGMLFLPSLVKFKGPQSVFVAE--SFLGFVWC 178
++L Q+ +RSR + + + L LPSL+ P +F+ E S + WC
Sbjct: 297 SLLCQYACRRDRSRELIQRVMNELVSS----LNLPSLMMAMIPFFIFLFEDESSVVLCWC 130
Query: 179 FIWFRYSSDPKSSASGAGESLLPVNKKIDRRVSDLKVGVEKNGGEGKKS 227
+ WF + + K + G K ++R+ ++K+ +++N K+S
Sbjct: 129 WCWFFSAGESK*NHLG--------TKSLNRKNGEIKIIIKENNE*RKES 7
>TC76991 similar to SP|Q9SRX2|R19A_ARATH 60S ribosomal protein L19-1.
[Mouse-ear cress] {Arabidopsis thaliana}, partial (96%)
Length = 850
Score = 30.0 bits (66), Expect = 1.9
Identities = 13/27 (48%), Positives = 20/27 (73%)
Frame = -1
Query: 259 LMNWLPTYFELGLKLSLHEMGSSKMLP 285
L N+L + EL +++SLH +GSS+ LP
Sbjct: 160 LTNFLMFWRELAMEISLHSLGSSQTLP 80
>TC81182
Length = 917
Score = 29.6 bits (65), Expect = 2.4
Identities = 17/60 (28%), Positives = 30/60 (49%)
Frame = -1
Query: 247 VNNFTFHYALYVLMNWLPTYFELGLKLSLHEMGSSKMLPYLNMFVFSNIGGVVADYLITR 306
++NF Y LMN + L L + L + SS +N+F F+ G +V D ++++
Sbjct: 563 ISNFNLSIRNYFLMNQEVLFPRLDLLICLSSLASSSFA--INVFSFTPFGNLVEDSVLSK 390
>TC93035
Length = 766
Score = 29.6 bits (65), Expect = 2.4
Identities = 20/63 (31%), Positives = 28/63 (43%)
Frame = -3
Query: 99 LILVTARLLVGVAQGFIFPSIHTVLAQWVPPHERSRSVSLTTSGMYLGAAFGMLFLPSLV 158
L L LL Q +F + V AQW+PP +SR LG + +PSL+
Sbjct: 320 LSLSVVCLLSTTGQVRVFLDLEDVEAQWLPPRSKSR----------LGRLLAIADVPSLL 171
Query: 159 KFK 161
F+
Sbjct: 170 AFQ 162
>TC82623 weakly similar to GP|17978937|gb|AAL47435.1 AT4g32850/T16I18_60
{Arabidopsis thaliana}, partial (6%)
Length = 721
Score = 29.3 bits (64), Expect = 3.2
Identities = 19/55 (34%), Positives = 30/55 (54%), Gaps = 2/55 (3%)
Frame = -1
Query: 78 LFSFLLWSLTCALLPLDPNKTLILVTARLLVG--VAQGFIFPSIHTVLAQWVPPH 130
L SF+ S T ++PL + + L++ L V V + PSIH++L VPP+
Sbjct: 499 LVSFICMSPTSIIVPLPESPSDDLISQLLSVSFVVEPSSLTPSIHSILI*GVPPN 335
>BM814764 similar to PIR|T10694|T106 hypothetical protein T16I18.80 -
Arabidopsis thaliana, partial (24%)
Length = 531
Score = 29.3 bits (64), Expect = 3.2
Identities = 19/83 (22%), Positives = 35/83 (41%), Gaps = 12/83 (14%)
Frame = +2
Query: 186 SDPKSSASGAGESLL---PVNKKIDRRVSDLKVGVEK---------NGGEGKKSGVGIPW 233
++P+++ A E LL P+ + + + D +G + N G+ K +G GI W
Sbjct: 218 AEPETTIKWAKEKLLKIDPIKRCLSYEIVDNNMGFKSYVATLKVLPNEGDAKSAGCGIEW 397
Query: 234 VKILTSLPVWAIVVNNFTFHYAL 256
+ + W + N Y L
Sbjct: 398 GFVCDPIEGWTLQDFNSYIEYCL 466
>TC90891
Length = 580
Score = 28.9 bits (63), Expect = 4.2
Identities = 10/15 (66%), Positives = 11/15 (72%)
Frame = -3
Query: 172 FLGFVWCFIWFRYSS 186
F+G WCF WFR SS
Sbjct: 212 FMGATWCFRWFRSSS 168
>BI311760 similar to GP|21536488|gb unknown {Arabidopsis thaliana}, partial
(30%)
Length = 641
Score = 28.9 bits (63), Expect = 4.2
Identities = 30/113 (26%), Positives = 50/113 (43%), Gaps = 9/113 (7%)
Frame = +2
Query: 44 STKGTILSTF-YYGYACSQVPGGYFAQKIGGRKILLFSFLLWSLTCALLPL--------D 94
ST T+ S+F ++G S +P Y K F+ W LT ALLP
Sbjct: 242 STLVTLYSSFSFFGRLLSAMPD-YIRNKF------YFARTGW-LTIALLPTPIAFILLAS 397
Query: 95 PNKTLILVTARLLVGVAQGFIFPSIHTVLAQWVPPHERSRSVSLTTSGMYLGA 147
+ L T L+G++ GFIF + V ++ P S + ++ + + +G+
Sbjct: 398 SESAMALNTGTALIGLSSGFIFAAAVAVTSELFGPDSLSVNHNILITNIPIGS 556
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.326 0.141 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,116,566
Number of Sequences: 36976
Number of extensions: 228032
Number of successful extensions: 1756
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 1733
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1750
length of query: 443
length of database: 9,014,727
effective HSP length: 99
effective length of query: 344
effective length of database: 5,354,103
effective search space: 1841811432
effective search space used: 1841811432
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148651.2


