

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148304.1 + phase: 0
(1290 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
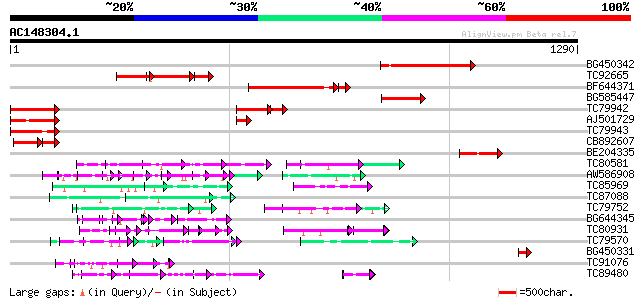
Score E
Sequences producing significant alignments: (bits) Value
BG450342 similar to GP|8927657|gb EST gb|N38213 comes from this ... 298 7e-81
TC92665 homologue to GP|15291583|gb|AAK93060.1 GH28569p {Drosoph... 194 3e-69
BF644371 weakly similar to GP|8927657|gb| EST gb|N38213 comes fr... 230 1e-65
BG585447 204 1e-52
TC79942 similar to GP|8927657|gb|AAF82148.1| EST gb|N38213 comes... 169 5e-42
AJ501729 similar to GP|8927657|gb| EST gb|N38213 comes from this... 152 6e-37
TC79943 homologue to GP|8927657|gb|AAF82148.1| EST gb|N38213 com... 144 3e-34
CB892607 similar to GP|8927657|gb| EST gb|N38213 comes from this... 85 5e-26
BE204335 weakly similar to GP|8927657|gb| EST gb|N38213 comes fr... 105 1e-22
TC80581 similar to GP|19683021|gb|AAL92644.1 hypothetical protei... 94 3e-19
AW586908 weakly similar to GP|2104814|emb| pleiotropic effects o... 75 1e-13
TC85969 similar to GP|21553908|gb|AAM62991.1 unknown {Arabidopsi... 59 1e-08
TC87088 similar to PIR|F86399|F86399 protein F17L21.22 [imported... 57 5e-08
TC79752 similar to GP|11994670|dbj|BAB02898. gene_id:K7P8.17~unk... 56 8e-08
BG644345 weakly similar to GP|19697333|gb putative protein poten... 55 1e-07
TC80931 similar to GP|14190783|gb|AAF65166.2 putative phloem tra... 55 1e-07
TC79570 similar to PIR|F86427|F86427 auxin response factor 6 (AR... 55 2e-07
BG450331 similar to GP|16649103|gb| Unknown protein {Arabidopsis... 55 2e-07
TC91076 similar to PIR|H96747|H96747 unknown protein T10D10.14 [... 54 5e-07
TC89480 similar to GP|20260684|gb|AAM13240.1 unknown protein {Ar... 50 7e-06
>BG450342 similar to GP|8927657|gb EST gb|N38213 comes from this gene.
{Arabidopsis thaliana}, partial (10%)
Length = 669
Score = 298 bits (764), Expect = 7e-81
Identities = 161/223 (72%), Positives = 181/223 (80%), Gaps = 6/223 (2%)
Frame = +2
Query: 843 VFQQHLTSGQNSTYSHQQMKQGSPFQVSSPQLFQAASPQIPHNSSPQVDQQTHLLSLTKV 902
VFQQHL S Q S Y HQQ+KQG PF VSSPQL QA SPQI +SSPQVDQQ HL S+TKV
Sbjct: 2 VFQQHLASSQRSAYPHQQLKQG-PFPVSSPQLLQATSPQISQHSSPQVDQQNHLPSVTKV 178
Query: 903 GTPLQSSNSPFGVPTPSPPMAPSPMLVDSEKPIPGVSS-SNAANVGQ----NAAAPAQSL 957
GTP+QS+NSPF +PTPSPP+APSPM D EKPI GVSS SN ANVG + AAPAQSL
Sbjct: 179 GTPMQSANSPFIIPTPSPPLAPSPMPGDFEKPISGVSSISNVANVGHQQTGSGAAPAQSL 358
Query: 958 AIGTPGISASPLLAEFSGPDGTFCNALGAPSGKST-ADHPIDRLIRAVQSMSTETLTAAV 1016
AIGTPGISASPLLAEF+GPDG NAL SGKST + P+DRLI+AV S++ L+AAV
Sbjct: 359 AIGTPGISASPLLAEFTGPDGAHGNALAPSSGKSTVTEEPMDRLIKAVSSLTPAALSAAV 538
Query: 1017 SDISSVVSMSDRISGSAPGNGSRAAVGEDLVSMTNCRLQARSF 1059
SDISSV+SM+DRI+GSAPGNGSRAAVGE LV+MTNCRL A +F
Sbjct: 539 SDISSVISMNDRIAGSAPGNGSRAAVGEXLVAMTNCRLXAXNF 667
>TC92665 homologue to GP|15291583|gb|AAK93060.1 GH28569p {Drosophila
melanogaster}, partial (4%)
Length = 668
Score = 194 bits (492), Expect(2) = 3e-69
Identities = 97/116 (83%), Positives = 99/116 (84%), Gaps = 7/116 (6%)
Frame = +1
Query: 312 MIQQQHQ-------QLAGPQSNATNGQHAQMLGQQNNVGDVQKSQRLHPQQNNLMNLQQR 364
+I +HQ L G NGQHAQMLGQQNNVGDVQKSQRLHPQQNNLMNLQQR
Sbjct: 190 LISNKHQ*FNSNISSLRGLSQMQRNGQHAQMLGQQNNVGDVQKSQRLHPQQNNLMNLQQR 369
Query: 365 QQQQQLMNHQNNLTNIHQQPGNNVQGLQQQQQFGTESGNQGIQTSHHSAQMLQQPK 420
QQQQQLMNHQNNLTNIHQQPGNNVQGLQQQQQFGTESGNQGIQTSHHSAQMLQQPK
Sbjct: 370 QQQQQLMNHQNNLTNIHQQPGNNVQGLQQQQQFGTESGNQGIQTSHHSAQMLQQPK 537
Score = 167 bits (422), Expect = 3e-41
Identities = 86/86 (100%), Positives = 86/86 (100%)
Frame = +3
Query: 243 PNQLLSSQQSVIPTSSAMQPSMVQSSLSSLPQNQQSNNVQQSTQSRLQQHSQIIRQQQQN 302
PNQLLSSQQSVIPTSSAMQPSMVQSSLSSLPQNQQSNNVQQSTQSRLQQHSQIIRQQQQN
Sbjct: 3 PNQLLSSQQSVIPTSSAMQPSMVQSSLSSLPQNQQSNNVQQSTQSRLQQHSQIIRQQQQN 182
Query: 303 SIVNQQQTPMIQQQHQQLAGPQSNAT 328
SIVNQQQTPMIQQQHQQLAGPQSNAT
Sbjct: 183 SIVNQQQTPMIQQQHQQLAGPQSNAT 260
Score = 88.2 bits (217), Expect(2) = 3e-69
Identities = 43/44 (97%), Positives = 44/44 (99%)
Frame = +2
Query: 420 KVSMQQQLQHNPSKLLPSQSQQSQTQASQQQLMSQIHNQPAQMQ 463
KVSMQQQLQHNPSK+LPSQSQQSQTQASQQQLMSQIHNQPAQMQ
Sbjct: 536 KVSMQQQLQHNPSKMLPSQSQQSQTQASQQQLMSQIHNQPAQMQ 667
Score = 38.5 bits (88), Expect = 0.017
Identities = 43/156 (27%), Positives = 65/156 (41%), Gaps = 8/156 (5%)
Frame = +1
Query: 377 LTNIHQQPGNNVQGLQQQQQFGTESGNQGIQTSHHSAQMLQQPK----VSMQQQLQHNPS 432
++N HQ +N+ L+ Q +Q + AQML Q V Q+L +
Sbjct: 193 ISNKHQ*FNSNISSLRGLSQ---------MQRNGQHAQMLGQQNNVGDVQKSQRLHPQQN 345
Query: 433 KLLPSQSQQSQTQA-SQQQLMSQIHNQPAQMQQQLGLQQQQNPLQRDMQQKLQA---SGS 488
L+ Q +Q Q Q + Q ++ IH QP Q GLQQQQ Q +Q S
Sbjct: 346 NLMNLQQRQQQQQLMNHQNNLTNIHQQPGNNVQ--GLQQQQQFGTESGNQGIQTSHHSAQ 519
Query: 489 LLQQQSVLDQQKQLYQSQRALPETSTTSVDSTTQTA 524
+LQQ + QS + + + T D++ A
Sbjct: 520 MLQQPKGFNAATIATQSIKNVAISVATVPDTSFSAA 627
Score = 37.0 bits (84), Expect = 0.050
Identities = 24/67 (35%), Positives = 38/67 (55%), Gaps = 3/67 (4%)
Frame = +3
Query: 159 QNISQNSNTQQPG--QNSVSNSIGQNSNVQSMFPGSQRQMSGRPQVVPQQQQQQSQNQQQ 216
Q++ S+ QP Q+S+S S+ QN ++ +Q ++ Q++ QQQQ NQQQ
Sbjct: 27 QSVIPTSSAMQPSMVQSSLS-SLPQNQQSNNVQQSTQSRLQQHSQIIRQQQQNSIVNQQQ 203
Query: 217 T-LYQQQ 222
T + QQQ
Sbjct: 204 TPMIQQQ 224
Score = 33.1 bits (74), Expect = 0.72
Identities = 22/73 (30%), Positives = 34/73 (46%), Gaps = 1/73 (1%)
Frame = +3
Query: 412 SAQMLQQPKVSMQQQLQHNPSKLLPSQSQQSQTQASQQQLMSQIHNQPAQMQQQLGL-QQ 470
S Q + +MQ + + LP Q + Q S Q + Q H+Q + QQQ + Q
Sbjct: 21 SQQSVIPTSSAMQPSMVQSSLSSLPQNQQSNNVQQSTQSRLQQ-HSQIIRQQQQNSIVNQ 197
Query: 471 QQNPLQRDMQQKL 483
QQ P+ + Q+L
Sbjct: 198 QQTPMIQQQHQQL 236
Score = 32.0 bits (71), Expect(2) = 0.003
Identities = 28/100 (28%), Positives = 37/100 (37%), Gaps = 12/100 (12%)
Frame = +1
Query: 663 SQMQPTNLQGSTSVQQNNIASL---------QNNSMSSLSTTQQNMLNTIQPS-NNLDSG 712
SQMQ QQNN+ + QNN M+ QQ L Q + N+
Sbjct: 247 SQMQRNGQHAQMLGQQNNVGDVQKSQRLHPQQNNLMNLQQRQQQQQLMNHQNNLTNIHQQ 426
Query: 713 QGNSVNSLQQVPVSSLQQNT--VNTQHTNINSLPSQGGVN 750
GN+V LQQ + + T H + L G N
Sbjct: 427 PGNNVQGLQQQQQFGTESGNQGIQTSHHSAQMLQQPKGFN 546
Score = 30.8 bits (68), Expect = 3.6
Identities = 25/71 (35%), Positives = 36/71 (50%), Gaps = 2/71 (2%)
Frame = +3
Query: 183 SNVQSMFPGSQRQMSGRPQ--VVPQQQQQQSQNQQQTLYQQQLLKHKLSQMQQQQQQQNL 240
S+ QS+ P S Q + Q QQS N QQ+ Q +L +H SQ+ +QQQQ ++
Sbjct: 18 SSQQSVIPTSSAMQPSMVQSSLSSLPQNQQSNNVQQST-QSRLQQH--SQIIRQQQQNSI 188
Query: 241 LQPNQLLSSQQ 251
+ Q QQ
Sbjct: 189 VNQQQTPMIQQ 221
Score = 28.1 bits (61), Expect(2) = 0.003
Identities = 19/43 (44%), Positives = 25/43 (57%), Gaps = 1/43 (2%)
Frame = +2
Query: 768 QQQLKHQQEQKMLQNQQFKQQYQQRQMMQRQQQQLH-QPAKQQ 809
QQQL+H KML +Q QQ Q + Q+ Q+H QPA+ Q
Sbjct: 548 QQQLQH-NPSKMLPSQ--SQQSQTQASQQQLMSQIHNQPAQMQ 667
>BF644371 weakly similar to GP|8927657|gb| EST gb|N38213 comes from this
gene. {Arabidopsis thaliana}, partial (4%)
Length = 673
Score = 230 bits (587), Expect(2) = 1e-65
Identities = 122/207 (58%), Positives = 156/207 (74%), Gaps = 2/207 (0%)
Frame = +3
Query: 544 ETYLPEINEIYQKISMKVHQFDSIPQQPKSDQIEKLKGYKTMFERMISILQIPKSSIQYG 603
E+YLPE++E+YQKI+ K+HQ DS+P QPKSDQ+EKLK +K M ER+I+ LQ+ KS+I
Sbjct: 3 ESYLPELSEMYQKIATKLHQHDSLPHQPKSDQLEKLKVFKMMLERLITFLQVSKSNISPS 182
Query: 604 VKEKLGSYEKQIAAAINQFRPRKAMSSLQPGQLPATHMALMPQSQSQVTSVQSHENQMNS 663
+KEKLGSYEKQI IN RPRK +SSLQPGQLP HM M Q+Q Q T VQSHENQMN+
Sbjct: 183 LKEKLGSYEKQIINFINTNRPRK-ISSLQPGQLPPPHMHSMSQTQPQATQVQSHENQMNT 359
Query: 664 QMQPTNLQGST-SVQQNNIASLQNNSMSSLSTTQQNMLNTIQPSNNLDSGQGNSVNSLQQ 722
Q+Q TN+QG ++QQNN+ S+Q++S+ +ST QQNM+NT+QPS +LD GQGN
Sbjct: 360 QLQTTNMQGPVPTMQQNNLTSMQHSSLXGVSTAQQNMMNTMQPSASLDLGQGN------- 518
Query: 723 VPVSSLQQNTVNT-QHTNINSLPSQGG 748
+SSLQQN+V Q TN++SL S G
Sbjct: 519 --MSSLQQNSVTAPQQTNVSSLSSXAG 593
Score = 39.7 bits (91), Expect(2) = 1e-65
Identities = 19/27 (70%), Positives = 20/27 (73%)
Frame = +1
Query: 748 GVNVIQPNLNTHQPGSNMLQQQQLKHQ 774
GVN+IQ NLN QPGS MLQ QLK Q
Sbjct: 592 GVNMIQQNLNPLQPGSGMLQHXQLKQQ 672
Score = 35.4 bits (80), Expect = 0.14
Identities = 29/110 (26%), Positives = 48/110 (43%), Gaps = 6/110 (5%)
Frame = +3
Query: 258 SAMQPSMVQSS-LSSLPQNQQSNNVQQSTQSRLQQHSQIIRQQQQNSIVNQQQTPMIQQQ 316
S++QP + + S+ Q Q QS ++ Q ++Q+ Q + QQ + Q
Sbjct: 255 SSLQPGQLPPPHMHSMSQTQPQATQVQSHEN--QMNTQLQTTNMQGPVPTMQQNNLTSMQ 428
Query: 317 HQQLAG---PQSNATNGQH--AQMLGQQNNVGDVQKSQRLHPQQNNLMNL 361
H L G Q N N A + Q N+ +Q++ PQQ N+ +L
Sbjct: 429 HSSLXGVSTAQQNMMNTMQPSASLDLGQGNMSSLQQNSVTAPQQTNVSSL 578
>BG585447
Length = 306
Score = 204 bits (520), Expect = 1e-52
Identities = 100/100 (100%), Positives = 100/100 (100%)
Frame = +2
Query: 846 QHLTSGQNSTYSHQQMKQGSPFQVSSPQLFQAASPQIPHNSSPQVDQQTHLLSLTKVGTP 905
QHLTSGQNSTYSHQQMKQGSPFQVSSPQLFQAASPQIPHNSSPQVDQQTHLLSLTKVGTP
Sbjct: 2 QHLTSGQNSTYSHQQMKQGSPFQVSSPQLFQAASPQIPHNSSPQVDQQTHLLSLTKVGTP 181
Query: 906 LQSSNSPFGVPTPSPPMAPSPMLVDSEKPIPGVSSSNAAN 945
LQSSNSPFGVPTPSPPMAPSPMLVDSEKPIPGVSSSNAAN
Sbjct: 182 LQSSNSPFGVPTPSPPMAPSPMLVDSEKPIPGVSSSNAAN 301
>TC79942 similar to GP|8927657|gb|AAF82148.1| EST gb|N38213 comes from this
gene. {Arabidopsis thaliana}, partial (9%)
Length = 879
Score = 169 bits (429), Expect = 5e-42
Identities = 84/112 (75%), Positives = 93/112 (83%)
Frame = +3
Query: 1 MDSNNWVPNQGTEATADTVDWRTQLQPDQRQRIVNKIMDTLRKHLPVSGSEGLLELRKIA 60
M +NNW PNQG E DT DWR QLQP+ RQRIVNKIMDTL++HLPVSG EGL EL KIA
Sbjct: 129 MVTNNWRPNQGAEPNMDTSDWRGQLQPESRQRIVNKIMDTLKRHLPVSGQEGLHELWKIA 308
Query: 61 QRFEDKIYTAATSQSDYLRKISMKMLTMENKSQNTMANNMLSNEGGPSNNLP 112
QRFE+KIYTAATSQSDYLRKIS+KMLTME KSQ T+ANN+ N+ GPSN P
Sbjct: 309 QRFEEKIYTAATSQSDYLRKISLKMLTMETKSQGTIANNIPPNQVGPSNQPP 464
Score = 108 bits (271), Expect(2) = 1e-29
Identities = 48/82 (58%), Positives = 66/82 (79%)
Frame = +3
Query: 516 SVDSTTQTAQPNGVDWQEEIYQKLQTMKETYLPEINEIYQKISMKVHQFDSIPQQPKSDQ 575
S+DS QT QPNG DWQEEIYQ ++ MKE+YLP+++E+ QKI+ K+ Q DS+PQQPKSD+
Sbjct: 474 SLDSVAQTGQPNGGDWQEEIYQNIKAMKESYLPKLSEMIQKIATKLQQHDSLPQQPKSDE 653
Query: 576 IEKLKGYKTMFERMISILQIPK 597
+E LK +K + ER+I+ LQ+ K
Sbjct: 654 LEMLKEFKMVLERLITFLQVSK 719
Score = 40.8 bits (94), Expect(2) = 1e-29
Identities = 23/38 (60%), Positives = 27/38 (70%)
Frame = +1
Query: 594 QIPKSSIQYGVKEKLGSYEKQIAAAINQFRPRKAMSSL 631
+ PKS+I +KEKLGS EKQI IN +RP K MSSL
Sbjct: 709 RFPKSNISPCLKEKLGSCEKQIINCINTYRPNK-MSSL 819
Score = 39.3 bits (90), Expect = 0.010
Identities = 16/39 (41%), Positives = 24/39 (61%)
Frame = +1
Query: 499 QKQLYQSQRALPETSTTSVDSTTQTAQPNGVDWQEEIYQ 537
+KQ+ +S+DST +T +P+G DWQEE+YQ
Sbjct: 763 EKQIINCINTYRPNKMSSLDSTAKTGKPSGGDWQEEVYQ 879
>AJ501729 similar to GP|8927657|gb| EST gb|N38213 comes from this gene.
{Arabidopsis thaliana}, partial (3%)
Length = 557
Score = 152 bits (385), Expect = 6e-37
Identities = 79/112 (70%), Positives = 87/112 (77%)
Frame = +1
Query: 1 MDSNNWVPNQGTEATADTVDWRTQLQPDQRQRIVNKIMDTLRKHLPVSGSEGLLELRKIA 60
M +NNW PNQG E DT DWR QLQP+ RQRIVNK+ HLPVSG EGL EL KIA
Sbjct: 127 MVTNNWRPNQGAEPNMDTSDWRGQLQPESRQRIVNKM------HLPVSGQEGLHELWKIA 288
Query: 61 QRFEDKIYTAATSQSDYLRKISMKMLTMENKSQNTMANNMLSNEGGPSNNLP 112
QRFE+KIYTAATSQSDYLRKIS+KMLTME KSQ T+ANN+ N+ GPSN P
Sbjct: 289 QRFEEKIYTAATSQSDYLRKISLKMLTMETKSQGTIANNIPPNQVGPSNQPP 444
Score = 54.3 bits (129), Expect = 3e-07
Identities = 22/34 (64%), Positives = 28/34 (81%)
Frame = +1
Query: 516 SVDSTTQTAQPNGVDWQEEIYQKLQTMKETYLPE 549
S+DS QT QPNG DWQEEIYQ ++ MK++YLP+
Sbjct: 454 SLDSVAQTGQPNGGDWQEEIYQNIKAMKQSYLPK 555
>TC79943 homologue to GP|8927657|gb|AAF82148.1| EST gb|N38213 comes from
this gene. {Arabidopsis thaliana}, partial (1%)
Length = 494
Score = 144 bits (362), Expect = 3e-34
Identities = 75/112 (66%), Positives = 83/112 (73%)
Frame = +1
Query: 1 MDSNNWVPNQGTEATADTVDWRTQLQPDQRQRIVNKIMDTLRKHLPVSGSEGLLELRKIA 60
M +NNW PNQG E DT DWR QLQP+ RQRIVNKIMDTL++HLPVSG EGL EL KIA
Sbjct: 79 MVTNNWRPNQGAEPNMDTSDWRGQLQPESRQRIVNKIMDTLKRHLPVSGQEGLHELWKIA 258
Query: 61 QRFEDKIYTAATSQSDYLRKISMKMLTMENKSQNTMANNMLSNEGGPSNNLP 112
QRFE+KIYTAATSQ MLTME KSQ T+ANN+ N+ GPSN P
Sbjct: 259 QRFEEKIYTAATSQ----------MLTMETKSQGTIANNIPPNQVGPSNQPP 384
Score = 25.4 bits (54), Expect(2) = 3.3
Identities = 10/13 (76%), Positives = 11/13 (83%)
Frame = +1
Query: 516 SVDSTTQTAQPNG 528
S+DS TQT QPNG
Sbjct: 394 SLDSVTQTGQPNG 432
Score = 23.9 bits (50), Expect(2) = 3.3
Identities = 9/18 (50%), Positives = 11/18 (61%)
Frame = +2
Query: 530 DWQEEIYQKLQTMKETYL 547
DWQEEIYQ ++ L
Sbjct: 437 DWQEEIYQNXXAIERELL 490
>CB892607 similar to GP|8927657|gb| EST gb|N38213 comes from this gene.
{Arabidopsis thaliana}, partial (1%)
Length = 782
Score = 85.1 bits (209), Expect(2) = 5e-26
Identities = 43/66 (65%), Positives = 49/66 (74%)
Frame = +3
Query: 9 NQGTEATADTVDWRTQLQPDQRQRIVNKIMDTLRKHLPVSGSEGLLELRKIAQRFEDKIY 68
NQG E D DWR Q + RQRIVNKI++TL+ HL VSG EGL EL KIA RFEDKI+
Sbjct: 87 NQGAELNMDPSDWRGQFPAESRQRIVNKILETLKSHLLVSGEEGLHELWKIA*RFEDKIF 266
Query: 69 TAATSQ 74
T+ATSQ
Sbjct: 267 TSATSQ 284
Score = 52.4 bits (124), Expect(2) = 5e-26
Identities = 27/42 (64%), Positives = 33/42 (78%), Gaps = 2/42 (4%)
Frame = +1
Query: 74 QSDYLRKISMKMLTMENKS--QNTMANNMLSNEGGPSNNLPD 113
QSDYL+KIS KMLTME +S QN +ANN+ SN+ GPSN P+
Sbjct: 385 QSDYLQKISSKMLTMETRSQVQNPVANNLPSNQVGPSNEPPN 510
>BE204335 weakly similar to GP|8927657|gb| EST gb|N38213 comes from this gene.
{Arabidopsis thaliana}, partial (6%)
Length = 537
Score = 105 bits (262), Expect = 1e-22
Identities = 57/97 (58%), Positives = 68/97 (69%)
Frame = -1
Query: 1024 SMSDRISGSAPGNGSRAAVGEDLVSMTNCRLQARSFITQDGGTTNGIRKFKRHIRGKTLD 1083
SM DRI+ SAPGNGS +VGEDLV+MTNC LQ R+F+ QDG NG K KR I D
Sbjct: 537 SMIDRIAASAPGNGSIVSVGEDLVAMTNCHLQDRNFLPQDG--VNGSSKMKRCINATPFD 364
Query: 1084 VGSSAGSMNDNLKQLSASEASQQESTATSNVKKPKAE 1120
V SSAG +ND++KQL+A EA E ATS +K+PK E
Sbjct: 363 VVSSAGCVNDSIKQLNAIEACDLEPIATSIIKRPKIE 253
>TC80581 similar to GP|19683021|gb|AAL92644.1 hypothetical protein
{Dictyostelium discoideum}, partial (5%)
Length = 850
Score = 94.4 bits (233), Expect = 3e-19
Identities = 98/276 (35%), Positives = 125/276 (44%)
Frame = +1
Query: 218 LYQQQLLKHKLSQMQQQQQQQNLLQPNQLLSSQQSVIPTSSAMQPSMVQSSLSSLPQNQQ 277
LY QQL +LSQ QQ + P S Q ++I Q V S S LP Q
Sbjct: 31 LYAQQL---QLSQQAPFNNQQLVGLPQ---SGQPAMIQNQLLKQIPAVSGSASLLPLQQP 192
Query: 278 SNNVQQSTQSRLQQHSQIIRQQQQNSIVNQQQTPMIQQQHQQLAGPQSNATNGQHAQMLG 337
+ Q ++ ++LQQ S + QQQ ++ QQ PM Q Q QQ QH Q+L
Sbjct: 193 QSQQQLASSAQLQQSSLTLNQQQLPQLM-QQPKPMGQPQLQQQL---------QHQQLLQ 342
Query: 338 QQNNVGDVQKSQRLHPQQNNLMNLQQRQQQQQLMNHQNNLTNIHQQPGNNVQGLQQQQQF 397
QQ +Q Q+ Q L + Q QQ +N Q L + QQP + Q L QQQQ
Sbjct: 343 QQ-----LQLQQQYQRLQQQLASSAQLQQNSLTLN-QQQLPLLMQQPKSMGQPLLQQQQQ 504
Query: 398 GTESGNQGIQTSHHSAQMLQQPKVSMQQQLQHNPSKLLPSQSQQSQTQASQQQLMSQIHN 457
+Q + Q LQQ +++ QLQ N S L Q QQ+ M Q
Sbjct: 505 QLLQQQLQLQQQY---QRLQQ-QLASSAQLQQNSSTL-----NQLPQLMQQQKSMGQPLL 657
Query: 458 QPAQMQQQLGLQQQQNPLQRDMQQKLQASGSLLQQQ 493
Q QQQL LQQQQ LQ+ QQ+ Q LLQQQ
Sbjct: 658 QQQLQQQQLLLQQQQLVLQQQQQQQQQQRTQLLQQQ 765
Score = 71.2 bits (173), Expect = 2e-12
Identities = 84/264 (31%), Positives = 114/264 (42%), Gaps = 14/264 (5%)
Frame = +1
Query: 153 LPQIPSQNISQNSNTQQPGQNSVSNSIGQNSNVQSMFPGSQRQMSGRPQVVPQQQQQQSQ 212
LPQ + QN +Q + G S + P SQ+Q++ Q+ QQ +
Sbjct: 94 LPQSGQPAMIQNQLLKQ-----IPAVSGSASLLPLQQPQSQQQLASSAQL---QQSSLTL 249
Query: 213 NQQQ-TLYQQQLLKHKLSQMQQQQQQQNLLQPNQLLSSQQSVIPTSSAMQPSMVQSSLSS 271
NQQQ QQ Q+QQQ Q Q LLQ L Q + A + Q+SL +
Sbjct: 250 NQQQLPQLMQQPKPMGQPQLQQQLQHQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSL-T 426
Query: 272 LPQNQQSNNVQQST---QSRLQQHSQIIRQQQQNSIVNQQQTPMIQQQHQQLAGPQSN-A 327
L Q Q +QQ Q LQQ Q + QQQ + QQQ +QQQ A Q N +
Sbjct: 427 LNQQQLPLLMQQPKSMGQPLLQQQQQQLLQQQ---LQLQQQYQRLQQQLASSAQLQQNSS 597
Query: 328 TNGQHAQMLGQQNNVG------DVQKSQRLHPQQNNLMNLQQRQQQQ---QLMNHQNNLT 378
T Q Q++ QQ ++G +Q+ Q L QQ ++ QQ+QQQQ QL+ Q
Sbjct: 598 TLNQLPQLMQQQKSMGQPLLQQQLQQQQLLLQQQQLVLQQQQQQQQQQRTQLLQQQMQAL 777
Query: 379 NIHQQPGNNVQGLQQQQQFGTESG 402
I G+ L Q T G
Sbjct: 778 RIPGPAGHKSLSLTGSQPEATAFG 849
Score = 67.0 bits (162), Expect = 4e-11
Identities = 97/311 (31%), Positives = 133/311 (42%), Gaps = 16/311 (5%)
Frame = +1
Query: 302 NSIVNQQQTPMIQQ---QHQQLAG-PQSNATNGQHAQMLGQQNNVGDVQKSQRLHPQQNN 357
N + QQ + QQ +QQL G PQS Q+L Q + V S L P
Sbjct: 22 NGALYAQQLQLSQQAPFNNQQLVGLPQSGQPAMIQNQLLKQ---IPAVSGSASLLP---- 180
Query: 358 LMNLQQRQQQQQLMN----HQNNLTNIHQQPGNNVQGLQQQQQFGTESGNQGIQTSHHSA 413
LQQ Q QQQL + Q++LT L QQQ
Sbjct: 181 ---LQQPQSQQQLASSAQLQQSSLT------------LNQQQL----------------P 267
Query: 414 QMLQQPKV----SMQQQLQHNPSKLLPSQSQ-QSQTQASQQQLMSQIHNQPAQMQQQ-LG 467
Q++QQPK +QQQLQH +LL Q Q Q Q Q QQQL S AQ+QQ L
Sbjct: 268 QLMQQPKPMGQPQLQQQLQHQ--QLLQQQLQLQQQYQRLQQQLASS-----AQLQQNSLT 426
Query: 468 LQQQQNPLQRDMQQKLQASGSLLQQQS--VLDQQKQLYQSQRALPETSTTSVDSTTQTAQ 525
L QQQ PL MQQ LLQQQ +L QQ QL Q + L + +S ++
Sbjct: 427 LNQQQLPLL--MQQPKSMGQPLLQQQQQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSST 600
Query: 526 PNGVDWQEEIYQKLQTMKETYLPEINEIYQKISMKVHQFDSIPQQPKSDQIEKLKGYKTM 585
N ++ Q +Q K P + + Q+ + + Q + QQ + Q ++ +
Sbjct: 601 LN------QLPQLMQQQKSMGQPLLQQQLQQQQLLLQQQQLVLQQQQQQQQQQ---RTQL 753
Query: 586 FERMISILQIP 596
++ + L+IP
Sbjct: 754 LQQQMQALRIP 786
Score = 59.3 bits (142), Expect = 9e-09
Identities = 54/184 (29%), Positives = 78/184 (42%), Gaps = 8/184 (4%)
Frame = +1
Query: 630 SLQPGQLPATHMALMPQSQSQVTSVQSHENQMNSQMQPTNLQGSTSVQQNNIASLQNNSM 689
+L QLP P Q Q+ H+ + Q+Q LQ Q +AS
Sbjct: 244 TLNQQQLPQLMQQPKPMGQPQLQQQLQHQQLLQQQLQ---LQQQYQRLQQQLASSAQLQQ 414
Query: 690 SSLSTTQQNMLNTIQPSNNLDSG--QGNSVNSLQQV-----PVSSLQQNTVNTQHTNINS 742
+SL+ QQ + +Q ++ Q LQQ LQQ ++ NS
Sbjct: 415 NSLTLNQQQLPLLMQQPKSMGQPLLQQQQQQLLQQQLQLQQQYQRLQQQLASSAQLQQNS 594
Query: 743 LPSQGGVNVIQPNLNTHQPG-SNMLQQQQLKHQQEQKMLQNQQFKQQYQQRQMMQRQQQQ 801
++Q + QP LQQQQL QQ+Q +LQ QQ +QQ Q+ Q++Q+Q Q
Sbjct: 595 STLNQLPQLMQQQKSMGQPLLQQQLQQQQLLLQQQQLVLQQQQQQQQQQRTQLLQQQMQA 774
Query: 802 LHQP 805
L P
Sbjct: 775 LRIP 786
Score = 44.3 bits (103), Expect = 3e-04
Identities = 64/246 (26%), Positives = 88/246 (35%), Gaps = 11/246 (4%)
Frame = +1
Query: 663 SQMQPTNLQGSTSVQQNNI-ASLQNNSMSSLSTTQ--QNMLNTIQPSNNLDSGQGNSVNS 719
SQ P N Q + Q+ A +QN + + ++L QP + + S
Sbjct: 55 SQQAPFNNQQLVGLPQSGQPAMIQNQLLKQIPAVSGSASLLPLQQPQSQ------QQLAS 216
Query: 720 LQQVPVSSLQQNTVNTQHTNINSLPSQGGVNVIQPNLNTHQPGSNMLQQQQLKHQQEQKM 779
Q+ SSL N Q + P G QP L +LQQQ QQ Q++
Sbjct: 217 SAQLQQSSLTLN--QQQLPQLMQQPKPMG----QPQLQQQLQHQQLLQQQLQLQQQYQRL 378
Query: 780 LQNQQFKQQYQQRQMMQRQQQ---QLHQPAKQ-QMSAQPQTHQLPQINQMNDMNDVKIRQ 835
Q Q QQ + QQQ + QP Q Q Q QL Q +++Q
Sbjct: 379 QQQLASSAQLQQNSLTLNQQQLPLLMQQPKSMGQPLLQQQQQQLLQQQLQLQQQYQRLQQ 558
Query: 836 GLGVKSGVFQQHLTSGQNSTYSHQQMKQGSPF---QVSSPQ-LFQAASPQIPHNSSPQVD 891
L + + Q T Q QQ G P Q+ Q L Q + Q
Sbjct: 559 QLASSAQLQQNSSTLNQLPQLMQQQKSMGQPLLQQQLQQQQLLLQQQQLVLQQQQQQQQQ 738
Query: 892 QQTHLL 897
Q+T LL
Sbjct: 739 QRTQLL 756
>AW586908 weakly similar to GP|2104814|emb| pleiotropic effects on cellular
differentiation and slug behaviour {Dictyostelium
discoideum}, partial (8%)
Length = 668
Score = 75.5 bits (184), Expect = 1e-13
Identities = 85/248 (34%), Positives = 102/248 (40%), Gaps = 2/248 (0%)
Frame = +2
Query: 211 SQNQQQTLYQQQLLKHKLSQMQQQQQQQNLLQPNQLLSSQQSVIPTSSAMQPSMVQSSLS 270
SQ QQQ Q LL L Q+QQQQQ+Q QL SQ +P QP + Q S
Sbjct: 23 SQQQQQQQKMQSLLPMPLDQLQQQQQRQ-----QQLAGSQN--LPQPQQQQPQLTQQS-P 178
Query: 271 SLPQNQQSNNVQQSTQSRLQQHSQIIRQQQQNSIVNQQQTPMIQQQHQQLAGPQSNATNG 330
PQ QQ + QQ Q+ + I Q N V QQ Q Q Q L+G + N
Sbjct: 179 QQPQQQQQH--QQPCQNTTMNNGTIGSNQIPNQCV-QQPVTYSQLQQQLLSGSMQSQQNL 349
Query: 331 QHAQMLG--QQNNVGDVQKSQRLHPQQNNLMNLQQRQQQQQLMNHQNNLTNIHQQPGNNV 388
Q A G + D Q Q++ QQ L LQ++QQQ QL
Sbjct: 350 QSAGKNGLMMTSLPQDSQFQQQIDQQQAGL--LQRQQQQNQLQQ---------------- 475
Query: 389 QGLQQQQQFGTESGNQGIQTSHHSAQMLQQPKVSMQQQLQHNPSKLLPSQSQQSQTQASQ 448
LQ QQ + G Q Q S Q + Q QQQLQ LL QQ Q Q Q
Sbjct: 476 SSLQLLQQSMLQRGPQQPQMS----QTVPQNISDQQQQLQ-----LLQKLQQQQQQQQQQ 628
Query: 449 QQLMSQIH 456
Q L + H
Sbjct: 629 QPLSTSSH 652
Score = 67.8 bits (164), Expect = 3e-11
Identities = 72/229 (31%), Positives = 93/229 (40%), Gaps = 14/229 (6%)
Frame = +2
Query: 282 QQSTQSRLQQHSQIIRQQQQNSIVNQ--QQTPMIQQQHQQLAGPQSNATNGQHAQMLGQQ 339
QQ S QQ Q QQ+ S++ Q QQ+ QQLAG Q+ Q L QQ
Sbjct: 2 QQPPASWSQQQQQ---QQKMQSLLPMPLDQLQQQQQRQQQLAGSQNLPQPQQQQPQLTQQ 172
Query: 340 NNVGDVQKSQRLHPQQNNLMNLQQRQQQQQLMNHQNNLTNIHQQPGNNVQG------LQQ 393
+ Q+ Q P QN MN N +Q P VQ LQQ
Sbjct: 173 SPQQPQQQQQHQQPCQNTTMN--------------NGTIGSNQIPNQCVQQPVTYSQLQQ 310
Query: 394 QQQFGTESGNQGIQTSHHSAQMLQQ-PKVSM-QQQLQHNPSKLLPSQSQQSQTQASQQQL 451
Q G+ Q +Q++ + M+ P+ S QQQ+ + LL Q QQ+Q Q S QL
Sbjct: 311 QLLSGSMQSQQNLQSAGKNGLMMTSLPQDSQFQQQIDQQQAGLLQRQQQQNQLQQSSLQL 490
Query: 452 MSQIHNQPAQMQQQLGLQQQQNPLQRDMQ----QKLQASGSLLQQQSVL 496
+ Q Q Q Q+ QN + Q QKLQ QQQ L
Sbjct: 491 LQQSMLQRGPQQPQMSQTVPQNISDQQQQLQLLQKLQQQQQQQQQQQPL 637
Score = 67.8 bits (164), Expect = 3e-11
Identities = 76/234 (32%), Positives = 100/234 (42%), Gaps = 19/234 (8%)
Frame = +2
Query: 155 QIPSQNISQNSNTQQPGQNSVSNSIGQNSNVQSMFPGSQRQMSGRPQVVPQQQQQQSQNQ 214
Q P + SQ QQ Q+ + + Q Q Q+Q++G Q +PQ QQQQ Q
Sbjct: 2 QQPPASWSQQQQQQQKMQSLLPMPLDQLQQQQQR----QQQLAGS-QNLPQPQQQQPQLT 166
Query: 215 QQTLYQQQLLKHKLSQMQQQQQQQNLLQPNQLLSSQQSVIPTSSAMQP----SMVQSSLS 270
QQ+ Q Q Q Q QQ QN N + S Q IP QP + Q LS
Sbjct: 167 QQSPQQPQ------QQQQHQQPCQNTTMNNGTIGSNQ--IPNQCVQQPVTYSQLQQQLLS 322
Query: 271 SLPQNQQS---------------NNVQQSTQSRLQQHSQIIRQQQQNSIVNQQQTPMIQQ 315
Q+QQ+ + Q Q QQ + RQQQQN + QQ + + Q
Sbjct: 323 GSMQSQQNLQSAGKNGLMMTSLPQDSQFQQQIDQQQAGLLQRQQQQNQL--QQSSLQLLQ 496
Query: 316 QHQQLAGPQSNATNGQHAQMLGQQNNVGDVQKSQRLHPQQNNLMNLQQRQQQQQ 369
Q GPQ Q +Q + Q N+ D Q+ +L L LQQ+QQQQQ
Sbjct: 497 QSMLQRGPQ----QPQMSQTVPQ--NISDQQQQLQL------LQKLQQQQQQQQ 622
Score = 66.2 bits (160), Expect = 8e-11
Identities = 63/175 (36%), Positives = 81/175 (46%), Gaps = 9/175 (5%)
Frame = +2
Query: 346 QKSQRLHPQQNNLMNLQQRQQQQQLMNHQNNLTNIHQQPGNNVQGLQQQQQF-------- 397
QK Q L P + + QQ+Q+QQQL QN QQP Q QQ QQ
Sbjct: 44 QKMQSLLPMPLDQLQ-QQQQRQQQLAGSQNLPQPQQQQPQLTQQSPQQPQQQQQHQQPCQ 220
Query: 398 GTESGNQGIQTSHHSAQMLQQPKVSMQQQLQHNPSKLLPSQSQQSQTQASQQQLMSQIHN 457
T N I ++ Q +QQP Q Q Q + QSQQ+ A + LM
Sbjct: 221 NTTMNNGTIGSNQIPNQCVQQPVTYSQLQQQLLSGSM---QSQQNLQSAGKNGLMMTSLP 391
Query: 458 QPAQMQQQLGLQQQQNPLQRDMQQ-KLQASGSLLQQQSVLDQQKQLYQSQRALPE 511
Q +Q QQQ+ QQQ LQR QQ +LQ S L QQS+L + Q Q + +P+
Sbjct: 392 QDSQFQQQID-QQQAGLLQRQQQQNQLQQSSLQLLQQSMLQRGPQQPQMSQTVPQ 553
Score = 60.8 bits (146), Expect = 3e-09
Identities = 68/220 (30%), Positives = 88/220 (39%), Gaps = 6/220 (2%)
Frame = +2
Query: 108 SNNLPDQGQQHPN------PLPNQHQPRQQLLSHNIQNNVAPQPNLSSVSTLPQIPSQNI 161
S NLP QQ P P Q Q QQ + NN N QIP+Q +
Sbjct: 122 SQNLPQPQQQQPQLTQQSPQQPQQQQQHQQPCQNTTMNNGTIGSN--------QIPNQCV 277
Query: 162 SQNSNTQQPGQNSVSNSIGQNSNVQSMFPGSQRQMSGRPQVVPQQQQQQSQNQQQTLYQQ 221
Q Q Q +S S+ N+QS +G+ ++ Q SQ QQQ QQ
Sbjct: 278 QQPVTYSQLQQQLLSGSMQSQQNLQS---------AGKNGLMMTSLPQDSQFQQQIDQQQ 430
Query: 222 QLLKHKLSQMQQQQQQQNLLQPNQLLSSQQSVIPTSSAMQPSMVQSSLSSLPQNQQSNNV 281
+ + Q+QQQQN LQ + L QQS++ QP M Q+ +PQN
Sbjct: 431 -------AGLLQRQQQQNQLQQSSLQLLQQSMLQRGP-QQPQMSQT----VPQNISDQQQ 574
Query: 282 QQSTQSRLQQHSQIIRQQQQNSIVNQQQTPMIQQQHQQLA 321
Q +LQQ QQQQ QQQ Q L+
Sbjct: 575 QLQLLQKLQQ------QQQQ------------QQQQQPLS 640
Score = 55.8 bits (133), Expect = 1e-07
Identities = 47/156 (30%), Positives = 63/156 (40%), Gaps = 9/156 (5%)
Frame = +2
Query: 112 PDQGQQHPNPLPNQHQPRQQLLSHNIQNNVAPQPNLSSVSTLPQIPSQNISQNSNTQQPG 171
P Q QQH P N + S+ I N QP S Q+ S ++ N Q G
Sbjct: 185 PQQQQQHQQPCQNTTMNNGTIGSNQIPNQCVQQPVTYS-QLQQQLLSGSMQSQQNLQSAG 361
Query: 172 QNSVS-NSIGQNSNVQSMFPGSQRQMSGRPQVVPQQQQQQSQNQQQTLYQQ--------Q 222
+N + S+ Q+S Q Q + R Q Q QQ Q QQ++ Q+ Q
Sbjct: 362 KNGLMMTSLPQDSQFQQQIDQQQAGLLQRQQQQNQLQQSSLQLLQQSMLQRGPQQPQMSQ 541
Query: 223 LLKHKLSQMQQQQQQQNLLQPNQLLSSQQSVIPTSS 258
+ +S QQQ Q LQ Q QQ + TSS
Sbjct: 542 TVPQNISDQQQQLQLLQKLQQQQQQQQQQQPLSTSS 649
Score = 54.3 bits (129), Expect = 3e-07
Identities = 55/174 (31%), Positives = 74/174 (41%), Gaps = 7/174 (4%)
Frame = +2
Query: 74 QSDYLRKISMKMLTMENKSQNTMANNMLSNEGGPSNNLPDQGQQHPNPLPNQHQPRQQLL 133
Q L + S + + + Q N ++N SN +P+Q Q P Q +QQLL
Sbjct: 149 QQPQLTQQSPQQPQQQQQHQQPCQNTTMNNGTIGSNQIPNQCVQQP---VTYSQLQQQLL 319
Query: 134 SHNIQNNVAPQPNLSS-------VSTLPQIPSQNISQNSNTQQPGQNSVSNSIGQNSNVQ 186
S ++Q+ Q NL S +++LPQ Q + QQ G + QN Q
Sbjct: 320 SGSMQS----QQNLQSAGKNGLMMTSLPQ--DSQFQQQIDQQQAGL--LQRQQQQNQLQQ 475
Query: 187 SMFPGSQRQMSGRPQVVPQQQQQQSQNQQQTLYQQQLLKHKLSQMQQQQQQQNL 240
S Q+ M R PQ Q QN Q QLL+ Q QQQQQQQ L
Sbjct: 476 SSLQLLQQSMLQRGPQQPQMSQTVPQNISDQQQQLQLLQKLQQQQQQQQQQQPL 637
Score = 45.8 bits (107), Expect = 1e-04
Identities = 58/210 (27%), Positives = 78/210 (36%), Gaps = 21/210 (10%)
Frame = +2
Query: 621 QFRPRKAMSSLQPGQLPATHMALMPQSQSQVTSVQSHENQMNSQMQPTNLQGSTSVQQNN 680
Q + ++ M SL P +P + Q Q Q+ Q+ Q Q T QQ
Sbjct: 29 QQQQQQKMQSLLP--MPLDQLQQQQQRQQQLAGSQNLPQPQQQQPQLTQQSPQQPQQQ-- 196
Query: 681 IASLQNNSMSSLSTTQQNMLNTIQPSNNLDSGQGNSVNSLQQVPVSSLQQNTVNTQHTNI 740
Q + +TT N N Q + + LQQ +S Q+ N Q
Sbjct: 197 ----QQHQQPCQNTTMNNGTIGSNQIPNQCVQQPVTYSQLQQQLLSGSMQSQQNLQSAGK 364
Query: 741 N-----SLPS----QGGVNVIQPNLNTHQPGSNMLQQQQLKHQQEQKMLQN--------- 782
N SLP Q ++ Q L Q N LQQ L+ Q Q MLQ
Sbjct: 365 NGLMMTSLPQDSQFQQQIDQQQAGLLQRQQQQNQLQQSSLQLLQ-QSMLQRGPQQPQMSQ 541
Query: 783 ---QQFKQQYQQRQMMQRQQQQLHQPAKQQ 809
Q Q QQ Q++Q+ QQQ Q +QQ
Sbjct: 542 TVPQNISDQQQQLQLLQKLQQQQQQQQQQQ 631
Score = 44.3 bits (103), Expect = 3e-04
Identities = 48/181 (26%), Positives = 71/181 (38%), Gaps = 22/181 (12%)
Frame = +2
Query: 417 QQPKVS--MQQQLQHNPSKLLPSQSQQSQTQASQQQLMSQIHNQPAQMQQQLGLQQQ--Q 472
QQP S QQQ Q LLP Q Q Q +QQ ++ N P QQQ L QQ Q
Sbjct: 2 QQPPASWSQQQQQQQKMQSLLPMPLDQLQQQQQRQQQLAGSQNLPQPQQQQPQLTQQSPQ 181
Query: 473 NPLQRDMQQK--------------LQASGSLLQQQSVLD--QQKQLYQSQRALPETSTTS 516
P Q+ Q+ Q +QQ QQ+ L S ++ +
Sbjct: 182 QPQQQQQHQQPCQNTTMNNGTIGSNQIPNQCVQQPVTYSQLQQQLLSGSMQSQQNLQSAG 361
Query: 517 VDSTTQTAQPNGVDWQEEIYQKLQTMKETYLPEINEIYQKISMKVHQ--FDSIPQQPKSD 574
+ T+ P +Q++I Q+ + + + N++ Q + Q PQQP+
Sbjct: 362 KNGLMMTSLPQDSQFQQQIDQQQAGLLQRQQQQ-NQLQQSSLQLLQQSMLQRGPQQPQMS 538
Query: 575 Q 575
Q
Sbjct: 539 Q 541
Score = 42.0 bits (97), Expect = 0.002
Identities = 47/162 (29%), Positives = 65/162 (40%), Gaps = 11/162 (6%)
Frame = +2
Query: 760 QPGSNMLQQQQLKHQQEQKMLQNQQFKQQYQQRQMMQRQQQQLHQPAKQQMSAQPQTHQL 819
QP ++ QQQQ + + + + QQ QQRQ Q L QP +QQ Q+ Q
Sbjct: 5 QPPASWSQQQQQQQKMQSLLPMPLDQLQQQQQRQQQLAGSQNLPQPQQQQPQLTQQSPQQ 184
Query: 820 PQINQMNDMNDVKIRQGLGVKSGVFQQHLTSGQNSTYS--HQQMKQGSPFQVSSPQLFQA 877
PQ Q + G + Q TYS QQ+ GS + S Q Q+
Sbjct: 185 PQQQQQHQQPCQNTTMNNGTIGSNQIPNQCVQQPVTYSQLQQQLLSGS---MQSQQNLQS 355
Query: 878 AS------PQIPHNS--SPQVD-QQTHLLSLTKVGTPLQSSN 910
A +P +S Q+D QQ LL + LQ S+
Sbjct: 356 AGKNGLMMTSLPQDSQFQQQIDQQQAGLLQRQQQQNQLQQSS 481
>TC85969 similar to GP|21553908|gb|AAM62991.1 unknown {Arabidopsis thaliana},
partial (31%)
Length = 2756
Score = 58.9 bits (141), Expect = 1e-08
Identities = 56/183 (30%), Positives = 79/183 (42%), Gaps = 3/183 (1%)
Frame = +2
Query: 645 PQSQSQVTSVQSH-ENQMNSQMQPTNLQGSTSVQQNNIASLQNNSMSSLSTTQQNMLNTI 703
P Q QV VQ +NQ +Q Q + Q T Q + S L +
Sbjct: 1277 PTPQPQVVQVQPQAQNQAQTQSQAQS-QAQTQSQAPQFQQKSSGGGSDLPSPD------- 1432
Query: 704 QPSNNLDSGQGNSVNSLQQVPVSSLQQNTVNTQHTNINSLPSQGGVNVIQPNLNTHQPGS 763
S + DS N+++ +Q PV +Q + T ++ N + P LN P S
Sbjct: 1433 --SVSSDSSLSNAMS--RQKPVVYQEQIQIQPGTTRVS--------NPVDPKLNLSDPHS 1576
Query: 764 NMLQQQQLKHQQEQKMLQNQQFKQQYQQRQ--MMQRQQQQLHQPAKQQMSAQPQTHQLPQ 821
+ QQ H Q+ L QQF+ Q QQ+Q + Q+QQQ HQP + Q QPQ Q Q
Sbjct: 1577 RIQVQQ---HVQDPGYLLQQQFELQQQQQQFELQQQQQQPQHQPQQSQPQHQPQQQQQLQ 1747
Query: 822 INQ 824
+Q
Sbjct: 1748 HHQ 1756
Score = 48.9 bits (115), Expect = 1e-05
Identities = 77/320 (24%), Positives = 111/320 (34%), Gaps = 57/320 (17%)
Frame = +2
Query: 98 NNMLSNEGGPSNNLPDQGQQHPNPLP----------NQHQPRQQLLSHNIQNNVAPQPNL 147
N + S++ + +P ++ P P P NQ Q + Q S + APQ
Sbjct: 1214 NRVFSDDERSDHGVPVGYRKPPTPQPQVVQVQPQAQNQAQTQSQAQSQAQTQSQAPQFQQ 1393
Query: 148 SSVSTLPQIPS-QNISQNSNTQ-----------------QPGQNSVSNSIGQNSNVQSMF 189
S +PS ++S +S+ QPG VSN + N+
Sbjct: 1394 KSSGGGSDLPSPDSVSSDSSLSNAMSRQKPVVYQEQIQIQPGTTRVSNPVDPKLNLSDPH 1573
Query: 190 PGSQRQMSGRPQVVPQQQQQQSQNQQQTLYQQQLLKHKLSQMQQQQQQQNLLQPNQLLSS 249
Q Q + QQQ + Q QQQ QQ + Q QQ Q Q Q QL
Sbjct: 1574 SRIQVQQHVQDPGYLLQQQFELQQQQQQFELQQQQQQPQHQPQQSQPQHQPQQQQQLQHH 1753
Query: 250 QQS-------------VIPTSSAMQPSMVQSS------LSSLPQNQQSNNVQQST----- 285
QQ IPT + PS Q + PQ + +VQQ+
Sbjct: 1754 QQQQFIHGGHYIHHNPAIPTYYPVYPSQQQPHHQVYYVPARQPQQGYNISVQQANMGESA 1933
Query: 286 ----QSRLQQHSQIIRQQQQNSIVNQ-QQTPMIQQQHQQLAGPQSNATNGQHAQMLGQQN 340
SR Q QQN+ N + P+ + + A + + Q Q+ Q+
Sbjct: 1934 TTIPSSRPQNPPNPTTLVQQNAAYNPIRSAPLPKTEMTAAAYRAATGGSPQFVQVPTSQH 2113
Query: 341 NVGDVQKSQRLHPQQNNLMN 360
V SQ HP Q+ N
Sbjct: 2114 QQQYVTYSQIHHPSQSMAPN 2173
Score = 47.0 bits (110), Expect = 5e-05
Identities = 54/206 (26%), Positives = 71/206 (34%), Gaps = 5/206 (2%)
Frame = +2
Query: 307 QQQTPMIQQQHQQLAGPQSNA-----TNGQHAQMLGQQNNVGDVQKSQRLHPQQNNLMNL 361
Q Q +Q Q Q A QS A T Q Q + + G S ++L N
Sbjct: 1286 QPQVVQVQPQAQNQAQTQSQAQSQAQTQSQAPQFQQKSSGGGSDLPSPDSVSSDSSLSNA 1465
Query: 362 QQRQQQQQLMNHQNNLTNIHQQPGNNVQGLQQQQQFGTESGNQGIQTSHHSAQMLQQPKV 421
RQ+ I QPG + + IQ H +Q P
Sbjct: 1466 MSRQKPVVYQEQ------IQIQPGTTRVSNPVDPKLNLSDPHSRIQVQQH----VQDPGY 1615
Query: 422 SMQQQLQHNPSKLLPSQSQQSQTQASQQQLMSQIHNQPAQMQQQLGLQQQQNPLQRDMQQ 481
+QQQ + QQ Q Q QQ Q +QP Q Q Q QQQQ LQ QQ
Sbjct: 1616 LLQQQFE----------LQQQQQQFELQQQQQQPQHQPQQSQPQHQPQQQQQ-LQHHQQQ 1762
Query: 482 KLQASGSLLQQQSVLDQQKQLYQSQR 507
+ G + + +Y SQ+
Sbjct: 1763 QFIHGGHYIHHNPAIPTYYPVYPSQQ 1840
>TC87088 similar to PIR|F86399|F86399 protein F17L21.22 [imported] -
Arabidopsis thaliana, partial (4%)
Length = 2997
Score = 57.0 bits (136), Expect = 5e-08
Identities = 78/278 (28%), Positives = 105/278 (37%), Gaps = 29/278 (10%)
Frame = +3
Query: 264 MVQSSLSSLPQNQQSNNVQQSTQSRLQQHSQIIRQQQQNSIVNQQQTPMIQQQHQQLAGP 323
M+Q Q QQ+ +QQ RLQQ QQQQ + QQQ +QQQ ++
Sbjct: 345 MLQKLQKERLQQQQAERLQQQQAERLQQQQAERLQQQQAERLQQQQAERLQQQ-TNISSH 521
Query: 324 QSNATNGQHAQML-----GQQNNVGDVQKSQRLHPQQNNLMNLQQRQQ----QQQLMNHQ 374
NG Q N G Q Q L LQ +Q+ QQQ M+HQ
Sbjct: 522 FPAYLNGSDLDRFPGFSPSQSNKSGIQQMMQNPGTDFERLFELQAQQRQLEIQQQDMHHQ 701
Query: 375 NNLTNIHQQP--GNNVQGLQQQQQFGTESGNQGIQTSHHSA---QMLQQPKVSMQ----Q 425
L + QP + VQ L +Q + + S H +L Q ++ Q
Sbjct: 702 QLLQQLKLQPQQQSQVQQLLLEQLMHQQMSDPNFGQSKHDPSRDNLLDQVQLRRYLHDLQ 881
Query: 426 QLQHNPSKLLPSQSQQSQT----------QASQQQLMSQI-HNQPAQMQQQLGLQQQQNP 474
Q H+ L PS Q Q QA +L+ Q H QQL QQ
Sbjct: 882 QNPHSLGHLDPSMEQFIQANIGLNAAQGRQADLSELLLQARHGNILPSDQQLRFQQD--- 1052
Query: 475 LQRDMQQKLQASGSLLQQQSVLDQQKQLYQSQRALPET 512
Q + Q L+QQ LD ++ +S R + ET
Sbjct: 1053-----QLQAQQLSMALKQQLGLDGERHFGRS-RPINET 1148
Score = 54.7 bits (130), Expect = 2e-07
Identities = 91/396 (22%), Positives = 149/396 (36%), Gaps = 20/396 (5%)
Frame = +3
Query: 90 NKSQNTMANNMLSNEGGPSNNLPDQGQQHPNPLPNQHQPRQQLLSHNIQNNVAPQPNLSS 149
N+ ++ + LS G NN L + +QQ + +Q A +
Sbjct: 261 NQRVGSLEDQFLSRIGPNFNNFDVADHLMLQKLQKERLQQQQ--AERLQQQQAERLQQQQ 434
Query: 150 VSTLPQIPSQNISQNSNTQQPGQNSVSNSIGQNSNVQSM--FPG---SQRQMSGRPQVVP 204
L Q ++ + Q + Q ++S+ N + FPG SQ SG Q++
Sbjct: 435 AERLQQQQAERLQQQQAERLQQQTNISSHFPAYLNGSDLDRFPGFSPSQSNKSGIQQMMQ 614
Query: 205 ------------QQQQQQSQNQQQTLYQQQLLKHKLSQMQQQQQQQNLLQPNQLLSSQQS 252
Q QQ+Q + QQQ ++ QQLL+ Q QQQ Q Q LL QL+ Q S
Sbjct: 615 NPGTDFERLFELQAQQRQLEIQQQDMHHQQLLQQLKLQPQQQSQVQQLLL-EQLMHQQMS 791
Query: 253 VIPTSSAMQPSMVQSSLSSLPQNQQSNNVQQSTQSRLQQHSQIIRQQQQNSIVNQQQTPM 312
P+ QS + +N+ Q R H QQN P
Sbjct: 792 --------DPNFGQSK-----HDPSRDNLLDQVQLRRYLH-----DLQQNPHSLGHLDPS 917
Query: 313 IQQQHQQLAGPQSNATNGQHA---QMLGQQNNVGDVQKSQRLHPQQNNLMNLQQRQQQQQ 369
++Q Q G NA G+ A ++L Q + + Q+L QQ+ L Q QQ
Sbjct: 918 MEQFIQANIG--LNAAQGRQADLSELLLQARHGNILPSDQQLRFQQDQL----QAQQLSM 1079
Query: 370 LMNHQNNLTNIHQQPGNNVQGLQQQQQFGTESGNQGIQTSHHSAQMLQQPKVSMQQQLQH 429
+ Q GL ++ FG + + + Q+++ P QL H
Sbjct: 1080ALKQQ--------------LGLDGERHFGRS------RPINETGQLVRNPS---NHQLGH 1190
Query: 430 NPSKLLPSQSQQSQTQASQQQLMSQIHNQPAQMQQQ 465
+ + +Q Q A+Q++ ++ + + Q+
Sbjct: 1191SAGFNVSEIHKQQQMLAAQEEQLNFLGRNHLEQNQE 1298
Score = 40.8 bits (94), Expect = 0.003
Identities = 50/196 (25%), Positives = 74/196 (37%), Gaps = 17/196 (8%)
Frame = +3
Query: 647 SQSQVTSVQSHENQMNSQMQPTNLQGSTSVQQNNIASLQNNSMSSLSTTQQNMLNTIQPS 706
+QS + E +N P GS ++ S + ++ ML +Q
Sbjct: 198 NQSSFRDTWTDEYGINRHFNPNQRVGSL---EDQFLSRIGPNFNNFDVADHLMLQKLQKE 368
Query: 707 NNLDSGQGNSVNSLQQVPVSSLQQNTVNT-QHTNINSLPSQGGVNVIQP-NLNTHQP--- 761
Q LQQ LQQ Q L Q + Q N+++H P
Sbjct: 369 RL----QQQQAERLQQQQAERLQQQQAERLQQQQAERLQQQQAERLQQQTNISSHFPAYL 536
Query: 762 -GSNM-----LQQQQLKHQQEQKMLQN------QQFKQQYQQRQMMQRQQQQLHQPAKQQ 809
GS++ Q Q+M+QN + F+ Q QQRQ+ +QQ HQ QQ
Sbjct: 537 NGSDLDRFPGFSPSQSNKSGIQQMMQNPGTDFERLFELQAQQRQLEIQQQDMHHQQLLQQ 716
Query: 810 MSAQPQTHQLPQINQM 825
+ QPQ Q Q+ Q+
Sbjct: 717 LKLQPQ--QQSQVQQL 758
Score = 35.4 bits (80), Expect = 0.14
Identities = 49/202 (24%), Positives = 82/202 (40%), Gaps = 7/202 (3%)
Frame = +3
Query: 638 ATHMALMPQSQSQVTSVQSHENQMNSQMQPTNLQGSTSVQQNNIASLQNNSMSSLSTTQQ 697
A H+ L + ++ Q+ E Q + Q + +QQ LQ L QQ
Sbjct: 333 ADHLMLQKLQKERLQQQQA-ERLQQQQAERLQQQQAERLQQQQAERLQQQQAERLQ--QQ 503
Query: 698 NMLNTIQPS----NNLDSGQGNSVNSLQQVPVSSLQQNTVNTQHTNINSLPSQGGVNVIQ 753
+++ P+ ++LD G S + + + + QN T + L +Q Q
Sbjct: 504 TNISSHFPAYLNGSDLDRFPGFSPSQSNKSGIQQMMQNP-GTDFERLFELQAQ------Q 662
Query: 754 PNLNTHQPGSNMLQQQQLKHQQ--EQKMLQNQQFKQQYQQRQMMQRQQQQLHQPAKQQMS 811
L +QQQ + HQQ +Q LQ QQ + Q QQ + Q QQ+ P Q
Sbjct: 663 RQLE--------IQQQDMHHQQLLQQLKLQPQQ-QSQVQQLLLEQLMHQQMSDPNFGQSK 815
Query: 812 AQP-QTHQLPQINQMNDMNDVK 832
P + + L Q+ ++D++
Sbjct: 816 HDPSRDNLLDQVQLRRYLHDLQ 881
Score = 29.6 bits (65), Expect = 7.9
Identities = 41/173 (23%), Positives = 68/173 (38%), Gaps = 1/173 (0%)
Frame = +3
Query: 646 QSQSQVTSVQSHENQMNSQMQPTNLQGSTSVQQNNIASLQNNSMSSLSTTQQNMLNTIQP 705
Q Q ++ H Q+ Q++ Q + VQQ + L + MS + Q
Sbjct: 660 QRQLEIQQQDMHHQQLLQQLK-LQPQQQSQVQQLLLEQLMHQQMSDPNFGQSKH------ 818
Query: 706 SNNLDSGQGNSVNSLQ-QVPVSSLQQNTVNTQHTNINSLPSQGGVNVIQPNLNTHQPGSN 764
D + N ++ +Q + + LQQN + H + PS LN Q
Sbjct: 819 ----DPSRDNLLDQVQLRRYLHDLQQNPHSLGHLD----PSMEQFIQANIGLNAAQGRQA 974
Query: 765 MLQQQQLKHQQEQKMLQNQQFKQQYQQRQMMQRQQQQLHQPAKQQMSAQPQTH 817
L + L+ + + +QQ + +QQ Q+ Q QQL KQQ+ + H
Sbjct: 975 DLSELLLQARHGNILPSDQQLR--FQQDQL---QAQQLSMALKQQLGLDGERH 1118
>TC79752 similar to GP|11994670|dbj|BAB02898. gene_id:K7P8.17~unknown
protein {Arabidopsis thaliana}, partial (2%)
Length = 2137
Score = 56.2 bits (134), Expect = 8e-08
Identities = 67/267 (25%), Positives = 104/267 (38%), Gaps = 2/267 (0%)
Frame = +1
Query: 153 LPQIPSQNISQNSNTQQPGQNSVSNSIGQNSNVQSM--FPGSQRQMSGRPQVVPQQQQQQ 210
+P++P Q ++Q ++ P + +S++ + S+ +P +Q S P Q
Sbjct: 532 VPELPMQ-VTQGNSQGIPAFSGMSSAFNSQTTPPSVQQYPVHAQQQSHLSNPHPHLQGPN 708
Query: 211 SQNQQQTLYQQQLLKHKLSQMQQQQQQQNLLQPNQLLSSQQSVIPTSSAMQPSMVQSSLS 270
N QQ Y + L K + Q QQQQQ Q LS+ ++IP A + S
Sbjct: 709 HPNNQQQAYIR-LAKER----QLQQQQQQRYLQQQQLSATNALIPHVQAQAQPPISSPQQ 873
Query: 271 SLPQNQQSNNVQQSTQSRLQQHSQIIRQQQQNSIVNQQQTPMIQQQHQQLAGPQSNATNG 330
+ Q Q N+ QQ + S S + Q+ QQ+ + Q + G +
Sbjct: 874 NSSQAQPQNSSQQVSVSPATPSSPLTPMSSQH---QQQKHDLPQPGFSRNPGSSVTSQAV 1044
Query: 331 QHAQMLGQQNNVGDVQKSQRLHPQQNNLMNLQQRQQQQQLMNHQNNLTNIHQQPGNNVQG 390
+ Q QQ Q+S R HP N + Q +QQ + L T IHQ NN
Sbjct: 1045KQRQRQAQQR---QYQQSARQHP--NQPQHAQAQQQAKILKGIGRGNTLIHQ---NNSVD 1200
Query: 391 LQQQQQFGTESGNQGIQTSHHSAQMLQ 417
G Q ++ QM Q
Sbjct: 1201PSHINGLSVAPGTQPVEKGDQITQMTQ 1281
Score = 55.8 bits (133), Expect = 1e-07
Identities = 85/385 (22%), Positives = 135/385 (34%), Gaps = 57/385 (14%)
Frame = +1
Query: 143 PQPNLSSVSTLPQIPSQNISQNSNTQQPGQNSVSNSIGQNSNVQSMFPGSQRQMSGRPQV 202
P PN SVS++ +PS +S S+ P S+ N++ S M ++ G P+
Sbjct: 13 PLPNHGSVSSV--LPSSGLS--SSNAPPSGTSLGNNLSSPSG--PMAASARDSRYGVPRG 174
Query: 203 VPQQQQQQSQNQQQTLYQQQLLKHKLSQMQQQ---------------------------- 234
VP +Q + QQ Y Q + + Q
Sbjct: 175 VPISADEQQRLQQ---YNQLISSRNMQQSSMSVPGSDRGARMLPGANGMGMMGGINRSIA 345
Query: 235 --------QQQQNLLQPNQLLSSQQSVIPTSSAMQPSMVQSSLSSLPQNQQSNNVQQSTQ 286
++L +LSS +P A Q + + ++ + +N Q
Sbjct: 346 MARPGFHGMTSSSMLSSGGMLSSSMVGMPGVGAGQGNSMMRPRDTVHMMRPGHNQGHQRQ 525
Query: 287 SRLQQHSQIIRQQQQNSI---------VNQQQTPMIQQQHQQLAGPQSNATNGQHAQMLG 337
+ + + Q I N Q TP QQ+ A QS+ +N H + G
Sbjct: 526 MMVPELPMQVTQGNSQGIPAFSGMSSAFNSQTTPPSVQQYPVHAQQQSHLSN-PHPHLQG 702
Query: 338 QQNNVGDVQKSQRLHPQQNNLMNLQQRQQQQQLMNHQNNL-TNIHQQPGNNVQGLQQ-QQ 395
+ Q RL ++ QQR QQQ ++ N L ++ Q + QQ
Sbjct: 703 PNHPNNQQQAYIRLAKERQLQQQQQQRYLQQQQLSATNALIPHVQAQAQPPISSPQQNSS 882
Query: 396 QFGTESGNQGIQTSHHSAQMLQQPKVSMQQQLQH---------NPSKLLPSQS-QQSQTQ 445
Q ++ +Q + S + P S QQ +H NP + SQ+ +Q Q Q
Sbjct: 883 QAQPQNSSQQVSVSPATPSSPLTPMSSQHQQQKHDLPQPGFSRNPGSSVTSQAVKQRQRQ 1062
Query: 446 ASQQQLMSQIHNQPAQMQQQLGLQQ 470
A Q+Q P Q Q QQ
Sbjct: 1063AQQRQYQQSARQHPNQPQHAQAQQQ 1137
Score = 45.8 bits (107), Expect = 1e-04
Identities = 71/284 (25%), Positives = 109/284 (38%), Gaps = 39/284 (13%)
Frame = +1
Query: 620 NQFRPRKAMSSLQPGQLPATHMALM-PQSQSQVTSVQSH--------ENQMNSQMQPTNL 670
+ RPR + ++PG +M P+ QVT S + NSQ P ++
Sbjct: 457 SMMRPRDTVHMMRPGHNQGHQRQMMVPELPMQVTQGNSQGIPAFSGMSSAFNSQTTPPSV 636
Query: 671 QGST--SVQQNNIAS----LQ------NNSMSSLSTTQQNMLNTIQPSNNLDSGQGNSVN 718
Q + QQ+++++ LQ N + + ++ L Q L Q ++ N
Sbjct: 637 QQYPVHAQQQSHLSNPHPHLQGPNHPNNQQQAYIRLAKERQLQQQQQQRYLQQQQLSATN 816
Query: 719 SL-------QQVPVSSLQQNTVNTQHTNINSLPSQGGV---NVIQPNLNTHQPGSNMLQQ 768
+L Q P+SS QQN+ Q N + S + + P + HQ + L Q
Sbjct: 817 ALIPHVQAQAQPPISSPQQNSSQAQPQNSSQQVSVSPATPSSPLTPMSSQHQQQKHDLPQ 996
Query: 769 QQLKHQQEQKMLQN--QQFKQQYQQRQMMQRQQQQLHQPAKQQMSAQPQTHQLPQINQ-- 824
+ +Q ++Q QQRQ Q +Q +QP Q AQ Q L I +
Sbjct: 997 PGFSRNPGSSVTSQAVKQRQRQAQQRQYQQSARQHPNQP--QHAQAQQQAKILKGIGRGN 1170
Query: 825 ----MNDMNDVKIRQGLGVKSGVFQQHLTSGQNSTYSHQQMKQG 864
N+ D GL V G Q + G T QM QG
Sbjct: 1171TLIHQNNSVDPSHINGLSVAPGT--QPVEKGDQIT----QMTQG 1284
Score = 44.3 bits (103), Expect = 3e-04
Identities = 58/230 (25%), Positives = 94/230 (40%), Gaps = 10/230 (4%)
Frame = +1
Query: 581 GYKTMFERMISILQIPKSSIQYGVKEKLGSYEKQIAAAINQFRPRKAMS----SLQPGQL 636
G+ +R + + ++P Q G + + ++ +A +Q P + Q L
Sbjct: 499 GHNQGHQRQMMVPELPMQVTQ-GNSQGIPAFSGMSSAFNSQTTPPSVQQYPVHAQQQSHL 675
Query: 637 PATHMALM----PQSQSQVTSVQSHENQMNSQMQPTNLQGSTSVQQNN-IASLQNNSMSS 691
H L P +Q Q + E Q+ Q Q LQ N I +Q +
Sbjct: 676 SNPHPHLQGPNHPNNQQQAYIRLAKERQLQQQQQQRYLQQQQLSATNALIPHVQAQAQPP 855
Query: 692 LSTTQQNMLNTIQPSNNLDSGQGNSVNSLQQVPVSSLQQNTVNTQHTNINSLPSQGGVNV 751
+S+ QQN S Q NS QQV VS T ++ T ++S Q ++
Sbjct: 856 ISSPQQN------------SSQAQPQNSSQQVSVSPA---TPSSPLTPMSSQHQQQKHDL 990
Query: 752 IQPNLNTHQPGSNMLQQQQLKHQQEQKMLQNQQFKQQY-QQRQMMQRQQQ 800
QP + + PGS++ Q + Q++ + Q QQ +Q+ Q Q Q QQQ
Sbjct: 991 PQPGFSRN-PGSSVTSQAVKQRQRQAQQRQYQQSARQHPNQPQHAQAQQQ 1137
>BG644345 weakly similar to GP|19697333|gb putative protein potential
transcriptional repressor Not4hp - Mus musculus, partial
(7%)
Length = 635
Score = 55.5 bits (132), Expect = 1e-07
Identities = 45/107 (42%), Positives = 54/107 (50%), Gaps = 4/107 (3%)
Frame = +1
Query: 155 QIPSQNISQNSNTQQPGQNSVS--NSIGQN--SNVQSMFPGSQRQMSGRPQVVPQQQQQQ 210
Q P Q Q+S Q PGQ S + GQ S + P SQ+Q + Q QQQQQQ
Sbjct: 196 QTPGQQ--QSSPFQTPGQQQASPFQTPGQQQPSPFSQITPFSQQQQQLQFQQQQQQQQQQ 369
Query: 211 SQNQQQTLYQQQLLKHKLSQMQQQQQQQNLLQPNQLLSSQQSVIPTS 257
Q QQQ QQQ Q+Q QQQQQ LLQ +Q QQ + T+
Sbjct: 370 LQQQQQQQLQQQ------QQLQLQQQQQQLLQQHQSSQQQQLYLFTN 492
Score = 48.5 bits (114), Expect = 2e-05
Identities = 45/123 (36%), Positives = 51/123 (40%), Gaps = 1/123 (0%)
Frame = +1
Query: 382 QQPGN-NVQGLQQQQQFGTESGNQGIQTSHHSAQMLQQPKVSMQQQLQHNPSKLLPSQSQ 440
QQP G QQ F T G Q Q P + QQ S++ P Q
Sbjct: 145 QQPSLFQTPGQQQASPFQTP-GQQQSSPFQTPGQQQASPFQTPGQQQPSPFSQITPFSQQ 321
Query: 441 QSQTQASQQQLMSQIHNQPAQMQQQLGLQQQQNPLQRDMQQKLQASGSLLQQQSVLDQQK 500
Q Q Q QQQ Q QQQL QQQQ LQ+ Q +LQ L QQ QQ+
Sbjct: 322 QQQLQFQQQQ---------QQQQQQL-QQQQQQQLQQQQQLQLQQQQQQLLQQHQSSQQQ 471
Query: 501 QLY 503
QLY
Sbjct: 472 QLY 480
Score = 47.0 bits (110), Expect = 5e-05
Identities = 46/148 (31%), Positives = 55/148 (37%), Gaps = 3/148 (2%)
Frame = +1
Query: 167 TQQPGQNSVSNSIGQNSNVQSMFPGSQRQMSGRPQVVPQQQQQ---QSQNQQQTLYQQQL 223
T Q Q S+ + GQ PG Q+ P P QQQ Q+ QQQ Q+
Sbjct: 133 TPQAQQPSLFQTPGQQQASPFQTPGQQQSS---PFQTPGQQQASPFQTPGQQQPSPFSQI 303
Query: 224 LKHKLSQMQQQQQQQNLLQPNQLLSSQQSVIPTSSAMQPSMVQSSLSSLPQNQQSNNVQQ 283
Q Q Q QQQ Q QL QQ + Q QQ +QQ
Sbjct: 304 TPFSQQQQQLQFQQQQQQQQQQLQQQQQQQL-------------------QQQQQLQLQQ 426
Query: 284 STQSRLQQHSQIIRQQQQNSIVNQQQTP 311
Q LQQH QQQQ + +TP
Sbjct: 427 QQQQLLQQHQS--SQQQQLYLFTNDKTP 504
Score = 46.6 bits (109), Expect = 6e-05
Identities = 50/147 (34%), Positives = 59/147 (40%), Gaps = 2/147 (1%)
Frame = +1
Query: 234 QQQQQNLLQPNQLLSSQQSVIPTSSAMQPSMVQSSLSSLPQNQQSNNVQQSTQSRLQQHS 293
Q QQ +L Q QQ P + P QSS P QQ++ Q Q + S
Sbjct: 139 QAQQPSLFQT----PGQQQASPFQT---PGQQQSSPFQTPGQQQASPFQTPGQQQPSPFS 297
Query: 294 QI--IRQQQQNSIVNQQQTPMIQQQHQQLAGPQSNATNGQHAQMLGQQNNVGDVQKSQRL 351
QI QQQQ QQQ QQQ QQL Q Q L QQ + +L
Sbjct: 298 QITPFSQQQQQLQFQQQQ----QQQQQQL--------QQQQQQQLQQQQQL-------QL 420
Query: 352 HPQQNNLMNLQQRQQQQQLMNHQNNLT 378
QQ L+ Q QQQQL N+ T
Sbjct: 421 QQQQQQLLQQHQSSQQQQLYLFTNDKT 501
Score = 46.6 bits (109), Expect = 6e-05
Identities = 42/116 (36%), Positives = 55/116 (47%), Gaps = 1/116 (0%)
Frame = +1
Query: 212 QNQQQTLYQQQLLKHKLSQMQQQQQQQNLLQPNQLLSSQQ-SVIPTSSAMQPSMVQSSLS 270
Q QQ +L+Q + + S Q QQQ+ P Q QQ S T QPS S ++
Sbjct: 139 QAQQPSLFQTPG-QQQASPFQTPGQQQS--SPFQTPGQQQASPFQTPGQQQPSPF-SQIT 306
Query: 271 SLPQNQQSNNVQQSTQSRLQQHSQIIRQQQQNSIVNQQQTPMIQQQHQQLAGPQSN 326
Q QQ QQ Q + QQ ++QQQQ + QQQ + QQQ Q L QS+
Sbjct: 307 PFSQQQQQLQFQQQQQQQQQQ----LQQQQQQQLQQQQQLQLQQQQQQLLQQHQSS 462
Score = 45.4 bits (106), Expect = 1e-04
Identities = 40/120 (33%), Positives = 50/120 (41%), Gaps = 8/120 (6%)
Frame = +1
Query: 204 PQQQQQ---QSQNQQQTLYQQQLLKHKLSQMQQQQQQQNLLQPNQLLSSQQ-----SVIP 255
PQ QQ Q+ QQQ Q + + S Q QQQ P Q QQ + P
Sbjct: 136 PQAQQPSLFQTPGQQQASPFQTPGQQQSSPFQTPGQQQ--ASPFQTPGQQQPSPFSQITP 309
Query: 256 TSSAMQPSMVQSSLSSLPQNQQSNNVQQSTQSRLQQHSQIIRQQQQNSIVNQQQTPMIQQ 315
S Q Q Q QQ +QQ Q +LQQ Q+ QQQQ ++ Q Q+ QQ
Sbjct: 310 FSQQQQQLQFQQQ-----QQQQQQQLQQQQQQQLQQQQQLQLQQQQQQLLQQHQSSQQQQ 474
Score = 41.6 bits (96), Expect = 0.002
Identities = 23/44 (52%), Positives = 29/44 (65%), Gaps = 2/44 (4%)
Frame = +1
Query: 768 QQQLKHQQEQKMLQNQQFKQQYQQRQMMQRQQQ--QLHQPAKQQ 809
QQQ + QQ+Q Q QQ QQ QQ Q+ Q+QQQ Q HQ ++QQ
Sbjct: 340 QQQQQQQQQQLQQQQQQQLQQQQQLQLQQQQQQLLQQHQSSQQQ 471
Score = 41.2 bits (95), Expect = 0.003
Identities = 55/179 (30%), Positives = 66/179 (36%), Gaps = 7/179 (3%)
Frame = +1
Query: 308 QQTPMIQQQHQQLAGPQSNATNGQHAQMLGQQNNVGDVQKSQRLHPQQNNLMNLQQ---- 363
QQ + Q QQ A P T GQ Q G Q S P Q Q
Sbjct: 145 QQPSLFQTPGQQQASPFQ--TPGQ--QQSSPFQTPGQQQASPFQTPGQQQPSPFSQITPF 312
Query: 364 RQQQQQLMNHQNNLTNIHQQPGNNVQGLQQQQQFGTESGNQGIQTSHHSAQMLQQPKVSM 423
QQQQQL Q Q Q LQQQQQ +Q Q+LQQ + S
Sbjct: 313 SQQQQQLQFQQQQQQQQQQLQQQQQQQLQQQQQ---------LQLQQQQQQLLQQHQSSQ 465
Query: 424 QQQLQHNPSKLLPSQSQQSQTQASQQQLMSQ---IHNQPAQMQQQLGLQQQQNPLQRDM 479
QQQL + P + S A L+ +H P + QQ Q Q+PL D+
Sbjct: 466 QQQLYLFTNDKTP--ATYSTKWADLHSLLPHPLLLHLLPCSLLQQC---QLQSPLCSDI 627
Score = 37.4 bits (85), Expect = 0.038
Identities = 38/131 (29%), Positives = 46/131 (35%), Gaps = 6/131 (4%)
Frame = +1
Query: 732 TVNTQHTNINSLPSQGGVNVIQ-PNLNTHQPGSNMLQQQQLK-----HQQEQKMLQNQQF 785
T Q ++ P Q + Q P P QQQ QQ Q F
Sbjct: 133 TPQAQQPSLFQTPGQQQASPFQTPGQQQSSPFQTPGQQQASPFQTPGQQQPSPFSQITPF 312
Query: 786 KQQYQQRQMMQRQQQQLHQPAKQQMSAQPQTHQLPQINQMNDMNDVKIRQGLGVKSGVFQ 845
QQ QQ Q Q+QQQQ Q +QQ Q Q Q Q+ Q + Q
Sbjct: 313 SQQQQQLQF-QQQQQQQQQQLQQQQQQQLQQQQQLQLQQQQQQ--------------LLQ 447
Query: 846 QHLTSGQNSTY 856
QH +S Q Y
Sbjct: 448 QHQSSQQQQLY 480
>TC80931 similar to GP|14190783|gb|AAF65166.2 putative phloem transcription
factor M1 {Apium graveolens}, partial (67%)
Length = 1193
Score = 55.5 bits (132), Expect = 1e-07
Identities = 37/99 (37%), Positives = 53/99 (53%), Gaps = 5/99 (5%)
Frame = +2
Query: 768 QQQLKHQQEQKML----QNQQFKQQYQQRQMMQRQQQQLHQPAKQQMS-AQPQTHQLPQI 822
QQQ++ QQ+Q+ L QNQQ Q QQ +Q+QQQQL Q +QQ+S Q Q QLPQ+
Sbjct: 569 QQQMQQQQQQQQLPQLQQNQQLSQIQQQIPQLQQQQQQLPQLQQQQLSQLQQQQQQLPQL 748
Query: 823 NQMNDMNDVKIRQGLGVKSGVFQQHLTSGQNSTYSHQQM 861
Q+ + +Q +G G G++ S Q+
Sbjct: 749 QQLQHQQLPQQQQMVGAGMGQTYVQGPGGRSQMVSQGQV 865
Score = 50.8 bits (120), Expect = 3e-06
Identities = 52/167 (31%), Positives = 72/167 (42%), Gaps = 2/167 (1%)
Frame = +2
Query: 241 LQPNQLLSSQQSVIPTSSAMQPSMVQSSLSSLPQNQQSNNVQQSTQS--RLQQHSQIIRQ 298
L N L+SS + ++ S + S PQ QQ QQ Q +LQQ+ Q+ +
Sbjct: 464 LSHNCLVSSSSNRCNSNKCNNTSRCNRNSSIFPQLQQQMQQQQQQQQLPQLQQNQQLSQ- 640
Query: 299 QQQNSIVNQQQTPMIQQQHQQLAGPQSNATNGQHAQMLGQQNNVGDVQKSQRLHPQQNNL 358
QQQ P +QQQ QQL PQ QQ + +Q+ Q+ PQ L
Sbjct: 641 -------IQQQIPQLQQQQQQL--PQL------------QQQQLSQLQQQQQQLPQLQQL 757
Query: 359 MNLQQRQQQQQLMNHQNNLTNIHQQPGNNVQGLQQQQQFGTESGNQG 405
+ QQ QQQQ++ T + Q PG Q + Q Q + N G
Sbjct: 758 QH-QQLPQQQQMVGAGMGQTYV-QGPGGRSQMVSQGQVSSQGATNIG 892
Score = 50.4 bits (119), Expect = 4e-06
Identities = 44/122 (36%), Positives = 58/122 (47%), Gaps = 2/122 (1%)
Frame = +2
Query: 158 SQNISQNSNTQQPGQNSVSNSIGQNSNVQSMFPGSQRQMSGRPQV--VPQQQQQQSQNQQ 215
S N +S++ + N +N+ N N S+FP Q+QM + Q +PQ Q QNQQ
Sbjct: 467 SHNCLVSSSSNRCNSNKCNNTSRCNRN-SSIFPQLQQQMQQQQQQQQLPQLQ----QNQQ 631
Query: 216 QTLYQQQLLKHKLSQMQQQQQQQNLLQPNQLLSSQQSVIPTSSAMQPSMVQSSLSSLPQN 275
+ QQQ + Q+QQQQQQ LQ QL QQ P + Q LPQ
Sbjct: 632 LSQIQQQ-----IPQLQQQQQQLPQLQQQQLSQLQQ-----QQQQLPQLQQLQHQQLPQQ 781
Query: 276 QQ 277
QQ
Sbjct: 782 QQ 787
Score = 45.8 bits (107), Expect = 1e-04
Identities = 34/77 (44%), Positives = 41/77 (53%), Gaps = 2/77 (2%)
Frame = +2
Query: 432 SKLLPSQSQQSQTQASQQQLMSQIHNQP-AQMQQQL-GLQQQQNPLQRDMQQKLQASGSL 489
S + P QQ Q Q QQQL NQ +Q+QQQ+ LQQQQ L + QQ+L L
Sbjct: 548 SSIFPQLQQQMQQQQQQQQLPQLQQNQQLSQIQQQIPQLQQQQQQLPQLQQQQL---SQL 718
Query: 490 LQQQSVLDQQKQLYQSQ 506
QQQ L Q +QL Q
Sbjct: 719 QQQQQQLPQLQQLQHQQ 769
Score = 45.1 bits (105), Expect = 2e-04
Identities = 26/73 (35%), Positives = 34/73 (45%)
Frame = +2
Query: 407 QTSHHSAQMLQQPKVSMQQQLQHNPSKLLPSQSQQSQTQASQQQLMSQIHNQPAQMQQQL 466
Q Q Q P++ QQL ++ Q QQ Q QQQ +SQ+ Q Q+ Q
Sbjct: 572 QQMQQQQQQQQLPQLQQNQQLSQIQQQIPQLQQQQQQLPQLQQQQLSQLQQQQQQLPQLQ 751
Query: 467 GLQQQQNPLQRDM 479
LQ QQ P Q+ M
Sbjct: 752 QLQHQQLPQQQQM 790
Score = 44.7 bits (104), Expect = 2e-04
Identities = 38/124 (30%), Positives = 50/124 (39%)
Frame = +2
Query: 317 HQQLAGPQSNATNGQHAQMLGQQNNVGDVQKSQRLHPQQNNLMNLQQRQQQQQLMNHQNN 376
H L SN N NN ++ + PQ M QQ+QQQ +
Sbjct: 470 HNCLVSSSSNRCNSNKC------NNTSRCNRNSSIFPQLQQQMQQQQQQQQLPQLQQNQQ 631
Query: 377 LTNIHQQPGNNVQGLQQQQQFGTESGNQGIQTSHHSAQMLQQPKVSMQQQLQHNPSKLLP 436
L+ I QQ + LQQQQQ + +Q S QQ ++ QQLQH + LP
Sbjct: 632 LSQIQQQ----IPQLQQQQQQLPQ-----LQQQQLSQLQQQQQQLPQLQQLQH---QQLP 775
Query: 437 SQSQ 440
Q Q
Sbjct: 776 QQQQ 787
Score = 44.7 bits (104), Expect = 2e-04
Identities = 34/80 (42%), Positives = 39/80 (48%), Gaps = 6/80 (7%)
Frame = +2
Query: 227 KLSQMQQQQQQQN---LLQPNQLLSSQQSVIPTSSAMQ---PSMVQSSLSSLPQNQQSNN 280
+L Q QQQQQQ LQ NQ LS Q IP Q P + Q LS L Q Q
Sbjct: 563 QLQQQMQQQQQQQQLPQLQQNQQLSQIQQQIPQLQQQQQQLPQLQQQQLSQLQQQQ---- 730
Query: 281 VQQSTQSRLQQHSQIIRQQQ 300
QQ Q + QH Q+ +QQQ
Sbjct: 731 -QQLPQLQQLQHQQLPQQQQ 787
Score = 42.0 bits (97), Expect = 0.002
Identities = 31/85 (36%), Positives = 36/85 (41%), Gaps = 2/85 (2%)
Frame = +2
Query: 151 STLPQIPSQNISQNSNTQQPG--QNSVSNSIGQNSNVQSMFPGSQRQMSGRPQVVPQQQQ 208
S PQ+ Q Q Q P QN Q S +Q P Q+Q PQ+ QQ
Sbjct: 551 SIFPQLQQQMQQQQQQQQLPQLQQNQ------QLSQIQQQIPQLQQQQQQLPQLQQQQLS 712
Query: 209 QQSQNQQQTLYQQQLLKHKLSQMQQ 233
Q Q QQQ QQL +L Q QQ
Sbjct: 713 QLQQQQQQLPQLQQLQHQQLPQQQQ 787
Score = 39.3 bits (90), Expect = 0.010
Identities = 34/95 (35%), Positives = 41/95 (42%), Gaps = 1/95 (1%)
Frame = +2
Query: 410 HHSAQMLQQPKVSMQQQLQHNPSKLLPSQSQQSQTQASQQQLMSQIHNQPAQMQQQLG-L 468
+ ++ + Q + MQQQ Q L Q SQ Q QL Q P QQQL L
Sbjct: 539 NRNSSIFPQLQQQMQQQQQQQQLPQLQQNQQLSQIQQQIPQLQQQQQQLPQLQQQQLSQL 718
Query: 469 QQQQNPLQRDMQQKLQASGSLLQQQSVLDQQKQLY 503
QQQQ L + Q+LQ QQQ V Q Y
Sbjct: 719 QQQQQQLPQ--LQQLQHQQLPQQQQMVGAGMGQTY 817
Score = 38.5 bits (88), Expect(2) = 1e-06
Identities = 30/82 (36%), Positives = 37/82 (44%), Gaps = 1/82 (1%)
Frame = +2
Query: 783 QQFKQQYQQRQMMQRQQ-QQLHQPAKQQMSAQPQTHQLPQINQMNDMNDVKIRQGLGVKS 841
QQ +QQ QQ+Q+ Q QQ QQL Q +Q Q Q QLPQ+ Q S
Sbjct: 572 QQMQQQQQQQQLPQLQQNQQLSQIQQQIPQLQQQQQQLPQLQQQQ-------------LS 712
Query: 842 GVFQQHLTSGQNSTYSHQQMKQ 863
+ QQ Q HQQ+ Q
Sbjct: 713 QLQQQQQQLPQLQQLQHQQLPQ 778
Score = 33.9 bits (76), Expect = 0.42
Identities = 24/84 (28%), Positives = 41/84 (48%)
Frame = +1
Query: 188 MFPGSQRQMSGRPQVVPQQQQQQSQNQQQTLYQQQLLKHKLSQMQQQQQQQNLLQPNQLL 247
+FPG + +PQ+ QQQQQQ + QQQ+ +H+ QMQ QQQ + +
Sbjct: 439 LFPGDM--VVFKPQLSGQQQQQQ-------MQQQQMQQHQ--QMQSQQQHLSTIAATDAT 585
Query: 248 SSQQSVIPTSSAMQPSMVQSSLSS 271
++ + T++ + S+ S
Sbjct: 586 TTTTATASTAATESAAFPDSTADS 657
Score = 33.5 bits (75), Expect = 0.55
Identities = 42/169 (24%), Positives = 62/169 (35%), Gaps = 1/169 (0%)
Frame = +2
Query: 688 SMSSLSTTQQNMLNTIQPSNNLDSGQGNSV-NSLQQVPVSSLQQNTVNTQHTNINSLPSQ 746
S + L ++ N N+ + +N + +S+ LQQ +QQ Q +
Sbjct: 467 SHNCLVSSSSNRCNSNKCNNTSRCNRNSSIFPQLQQ----QMQQQQQQQQLPQLQQNQQL 634
Query: 747 GGVNVIQPNLNTHQPGSNMLQQQQLKHQQEQKMLQNQQFKQQYQQRQMMQRQQQQLHQPA 806
+ P L Q LQQQQL Q+ QQ Q Q+ Q Q QQL Q
Sbjct: 635 SQIQQQIPQLQQQQQQLPQLQQQQLSQLQQ----------QQQQLPQLQQLQHQQLPQQQ 784
Query: 807 KQQMSAQPQTHQLPQINQMNDMNDVKIRQGLGVKSGVFQQHLTSGQNST 855
+ + QT+ QG G +S + Q S Q +T
Sbjct: 785 QMVGAGMGQTYV----------------QGPGGRSQMVSQGQVSSQGAT 883
Score = 33.5 bits (75), Expect(2) = 1e-06
Identities = 43/182 (23%), Positives = 74/182 (40%), Gaps = 23/182 (12%)
Frame = +1
Query: 624 PRKAMSSLQP--GQLPATHMALMPQS-QSQVTSVQSHENQMNSQMQP-----TNLQG-ST 674
P++ S +QP A +M L Q+ SQ ++ E ++ Q Q T L+G +
Sbjct: 13 PQQVQSGMQPL*NNAAAANMTLTQQTASSQSKYIKVWEGSLSGQRQGQPVFITKLEGYRS 192
Query: 675 SVQQNNIASLQNNSMSSLSTTQQNMLNTIQ--------------PSNNLDSGQGNSVNSL 720
S +A+ M + Q+ +N Q P L Q + ++
Sbjct: 193 SSASETLAANWPPVMQIVRLISQDHMNNKQYVGKADFLVFRAMNPHGFLGQLQEKKLCAV 372
Query: 721 QQVPVSSLQQNTVNTQHTNINSLPSQGGVNVIQPNLNTHQPGSNMLQQQQLKHQQEQKML 780
Q+P +L + V+ + + + G + V +P L+ Q M QQQ +HQQ Q
Sbjct: 373 IQLPSQTLLLS-VSDKACRLIGMLFPGDMVVFKPQLSGQQQQQQMQQQQMQQHQQMQSQQ 549
Query: 781 QN 782
Q+
Sbjct: 550 QH 555
>TC79570 similar to PIR|F86427|F86427 auxin response factor 6 (ARF6)
[imported] - Arabidopsis thaliana, partial (15%)
Length = 1143
Score = 55.1 bits (131), Expect = 2e-07
Identities = 68/272 (25%), Positives = 108/272 (39%), Gaps = 4/272 (1%)
Frame = +2
Query: 661 MNSQMQPTNLQGSTSVQQNNIASLQNNSMSSLSTTQQNMLNTIQPSNNLDSGQGNSVNSL 720
M +++ P+ + + Q +A+ M +L ++Q+ + +Q + GN+
Sbjct: 251 MQARLDPSMMNMQPDMYQA-MAAAALQDMRTLDPSKQHPASLLQFQQPQNFSNGNA---- 415
Query: 721 QQVPVSSLQQNTVNTQHTNINSLPSQGGVNVIQPNLNTHQPGSNMLQQQQLKHQQEQKML 780
+L QN + Q + P Q + + Q + Q QQL Q
Sbjct: 416 ------ALMQNQMLQQSQPHQAFPKN------QESQHPSQSQAQTQQFQQLLQHQHSFTN 559
Query: 781 QNQQFKQQYQQRQMMQRQQQQLHQPAKQQMSAQPQTHQLPQINQMNDMNDVKIRQGLGVK 840
QN +QQ QQ+Q Q+QQQQ HQ +QQ S Q Q Q+ M++ V
Sbjct: 560 QNYHMQQQQQQQQQQQQQQQQQHQQQQQQQSQQQQ-----QVVDHQQMSNAVSTMSQFVS 724
Query: 841 SGVFQQHLTSGQNSTYSHQQMKQGSPFQVSSPQLFQAASPQIPHN--SSPQVDQQTHLLS 898
+ Q S+ HQQ S SP HN S D+ +HLL+
Sbjct: 725 APPSQSTQPMQAISSIGHQQNFSDSNGNPVSPL----------HNMLGSFSNDETSHLLN 874
Query: 899 LTKVGT--PLQSSNSPFGVPTPSPPMAPSPML 928
+ + P+QSS + PS +A P+L
Sbjct: 875 FPRPNSWVPVQSSTA-----RPSKRVAVDPLL 955
Score = 55.1 bits (131), Expect = 2e-07
Identities = 59/179 (32%), Positives = 75/179 (40%), Gaps = 2/179 (1%)
Frame = +2
Query: 350 RLHPQQNNLMNLQQRQQQQQLMNHQNNLTNIHQQPGNNVQGLQQQQQFGTESGNQGIQTS 409
RL P N+ + + L Q P + +Q QQ Q F +GN + +
Sbjct: 260 RLDPSMMNMQPDMYQAMAAAALQDMRTLDPSKQHPASLLQ-FQQPQNFS--NGNAALMQN 430
Query: 410 HHSAQMLQQPKVSMQQQLQHNPSKLLPSQSQQSQTQASQQQLMSQ--IHNQPAQMQQQLG 467
QMLQQ + Q N PSQSQ +QTQ QQ L Q NQ MQQQ
Sbjct: 431 ----QMLQQSQP--HQAFPKNQESQHPSQSQ-AQTQQFQQLLQHQHSFTNQNYHMQQQQQ 589
Query: 468 LQQQQNPLQRDMQQKLQASGSLLQQQSVLDQQKQLYQSQRALPETSTTSVDSTTQTAQP 526
QQQQ Q+ Q+ Q S QQQ V+D Q Q A+ S +Q+ QP
Sbjct: 590 QQQQQQQQQQQQHQQQQQQQS-QQQQQVVDHQ----QMSNAVSTMSQFVSAPPSQSTQP 751
Score = 53.9 bits (128), Expect = 4e-07
Identities = 53/175 (30%), Positives = 72/175 (40%), Gaps = 8/175 (4%)
Frame = +2
Query: 113 DQGQQHPNPLPNQHQPRQQLLSHN---IQNNVAPQPNLSSVSTLPQIPSQNISQNSNTQQ 169
D +QHP L QP Q + N +QN + Q P Q +N +Q
Sbjct: 344 DPSKQHPASLLQFQQP-QNFSNGNAALMQNQMLQQSQ----------PHQAFPKNQESQH 490
Query: 170 PGQNSVSNSIGQNSNVQSMFPGSQRQMSGRPQVVPQQQQQQSQNQQQTLYQQQLLKHKLS 229
P Q+ Q Q + Q + + + QQQQQQ Q QQQ QQQ +H+
Sbjct: 491 PSQSQA-----QTQQFQQLLQ-HQHSFTNQNYHMQQQQQQQQQQQQQ---QQQ--QHQQQ 637
Query: 230 QMQQQQQQQNLLQPNQLLS-----SQQSVIPTSSAMQPSMVQSSLSSLPQNQQSN 279
Q QQ QQQQ ++ Q+ + SQ P S + QP SS+ SN
Sbjct: 638 QQQQSQQQQQVVDHQQMSNAVSTMSQFVSAPPSQSTQPMQAISSIGHQQNFSDSN 802
Score = 52.0 bits (123), Expect = 1e-06
Identities = 69/278 (24%), Positives = 103/278 (36%), Gaps = 12/278 (4%)
Frame = +2
Query: 250 QQSVIPTSSAMQPSMVQSSLSSLPQNQQSNNVQQSTQSRLQQHSQIIRQQQQNSIVNQQQ 309
Q + P+ MQP M Q+ ++ Q+ ++ + + + L Q Q N+ + Q Q
Sbjct: 254 QARLDPSMMNMQPDMYQAMAAAALQDMRTLDPSKQHPASLLQFQQPQNFSNGNAALMQNQ 433
Query: 310 TPMIQQQHQQLAGP---------QSNATNGQHAQMLGQQN---NVGDVQKSQRLHPQQNN 357
M+QQ A P QS A Q Q+L Q+ N + Q+ QQ
Sbjct: 434 --MLQQSQPHQAFPKNQESQHPSQSQAQTQQFQQLLQHQHSFTNQNYHMQQQQQQQQQQQ 607
Query: 358 LMNLQQRQQQQQLMNHQNNLTNIHQQPGNNVQGLQQQQQFGTESGNQGIQTSHHSAQMLQ 417
QQ QQQQQ + Q HQQ N V + QF + +Q Q
Sbjct: 608 QQQQQQHQQQQQQQSQQQQQVVDHQQMSNAVSTM---SQFVSAPPSQSTQP--------M 754
Query: 418 QPKVSMQQQLQHNPSKLLPSQSQQSQTQASQQQLMSQIHNQPAQMQQQLGLQQQQNPLQR 477
Q S+ Q + S P + + S + N P P +R
Sbjct: 755 QAISSIGHQQNFSDSNGNPVSPLHNMLGSFSNDETSHLLNFPRPNSWVPVQSSTARPSKR 934
Query: 478 DMQQKLQASGSLLQQQSVLDQQKQLYQSQRALPETSTT 515
L +SG+ VL Q +QL QSQ + + + T
Sbjct: 935 VAVDPLLSSGA---SHYVLPQVEQLGQSQTTMSQNAIT 1039
Score = 48.1 bits (113), Expect = 2e-05
Identities = 46/183 (25%), Positives = 70/183 (38%), Gaps = 4/183 (2%)
Frame = +2
Query: 114 QGQQHPNPLPNQHQPRQQLLSHNIQNNVAPQPNLSSVSTLPQIPSQNISQNSNTQQPGQN 173
Q + P+ + Q Q + + +Q+ P+ ++L Q N N
Sbjct: 254 QARLDPSMMNMQPDMYQAMAAAALQDMRTLDPSKQHPASLLQFQQPQNFSNGNAAL---- 421
Query: 174 SVSNSIGQNSNVQSMFPGSQR-QMSGRPQVVPQQQQQQSQNQQQTL---YQQQLLKHKLS 229
+ N + Q S FP +Q Q + Q QQ QQ Q+Q Y Q + +
Sbjct: 422 -MQNQMLQQSQPHQAFPKNQESQHPSQSQAQTQQFQQLLQHQHSFTNQNYHMQQQQQQQQ 598
Query: 230 QMQQQQQQQNLLQPNQLLSSQQSVIPTSSAMQPSMVQSSLSSLPQNQQSNNVQQSTQSRL 289
Q QQQQQQQ+ Q Q QQ V+ S S P +Q + +Q +
Sbjct: 599 QQQQQQQQQHQQQQQQQSQQQQQVVDHQQMSNAVSTMSQFVSAPPSQSTQPMQAISSIGH 778
Query: 290 QQH 292
QQ+
Sbjct: 779 QQN 787
Score = 47.4 bits (111), Expect = 4e-05
Identities = 52/187 (27%), Positives = 69/187 (36%), Gaps = 10/187 (5%)
Frame = +2
Query: 168 QQPGQNSVSNSIGQNSNVQSMFPGSQRQMSGRPQVVPQQQQQQ--SQNQQQTLYQQQLLK 225
QQP S N+ ++ Q S Q P+ Q+ Q SQ+Q QT QQLL+
Sbjct: 383 QQPQNFSNGNA--------ALMQNQMLQQSQPHQAFPKNQESQHPSQSQAQTQQFQQLLQ 538
Query: 226 HKLS-------QMQQQQQQQNLLQPNQLLSSQQSVIPTSSAMQPSMVQSSLSSLPQNQQS 278
H+ S QQQQQQQ QQ
Sbjct: 539 HQHSFTNQNYHMQQQQQQQQ-------------------------------------QQQ 607
Query: 279 NNVQQSTQSRLQQHSQIIRQQQQNSIVNQQQTPMIQQQHQQLAGPQSNATNGQHA-QMLG 337
QQ Q + QQ S QQQQ + +QQ + + Q ++ P S +T A +G
Sbjct: 608 QQQQQQHQQQQQQQS----QQQQQVVDHQQMSNAVSTMSQFVSAPPSQSTQPMQAISSIG 775
Query: 338 QQNNVGD 344
Q N D
Sbjct: 776 HQQNFSD 796
Score = 44.7 bits (104), Expect = 2e-04
Identities = 52/191 (27%), Positives = 72/191 (37%), Gaps = 4/191 (2%)
Frame = +2
Query: 93 QNTMANNMLSNEGGPSNNLPDQGQQHPNPLPNQHQPRQQLLSHNIQNNVAPQPNLSSVST 152
QN M ++ P N Q QHP+ Q Q QQLL H
Sbjct: 425 QNQMLQQSQPHQAFPKN----QESQHPSQSQAQTQQFQQLLQH----------------- 541
Query: 153 LPQIPSQNISQNSNTQQPGQNSVSNSIGQNSNVQSMFPGSQRQMSGRPQVVPQQQQQQSQ 212
Q+S +N QN ++Q Q+Q + Q QQQQQQSQ
Sbjct: 542 -------------------QHSFTN---QNYHMQQQQQQQQQQQQQQQQQHQQQQQQQSQ 655
Query: 213 NQQQTLYQQQLLKHKLSQMQQ--QQQQQNLLQPNQLLSS--QQSVIPTSSAMQPSMVQSS 268
QQQ + QQ + + +S M Q QP Q +SS Q S+ S + +
Sbjct: 656 QQQQVVDHQQ-MSNAVSTMSQFVSAPPSQSTQPMQAISSIGHQQNFSDSNGNPVSPLHNM 832
Query: 269 LSSLPQNQQSN 279
L S ++ S+
Sbjct: 833 LGSFSNDETSH 865
>BG450331 similar to GP|16649103|gb| Unknown protein {Arabidopsis thaliana},
partial (12%)
Length = 433
Score = 54.7 bits (130), Expect = 2e-07
Identities = 24/30 (80%), Positives = 28/30 (93%)
Frame = +2
Query: 1158 AEWVHGTIVKCSFIPVSLSPSLKSQYVSLQ 1187
+EWV+ TIVKCSF+PVSL+PSLKSQY SLQ
Sbjct: 71 SEWVYETIVKCSFVPVSLNPSLKSQYASLQ 160
>TC91076 similar to PIR|H96747|H96747 unknown protein T10D10.14 [imported] -
Arabidopsis thaliana, partial (9%)
Length = 834
Score = 53.5 bits (127), Expect = 5e-07
Identities = 56/191 (29%), Positives = 83/191 (43%), Gaps = 4/191 (2%)
Frame = +3
Query: 105 GGPSNNLPDQGQQHPNPLPNQHQPRQQLLSHNIQNNVAPQPNLSSVSTLPQIPSQNISQN 164
G S+ P++ QQ+ P+P Q S+N +A N S LPQ ++ N
Sbjct: 246 GSQSSLPPNEMQQYGEPIPGQ--------SNNEAAKLAGGSNASL--NLPQ----SLVAN 383
Query: 165 SNTQQPGQNSVSNSIGQNSNVQSMFPGSQRQMSGRPQVVPQQQQQQSQNQQQTLYQQQLL 224
+ PG N G + Q++ G M+ RPQ + QQ Q QQQ QQ
Sbjct: 384 ARMLPPG-----NPQGLQMS-QALLSGVS--MAQRPQQLDSQQAVLQQQQQQQQLQQN-- 533
Query: 225 KHKLSQMQQQQQQQNLLQPNQLLS----SQQSVIPTSSAMQPSMVQSSLSSLPQNQQSNN 280
+H L Q Q Q Q++LL NQL Q S +P + + + L Q Q
Sbjct: 534 QHSLLQQQNPQFQRSLLSANQLSHLNGVGQNSNMPLGNHLLNKASPLQIQMLQQQHQQQQ 713
Query: 281 VQQSTQSRLQQ 291
+QQ+ Q ++Q+
Sbjct: 714 LQQNQQPQMQR 746
Score = 48.5 bits (114), Expect = 2e-05
Identities = 56/235 (23%), Positives = 91/235 (37%), Gaps = 1/235 (0%)
Frame = +3
Query: 138 QNNVAPQPNLSSVSTLPQIPSQNISQNSNTQQPGQNSVS-NSIGQNSNVQSMFPGSQRQM 196
++++ PN L S I + ++ + V N + QS P ++ Q
Sbjct: 105 EDHLPTLPNTHFADLLADQFSSQIEHDGYVKEDDRIQVRPNLVNLPLGSQSSLPPNEMQQ 284
Query: 197 SGRPQVVPQQQQQQSQNQQQTLYQQQLLKHKLSQMQQQQQQQNLLQPNQLLSSQQSVIPT 256
G P +P Q ++ L L + N P L SQ +
Sbjct: 285 YGEP--IPGQSNNEAAKLAGGSNASLNLPQSLVANARMLPPGN---PQGLQMSQALLSGV 449
Query: 257 SSAMQPSMVQSSLSSLPQNQQSNNVQQSTQSRLQQHSQIIRQQQQNSIVNQQQTPMIQQQ 316
S A +P + S + L Q QQ +QQ+ S LQQ + Q Q S+++ Q
Sbjct: 450 SMAQRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQQQNP----QFQRSLLSANQL-----S 602
Query: 317 HQQLAGPQSNATNGQHAQMLGQQNNVGDVQKSQRLHPQQNNLMNLQQRQQQQQLM 371
H G SN G H +L + + + +Q Q+ H QQ N Q + Q++ +M
Sbjct: 603 HLNGVGQNSNMPLGNH--LLNKASPL-QIQMLQQQHQQQQLQQNQQPQMQRKMMM 758
Score = 46.6 bits (109), Expect = 6e-05
Identities = 56/202 (27%), Positives = 76/202 (36%), Gaps = 11/202 (5%)
Frame = +3
Query: 147 LSSVSTLPQIPSQNISQNSNTQQPGQ-NSVSNSIGQNSNV-----QSMFPGSQRQMSGRP 200
L S S+LP N Q PGQ N+ + + SN QS+ ++ G P
Sbjct: 243 LGSQSSLPP----NEMQQYGEPIPGQSNNEAAKLAGGSNASLNLPQSLVANARMLPPGNP 410
Query: 201 QVVPQQQQ-----QQSQNQQQTLYQQQLLKHKLSQMQQQQQQQNLLQPNQLLSSQQSVIP 255
Q + Q +Q QQ QQ +L+ + Q Q QQ Q +LLQ
Sbjct: 411 QGLQMSQALLSGVSMAQRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQ------------- 551
Query: 256 TSSAMQPSMVQSSLSSLPQNQQSNNVQQSTQSRLQQHSQIIRQQQQNSIVNQQQTPMIQQ 315
Q Q SL S Q N V Q++ L H + + Q M+QQ
Sbjct: 552 ----QQNPQFQRSLLSANQLSHLNGVGQNSNMPLGNHLL--------NKASPLQIQMLQQ 695
Query: 316 QHQQLAGPQSNATNGQHAQMLG 337
QHQQ Q+ Q M+G
Sbjct: 696 QHQQQQLQQNQQPQMQRKMMMG 761
Score = 46.6 bits (109), Expect = 6e-05
Identities = 37/139 (26%), Positives = 60/139 (42%), Gaps = 9/139 (6%)
Frame = +3
Query: 245 QLLSSQQSVIPTSSAMQPSMVQSSLSSLPQNQQSNNVQQSTQSRLQQHSQIIRQQQQNSI 304
Q L + ++P + M Q+ LS + Q+ +L +++QQQQ
Sbjct: 366 QSLVANARMLPPGNPQGLQMSQALLSGVSMAQRPQ--------QLDSQQAVLQQQQQQQQ 521
Query: 305 VNQQQTPMIQQQHQQLAGPQSNATNGQHAQMLGQQNNVGDVQKSQRLHPQQNNLMN---- 360
+ Q Q ++QQQ+ Q +A H +GQ +N+ P N+L+N
Sbjct: 522 LQQNQHSLLQQQNPQFQRSLLSANQLSHLNGVGQNSNM----------PLGNHLLNKASP 671
Query: 361 -----LQQRQQQQQLMNHQ 374
LQQ+ QQQQL +Q
Sbjct: 672 LQIQMLQQQHQQQQLQQNQ 728
Score = 35.0 bits (79), Expect = 0.19
Identities = 50/173 (28%), Positives = 67/173 (37%)
Frame = +3
Query: 334 QMLGQQNNVGDVQKSQRLHPQQNNLMNLQQRQQQQQLMNHQNNLTNIHQQPGNNVQGLQQ 393
Q Q + G V++ R+ + N L+NL Q N QQ G + G
Sbjct: 159 QFSSQIEHDGYVKEDDRIQVRPN-LVNLPLGSQSSLPPNEM-------QQYGEPIPG--- 305
Query: 394 QQQFGTESGNQGIQTSHHSAQMLQQPKVSMQQQLQHNPSKLLPSQSQQSQTQASQQQLMS 453
+S N+ + + S L P Q L N L P Q Q Q L+S
Sbjct: 306 ------QSNNEAAKLAGGSNASLNLP-----QSLVANARMLPPGNPQGLQMS---QALLS 443
Query: 454 QIHNQPAQMQQQLGLQQQQNPLQRDMQQKLQASGSLLQQQSVLDQQKQLYQSQ 506
+ AQ QQL QQ Q+ QQ Q SLLQQQ+ Q+ L +Q
Sbjct: 444 GV--SMAQRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQQQNPQFQRSLLSANQ 596
Score = 31.6 bits (70), Expect = 2.1
Identities = 56/229 (24%), Positives = 81/229 (34%), Gaps = 26/229 (11%)
Frame = +3
Query: 636 LPATHMA--LMPQSQSQVTSVQSHENQMNSQMQPTNLQGSTSVQQNNIASLQNNSMSSLS 693
LP TH A L Q SQ+ + Q++P N+ +L S SSL
Sbjct: 123 LPNTHFADLLADQFSSQIEHDGYVKEDDRIQVRP------------NLVNLPLGSQSSLP 266
Query: 694 TTQQNMLNTIQP--SNN----LDSGQGNSVNSLQQVPVSSLQQNTVNTQHTNINSLPSQG 747
+ P SNN L G S+N Q + ++ N Q + S
Sbjct: 267 PNEMQQYGEPIPGQSNNEAAKLAGGSNASLNLPQSLVANARMLPPGNPQGLQM-SQALLS 443
Query: 748 GVNVIQPNLNTHQPGSNMLQQQQLKHQQEQKMLQNQQFKQQYQQRQMMQRQQQQLH---- 803
GV++ Q + + QQQQ + Q+ + QQ Q+Q+ + Q L+
Sbjct: 444 GVSMAQRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQQQNPQFQRSLLSANQLSHLNGVGQ 623
Query: 804 --------------QPAKQQMSAQPQTHQLPQINQMNDMNDVKIRQGLG 838
P + QM Q Q Q NQ M K+ GLG
Sbjct: 624 NSNMPLGNHLLNKASPLQIQMLQQQHQQQQLQQNQQPQMQR-KMMMGLG 767
>TC89480 similar to GP|20260684|gb|AAM13240.1 unknown protein {Arabidopsis
thaliana}, partial (43%)
Length = 1068
Score = 49.7 bits (117), Expect = 7e-06
Identities = 28/73 (38%), Positives = 39/73 (53%)
Frame = +1
Query: 757 NTHQPGSNMLQQQQLKHQQEQKMLQNQQFKQQYQQRQMMQRQQQQLHQPAKQQMSAQPQT 816
+ P + QQQQ HQQ+Q+ Q + Q Q Q Q+QQQQL+Q +QQ Q Q
Sbjct: 139 SNQDPNLYLQQQQQFLHQQQQQPFQ----QSQTTQSQFQQQQQQQLYQQQQQQRILQQQQ 306
Query: 817 HQLPQINQMNDMN 829
QL Q Q +++
Sbjct: 307 QQLQQPQQQQNLH 345
Score = 48.5 bits (114), Expect = 2e-05
Identities = 49/190 (25%), Positives = 82/190 (42%)
Frame = +1
Query: 164 NSNTQQPGQNSVSNSIGQNSNVQSMFPGSQRQMSGRPQVVPQQQQQQSQNQQQTLYQQQL 223
NSN Q Q+ Q++ +++ Q+Q + Q P QQ Q +Q+Q Q QQQL
Sbjct: 100 NSNPQIQSQSQ-----SQSNQDPNLYLQQQQQFLHQQQQQPFQQSQTTQSQFQQQQQQQL 264
Query: 224 LKHKLSQMQQQQQQQNLLQPNQLLSSQQSVIPTSSAMQPSMVQSSLSSLPQNQQSNNVQQ 283
+ + Q QQQQQ L QP Q + QS+ +S + +L+ + ++ +
Sbjct: 265 YQQQQQQRILQQQQQQLQQPQQQQNLHQSL---ASHFHLLNLVENLAEVVEHGNPDQQSD 435
Query: 284 STQSRLQQHSQIIRQQQQNSIVNQQQTPMIQQQHQQLAGPQSNATNGQHAQMLGQQNNVG 343
+ + L H + QQ NSI T + + Q+ +S Q ++ N
Sbjct: 436 ALITELSNHFEKC-QQLLNSISASISTKAMTVEGQKKKLEESEQLLNQRRDLIA--NYTK 606
Query: 344 DVQKSQRLHP 353
V++ R P
Sbjct: 607 SVEELVRSEP 636
Score = 47.8 bits (112), Expect = 3e-05
Identities = 51/187 (27%), Positives = 79/187 (41%)
Frame = +1
Query: 272 LPQNQQSNNVQQSTQSRLQQHSQIIRQQQQNSIVNQQQTPMIQQQHQQLAGPQSNATNGQ 331
+P + +Q +QS+ Q + QQQQ + QQQ P QS T Q
Sbjct: 88 IPTPNSNPQIQSQSQSQSNQDPNLYLQQQQQFLHQQQQQPF----------QQSQTTQSQ 237
Query: 332 HAQMLGQQNNVGDVQKSQRLHPQQNNLMNLQQRQQQQQLMNHQNNLTNIHQQPGNNVQGL 391
Q QQ + Q+ QR+ QQ LQQ QQQQ L HQ+ ++ H N V+ L
Sbjct: 238 FQQQ--QQQQLYQQQQQQRILQQQQ--QQLQQPQQQQNL--HQSLASHFHLL--NLVENL 393
Query: 392 QQQQQFGTESGNQGIQTSHHSAQMLQQPKVSMQQQLQHNPSKLLPSQSQQSQTQASQQQL 451
+ E GN Q+ ++ QQL ++ S + +++ + Q + +
Sbjct: 394 AEV----VEHGNPDQQSDALITELSNH--FEKCQQLLNSISASISTKAMTVEGQKKKLEE 555
Query: 452 MSQIHNQ 458
Q+ NQ
Sbjct: 556 SEQLLNQ 576
Score = 47.0 bits (110), Expect = 5e-05
Identities = 39/100 (39%), Positives = 48/100 (48%)
Frame = +1
Query: 143 PQPNLSSVSTLPQIPSQNISQNSNTQQPGQNSVSNSIGQNSNVQSMFPGSQRQMSGRPQV 202
P PN + PQI SQ SQ+ + Q P + Q F SQ S
Sbjct: 91 PTPNSN-----PQIQSQ--SQSQSNQDPNLYLQQQQQFLHQQQQQPFQQSQTTQSQ---- 237
Query: 203 VPQQQQQQSQNQQQTLYQQQLLKHKLSQMQQQQQQQNLLQ 242
QQQQQQ QQQ QQ++L+ + Q+QQ QQQQNL Q
Sbjct: 238 FQQQQQQQLYQQQQ---QQRILQQQQQQLQQPQQQQNLHQ 348
Score = 45.4 bits (106), Expect = 1e-04
Identities = 27/73 (36%), Positives = 37/73 (49%)
Frame = +1
Query: 759 HQPGSNMLQQQQLKHQQEQKMLQNQQFKQQYQQRQMMQRQQQQLHQPAKQQMSAQPQTHQ 818
HQ QQ Q Q Q+ Q Q ++QQ QQR + Q QQQQL QP +QQ Q
Sbjct: 187 HQQQQQPFQQSQTTQSQFQQQQQQQLYQQQQQQRILQQ-QQQQLQQPQQQQNLHQSLASH 363
Query: 819 LPQINQMNDMNDV 831
+N + ++ +V
Sbjct: 364 FHLLNLVENLAEV 402
Score = 45.1 bits (105), Expect = 2e-04
Identities = 48/162 (29%), Positives = 68/162 (41%), Gaps = 1/162 (0%)
Frame = +1
Query: 419 PKVSMQQQLQHNPSKLLPSQSQQSQTQASQQQLMSQIHNQPAQMQQQLGLQQQQNPLQRD 478
P++ Q Q Q N L Q QQ QQQ Q +Q QQQ QQQQ Q+
Sbjct: 109 PQIQSQSQSQSNQDPNLYLQQQQQFLHQQQQQPFQQSQTTQSQFQQQ---QQQQLYQQQQ 279
Query: 479 MQQKLQASGSLLQQQSVLDQQKQLYQSQRALPETSTTSVDSTTQTAQPNGVDWQEE-IYQ 537
Q+ LQ LQQ QQ+ L+QS A V++ + + D Q + +
Sbjct: 280 QQRILQQQQQQLQQP---QQQQNLHQS-LASHFHLLNLVENLAEVVEHGNPDQQSDALIT 447
Query: 538 KLQTMKETYLPEINEIYQKISMKVHQFDSIPQQPKSDQIEKL 579
+L E +N I IS K + Q+ K ++ E+L
Sbjct: 448 ELSNHFEKCQQLLNSISASISTKAMTVEG--QKKKLEESEQL 567
Score = 42.4 bits (98), Expect = 0.001
Identities = 28/74 (37%), Positives = 38/74 (50%)
Frame = +1
Query: 434 LLPSQSQQSQTQASQQQLMSQIHNQPAQMQQQLGLQQQQNPLQRDMQQKLQASGSLLQQQ 493
++P+ + Q Q+ Q +Q N Q QQQ QQQQ P Q+ Q + S QQQ
Sbjct: 85 MIPTPNSNPQIQSQSQSQSNQDPNLYLQQQQQFLHQQQQQPFQQS-----QTTQSQFQQQ 249
Query: 494 SVLDQQKQLYQSQR 507
QQ+QLYQ Q+
Sbjct: 250 ----QQQQLYQQQQ 279
Score = 42.0 bits (97), Expect = 0.002
Identities = 36/88 (40%), Positives = 45/88 (50%), Gaps = 4/88 (4%)
Frame = +1
Query: 399 TESGNQGIQTSHHSAQMLQQPKVSMQQQLQ--HNPSKLLPSQSQQSQTQAS--QQQLMSQ 454
T + N IQ+ S Q Q P + +QQQ Q H + QQSQT S QQQ Q
Sbjct: 94 TPNSNPQIQSQSQS-QSNQDPNLYLQQQQQFLHQQQQ---QPFQQSQTTQSQFQQQQQQQ 261
Query: 455 IHNQPAQMQQQLGLQQQQNPLQRDMQQK 482
++ Q QQQ LQQQQ LQ+ QQ+
Sbjct: 262 LYQ---QQQQQRILQQQQQQLQQPQQQQ 336
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.308 0.122 0.330
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,358,118
Number of Sequences: 36976
Number of extensions: 432144
Number of successful extensions: 6199
Number of sequences better than 10.0: 218
Number of HSP's better than 10.0 without gapping: 3624
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4712
length of query: 1290
length of database: 9,014,727
effective HSP length: 107
effective length of query: 1183
effective length of database: 5,058,295
effective search space: 5983962985
effective search space used: 5983962985
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC148304.1


