

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148227.2 + phase: 0 /pseudo
(1075 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
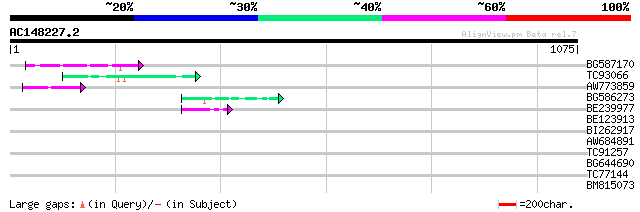
Score E
Sequences producing significant alignments: (bits) Value
BG587170 similar to PIR|F86470|F8 probable retroelement polyprot... 76 8e-14
TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative ret... 59 1e-08
AW773859 52 2e-06
BG586273 weakly similar to PIR|F86470|F8 probable retroelement p... 45 1e-04
BE239977 weakly similar to GP|23237899|db polyprotein-like {Oryz... 45 2e-04
BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sati... 41 0.002
BI262917 weakly similar to GP|19920130|g Putative retroelement {... 37 0.031
AW684891 35 0.16
TC91257 similar to GP|21434|emb|CAA36616.1|| ORF4 {Solanum tuber... 34 0.27
BG644690 weakly similar to GP|18542179|gb putative pol protein {... 33 0.45
TC77144 similar to GP|20160594|dbj|BAB89541. contains ESTs AU031... 31 2.9
BM815073 weakly similar to GP|18033111|gb functional candidate r... 30 5.0
>BG587170 similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 718
Score = 75.9 bits (185), Expect = 8e-14
Identities = 61/232 (26%), Positives = 104/232 (44%), Gaps = 8/232 (3%)
Frame = -3
Query: 31 SGKTIVSAKKNGGLYYLEDELETGHQLGQISSFSESFFVSNNKDDVMLWHLRLGHPSFKY 90
S + I G LY LE +L +S++ SF S++ + LWH RLGHP +
Sbjct: 704 SSQLIGEGVTKGDLYMLE-------KLDPVSNYKCSFTSSSSLNKDALWHARLGHPHGRA 546
Query: 91 LKIVFPKLFFGHDFSSFQCEIYEFSKQHRSSFPVQTYKPSKLFSIIHSDVWGPNRINSYP 150
L ++ P G F + CE K ++ FP + F +I++D+W +
Sbjct: 545 LNLMLP----GVVFENKNCEACILGKHCKNVFPRTSTVYENCFDLIYTDLWTAPSL---- 390
Query: 151 TKDGLSPLLMIILDFIGYIY*KKNQR*DKLSRILLN*CRLSLIPLYKFSELTMELNIL-- 208
++D + I + Y + D++ N Y + ++ IL
Sbjct: 389 SRDNHKYFVTFIDEKSKYTWLTLIPSKDRVIDAFKN------FQAYVTNHYHAKIKILRS 228
Query: 209 ------TQF*ETFFFMENGIVQQSTCVSSPQQNGITERKNRHLLEMARALLF 254
T + +GI+ Q++C +PQQNG+ +RKN+HL+E+AR+L+F
Sbjct: 227 DNGGEYTSYAFKSHLDHHGILHQTSCPYTPQQNGVAKRKNKHLMEVARSLMF 72
>TC93066 weakly similar to GP|19920130|gb|AAM08562.1 Putative retroelement
{Oryza sativa} [Oryza sativa (japonica cultivar-group)],
partial (10%)
Length = 823
Score = 58.9 bits (141), Expect = 1e-08
Identities = 73/282 (25%), Positives = 113/282 (39%), Gaps = 21/282 (7%)
Frame = +1
Query: 101 GHDFSSFQCEIYEFSKQHRSSFPVQTYKPSKLFSIIHSDVWGPNRINSYPTKDGLSPLLM 160
G D F + F + + SF T++ + IHSD+WGP+++ SY G ++
Sbjct: 1 GIDKLEFCKHLLFFGNRKKVSFSTATHRTKGILDYIHSDLWGPSKVTSY---GGRRYMMT 171
Query: 161 IILDFIGYIY*KKNQR*DKLSRILLN*CRLSLIPLYKFSELTMEL-------NILTQF*E 213
II DF ++ + + P +K + +E ++T *
Sbjct: 172 IIDDFPRKVW------------VYFLRYKNETFPTFKKWRILVETQTGKNVKKLITDN*L 315
Query: 214 TF-------FFMENGIVQQSTCVSSPQQNGITERKNRHLLEMARALLFFH--*SSKILMG 264
F F +GI + T +PQQNG+ ER R LLE AR +L *+ + L
Sbjct: 316 EFCSSDFNEFCTNHGIARHKTIPRNPQQNGVAERMIRTLLERARCMLSNAGL*N*RDLWV 495
Query: 265 *GCINCCTLNKLYVISC--FK-P*DSSRNLLKILSKCPYFSRFAFKNIRMHYFCP*TQTN 321
C L S FK P D L S F A+ + P
Sbjct: 496 EAASTACHLVNRSPHSALDFKVPEDIWSGNLVDYSNLRIFGCPAYALVNDGKLAP----- 660
Query: 322 RQT*TRASKRVFVGYSPTRKGYK--CLDLNSKRFLVTMDVTF 361
RA + +F+ Y+ KGY+ C D S++ +++ DVTF
Sbjct: 661 -----RAGECIFLSYASESKGYRLWCSDPKSQKLILSRDVTF 771
>AW773859
Length = 538
Score = 51.6 bits (122), Expect = 2e-06
Identities = 35/121 (28%), Positives = 54/121 (43%), Gaps = 2/121 (1%)
Frame = -3
Query: 25 LFQDSISGKTIVSAKKNGGLYYL--EDELETGHQLGQISSFSESFFVSNNKDDVMLWHLR 82
L Q+ S K I S + GLY+L +D Q+ + F+ LWH R
Sbjct: 383 LIQEKRSLKMIGSGELFEGLYFLTTQDTPSATIASSQVQPQPQPTFLPQEA----LWHFR 216
Query: 83 LGHPSFKYLKIVFPKLFFGHDFSSFQCEIYEFSKQHRSSFPVQTYKPSKLFSIIHSDVWG 142
LGH S + L + F + C+I +S+ + F + T + SK + + H D+WG
Sbjct: 215 LGHLSNRKLLSLHSNFPFITIDQNSVCDICHYSRHKKLPFQLSTNRASKCYELFHFDIWG 36
Query: 143 P 143
P
Sbjct: 35 P 33
>BG586273 weakly similar to PIR|F86470|F8 probable retroelement polyprotein
[imported] - Arabidopsis thaliana, partial (7%)
Length = 705
Score = 45.4 bits (106), Expect = 1e-04
Identities = 47/200 (23%), Positives = 78/200 (38%), Gaps = 7/200 (3%)
Frame = -2
Query: 327 RASKRVFVGYSPTRKGYKCLDLNSKRFLVTMDVTFF*K*TFF-------FRTIIFKGGNQ 379
R+ K +F+GYS T+KGYKC D ++R LV+ DV F + ++ R +
Sbjct: 545 RSRKAMFIGYSTTQKGYKCYDPEARRVLVSRDVKFIEERGYYEEKNQEDLRDLTSDKAGV 366
Query: 380 MKIHLIFFEDLILFENMFMSHSSRPFVSKENAPDNVSEHTPSMSEDVTKLVATNQNSNND 439
+++ L E L + N S SR N P ++ TP + + T +N
Sbjct: 365 LRVIL---EGLGIKMNQDQSTRSRQPEESSNEPRRAAQ-TPHLDHEGGSEPETQEN---- 210
Query: 440 SLEPNDNQELIQMSLHEHPYNETERKFGEVEGTWKGIIYGRRNHDKVVEDLIPQHSHESE 499
QE + +S +N GE + G H ++ P E +
Sbjct: 209 ------GQEGVGLSEEGEEFNSGSHHQGEQD-------QGDEEHHSEAQE--PHIQDEQD 75
Query: 500 PRENQPIKTNKGKGKISPFS 519
+ QP+ + + P S
Sbjct: 74 QVQEQPVLRRSTRIRKDPSS 15
>BE239977 weakly similar to GP|23237899|db polyprotein-like {Oryza sativa
(japonica cultivar-group)}, partial (2%)
Length = 514
Score = 44.7 bits (104), Expect = 2e-04
Identities = 31/95 (32%), Positives = 46/95 (47%)
Frame = -2
Query: 327 RASKRVFVGYSPTRKGYKCLDLNSKRFLVTMDVTFF*K*TFFFRTIIFKGGNQMKIHLIF 386
RA K VF+ YS T+KGY+C S+++ V+ DVTF + ++F + GG +
Sbjct: 393 RALKCVFIRYSSTQKGYRCYHPPSRKYFVSRDVTFHEQESYFGQN*SSSGGKYKE----- 229
Query: 387 FEDLILFENMFMSHSSRPFVSKENAPDNVSEHTPS 421
E L+L + F P + E DNV S
Sbjct: 228 DESLLLLDLTF-----GPEIEVETGGDNVETDVDS 139
>BE123913 weakly similar to GP|22093573|d polyprotein {Oryza sativa (japonica
cultivar-group)}, partial (8%)
Length = 503
Score = 41.2 bits (95), Expect = 0.002
Identities = 30/66 (45%), Positives = 40/66 (60%)
Frame = +2
Query: 752 RKLLF*LFMLMI*FLLEMM*QRWRD*RETSQQSLKSKTWDL*STFLALK*LDQRKEYRYA 811
RKL * MLMI F E+M + RD*R + +SL+SKTW++ + L +*L RK R+
Sbjct: 110 RKLFL*C-MLMIFF*QEIMVNKLRD*RTS*PRSLRSKTWEISNISLGWR*LGGRKVVRFP 286
Query: 812 TEVCS* 817
E S*
Sbjct: 287 NENTS* 304
>BI262917 weakly similar to GP|19920130|g Putative retroelement {Oryza
sativa} [Oryza sativa (japonica cultivar-group)],
partial (8%)
Length = 426
Score = 37.4 bits (85), Expect = 0.031
Identities = 35/128 (27%), Positives = 53/128 (41%), Gaps = 6/128 (4%)
Frame = +1
Query: 226 STCVSSPQQNGITERKNRHLLEMARALLFFH*SSKILMG*GCINCCTLNKLYVISCFKP* 285
S +PQQNG+ ER NR LLE RA+L +K C YVI+
Sbjct: 70 SXVAHTPQQNGVAERMNRTLLERTRAMLKTAGMAKSFWAEAVKTAC-----YVIN----- 219
Query: 286 DSSRNLLKILSKCPYFSRFAFKNIRMHYF-CP-----*TQTNRQT*TRASKRVFVGYSPT 339
S ++ + + + +H F CP +Q + ++ K +F+GY+
Sbjct: 220 RSPSTVIDLKTPMEMWKGKPVDYSSLHVFGCPVYVMYNSQERTKLDPKSRKCIFLGYADN 399
Query: 340 RKGYKCLD 347
KGY D
Sbjct: 400 VKGYXLWD 423
>AW684891
Length = 539
Score = 35.0 bits (79), Expect = 0.16
Identities = 13/24 (54%), Positives = 19/24 (79%)
Frame = -3
Query: 216 FFMENGIVQQSTCVSSPQQNGITE 239
+ ENG+ Q+ CV+SPQQNG++E
Sbjct: 195 YLEENGVSQELKCVNSPQQNGVSE 124
>TC91257 similar to GP|21434|emb|CAA36616.1|| ORF4 {Solanum tuberosum},
partial (8%)
Length = 854
Score = 34.3 bits (77), Expect = 0.27
Identities = 17/36 (47%), Positives = 21/36 (58%)
Frame = +2
Query: 845 GHTCRHRKIPKVSWEIDLLGTYKA*HCFSR*CC*SI 880
G R RK+P+VSWE +L K *+ CC*SI
Sbjct: 737 GDLFRSRKVPEVSWEAELFDHDKT*YFICSQCC*SI 844
>BG644690 weakly similar to GP|18542179|gb putative pol protein {Zea mays},
partial (22%)
Length = 629
Score = 33.5 bits (75), Expect = 0.45
Identities = 23/51 (45%), Positives = 31/51 (60%), Gaps = 1/51 (1%)
Frame = -3
Query: 675 M*KMHFLMVI*KKKCTWMVLQVLKKNLGQ-RCAN*KNLFMV*SSLQELGLK 724
M*++H LM I K++C L LK Q C++* +MV*S LQE G+K
Sbjct: 270 M*RVHLLMEISKRRCLSSNLLDLKMQRYQIMCSD*IRHYMV*SKLQEHGMK 118
>TC77144 similar to GP|20160594|dbj|BAB89541. contains ESTs AU031508(E61760)
AU068322(C20031) AU068323(C20031)~similar to Arabidopsis
thaliana, partial (46%)
Length = 817
Score = 30.8 bits (68), Expect = 2.9
Identities = 17/73 (23%), Positives = 34/73 (46%)
Frame = +3
Query: 370 RTIIFKGGNQMKIHLIFFEDLILFENMFMSHSSRPFVSKENAPDNVSEHTPSMSEDVTKL 429
++I G + K + E+M SSRP ++ P+N E T +SED ++
Sbjct: 96 KSIFIMGRGKFKAKPTGRRNFSTHEDMVAGTSSRPKTFRQKEPENEDEATREVSEDESEE 275
Query: 430 VATNQNSNNDSLE 442
+ ++N+ ++
Sbjct: 276 ESEDENTKTKGVQ 314
>BM815073 weakly similar to GP|18033111|gb functional candidate resistance
protein KR1 {Glycine max}, partial (4%)
Length = 516
Score = 30.0 bits (66), Expect = 5.0
Identities = 15/51 (29%), Positives = 26/51 (50%), Gaps = 6/51 (11%)
Frame = +3
Query: 536 LHQAPYV*VRIIFKFILLFFCIYLSFVLYRNSKEC------TGSSKCSKVE 580
+H P+V + +F +LF+ SF+++ N C T +S C K+E
Sbjct: 30 IH*LPHVEACYLVRFCILFYIYLTSFLIFHNDFSC*TC*FFTNNSICMKME 182
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.355 0.158 0.554
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 36,562,825
Number of Sequences: 36976
Number of extensions: 564251
Number of successful extensions: 6415
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 1939
Number of HSP's successfully gapped in prelim test: 309
Number of HSP's that attempted gapping in prelim test: 4377
Number of HSP's gapped (non-prelim): 2453
length of query: 1075
length of database: 9,014,727
effective HSP length: 106
effective length of query: 969
effective length of database: 5,095,271
effective search space: 4937317599
effective search space used: 4937317599
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.6 bits)
S2: 64 (29.3 bits)
Medicago: description of AC148227.2


