

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
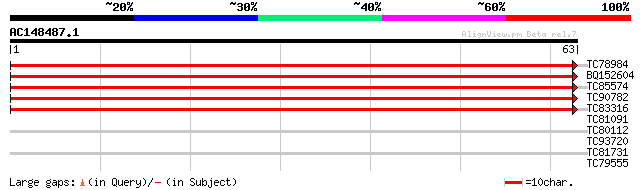
Query= AC148487.1 - phase: 1
(63 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC78984 similar to GP|22759723|dbj|BAC10910. protease inhibitor ... 134 9e-33
BQ152604 similar to GP|22759723|dbj| protease inhibitor 2 {Zinni... 115 3e-27
TC85574 homologue to PIR|T09390|T09390 21K protein precursor - a... 107 9e-25
TC90782 weakly similar to GP|603890|emb|CAA87073.1| pathogenesis... 94 1e-20
TC83316 weakly similar to GP|603890|emb|CAA87073.1| pathogenesis... 69 5e-13
TC81091 similar to GP|4768916|gb|AAD29679.1| CLC-Nt2 protein {Ni... 27 1.2
TC80112 weakly similar to GP|20161371|dbj|BAB90295. hypothetical... 27 1.2
TC93720 similar to GP|21743320|dbj|BAC03314. putative nematode r... 25 5.9
TC81731 weakly similar to GP|9965103|gb|AAG09951.1| resistance p... 25 5.9
TC79555 homologue to GP|10567859|gb|AAG18593.1 L8019.5 {Leishman... 25 7.7
>TC78984 similar to GP|22759723|dbj|BAC10910. protease inhibitor 2 {Zinnia
elegans}, partial (74%)
Length = 545
Score = 134 bits (336), Expect = 9e-33
Identities = 63/63 (100%), Positives = 63/63 (100%)
Frame = +1
Query: 1 KSSWPELVGVEGKVAEATIQRENPLVDAIIVPEGSSVPFDFRCDRVWVWINKDEIVYQVP 60
KSSWPELVGVEGKVAEATIQRENPLVDAIIVPEGSSVPFDFRCDRVWVWINKDEIVYQVP
Sbjct: 115 KSSWPELVGVEGKVAEATIQRENPLVDAIIVPEGSSVPFDFRCDRVWVWINKDEIVYQVP 294
Query: 61 TIG 63
TIG
Sbjct: 295 TIG 303
>BQ152604 similar to GP|22759723|dbj| protease inhibitor 2 {Zinnia elegans},
partial (74%)
Length = 477
Score = 115 bits (288), Expect = 3e-27
Identities = 53/63 (84%), Positives = 60/63 (95%)
Frame = +3
Query: 1 KSSWPELVGVEGKVAEATIQRENPLVDAIIVPEGSSVPFDFRCDRVWVWINKDEIVYQVP 60
KSSWPELVGVEGKVAEATI+RENPLV+AIIVPEGS+V DFRCDRVWVW++KD IV++VP
Sbjct: 105 KSSWPELVGVEGKVAEATIERENPLVNAIIVPEGSAVILDFRCDRVWVWVDKDGIVFKVP 284
Query: 61 TIG 63
TIG
Sbjct: 285 TIG 293
>TC85574 homologue to PIR|T09390|T09390 21K protein precursor - alfalfa,
partial (96%)
Length = 1211
Score = 107 bits (267), Expect = 9e-25
Identities = 52/63 (82%), Positives = 56/63 (88%)
Frame = -1
Query: 1 KSSWPELVGVEGKVAEATIQRENPLVDAIIVPEGSSVPFDFRCDRVWVWINKDEIVYQVP 60
K+SWPELVGVEGKVAEATIQ ENPLV+AIIVPEGS V DFRCDRV VW++KD IVYQVP
Sbjct: 389 KNSWPELVGVEGKVAEATIQSENPLVNAIIVPEGSFVTADFRCDRVRVWVDKDGIVYQVP 210
Query: 61 TIG 63
IG
Sbjct: 209 IIG 201
>TC90782 weakly similar to GP|603890|emb|CAA87073.1| pathogenesis-related
protein PR-6 type {Sambucus nigra}, partial (81%)
Length = 461
Score = 93.6 bits (231), Expect = 1e-20
Identities = 49/63 (77%), Positives = 52/63 (81%)
Frame = +2
Query: 1 KSSWPELVGVEGKVAEATIQRENPLVDAIIVPEGSSVPFDFRCDRVWVWINKDEIVYQVP 60
KSSWPELVGVEGKVAEATIQREN V AIIV EGSSV D R DRV VW++K+ IV QVP
Sbjct: 68 KSSWPELVGVEGKVAEATIQRENASVKAIIVLEGSSVTDDLRFDRVRVWVDKEGIVTQVP 247
Query: 61 TIG 63
TIG
Sbjct: 248 TIG 256
>TC83316 weakly similar to GP|603890|emb|CAA87073.1| pathogenesis-related
protein PR-6 type {Sambucus nigra}, partial (81%)
Length = 320
Score = 68.6 bits (166), Expect = 5e-13
Identities = 33/63 (52%), Positives = 42/63 (66%)
Frame = +2
Query: 1 KSSWPELVGVEGKVAEATIQRENPLVDAIIVPEGSSVPFDFRCDRVWVWINKDEIVYQVP 60
K +WPELVGV G+ A I EN LV A +PE S DFRCDRV V+++K ++V +VP
Sbjct: 17 KKTWPELVGVNGEAAAQIIMSENSLVTASTLPEDSVFTADFRCDRVRVFVDKQDVVTRVP 196
Query: 61 TIG 63
IG
Sbjct: 197KIG 205
>TC81091 similar to GP|4768916|gb|AAD29679.1| CLC-Nt2 protein {Nicotiana
tabacum}, partial (13%)
Length = 662
Score = 27.3 bits (59), Expect = 1.2
Identities = 14/39 (35%), Positives = 20/39 (50%)
Frame = +3
Query: 1 KSSWPELVGVEGKVAEATIQRENPLVDAIIVPEGSSVPF 39
K +W EL EGK+ E I RE + + P ++ PF
Sbjct: 18 KFTWVELAEREGKIEEVAITREEMEMFVDLHPLTNTTPF 134
>TC80112 weakly similar to GP|20161371|dbj|BAB90295. hypothetical
protein~similar to Arabidopsis thaliana chromosome 1
NM_102624.1, partial (31%)
Length = 2068
Score = 27.3 bits (59), Expect = 1.2
Identities = 13/35 (37%), Positives = 20/35 (57%), Gaps = 6/35 (17%)
Frame = -3
Query: 24 PLVDAII-----VPEGS-SVPFDFRCDRVWVWINK 52
P V A++ +P S +P+ F +RVWVW N+
Sbjct: 1475 PFVSAVVPHL*YIPISSVPLPWSFEANRVWVWSNQ 1371
>TC93720 similar to GP|21743320|dbj|BAC03314. putative nematode
resistance-like protein {Oryza sativa (japonica
cultivar-group)}, partial (3%)
Length = 457
Score = 25.0 bits (53), Expect = 5.9
Identities = 10/15 (66%), Positives = 11/15 (72%)
Frame = -3
Query: 2 SSWPELVGVEGKVAE 16
SSWP L G G+VAE
Sbjct: 398 SSWPRLCGAFGRVAE 354
>TC81731 weakly similar to GP|9965103|gb|AAG09951.1| resistance protein LM6
{Glycine max}, partial (7%)
Length = 577
Score = 25.0 bits (53), Expect = 5.9
Identities = 13/43 (30%), Positives = 22/43 (50%)
Frame = -3
Query: 1 KSSWPELVGVEGKVAEATIQRENPLVDAIIVPEGSSVPFDFRC 43
+++W L G+ GK+ + + + + EG S PFDF C
Sbjct: 191 QTTWKNL-GISGKLCVSNYKN----FQILHLSEGFSTPFDFSC 78
>TC79555 homologue to GP|10567859|gb|AAG18593.1 L8019.5 {Leishmania major},
partial (1%)
Length = 1410
Score = 24.6 bits (52), Expect = 7.7
Identities = 8/14 (57%), Positives = 10/14 (71%)
Frame = -2
Query: 40 DFRCDRVWVWINKD 53
DF +RVWVW+ D
Sbjct: 482 DFMVERVWVWVMVD 441
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.139 0.441
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,203,051
Number of Sequences: 36976
Number of extensions: 22324
Number of successful extensions: 114
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 113
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 114
length of query: 63
length of database: 9,014,727
effective HSP length: 39
effective length of query: 24
effective length of database: 7,572,663
effective search space: 181743912
effective search space used: 181743912
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 51 (24.3 bits)
Medicago: description of AC148487.1


