

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147472.2 - phase: 0
(363 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
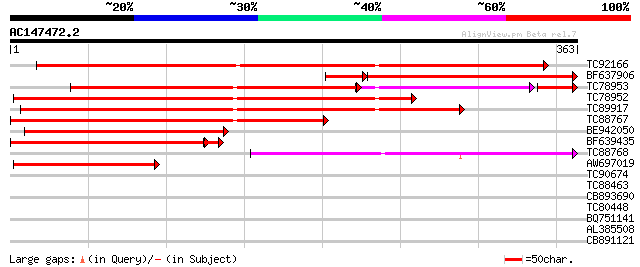
Score E
Sequences producing significant alignments: (bits) Value
TC92166 weakly similar to GP|21554042|gb|AAM63123.1 AX110P-like ... 345 1e-95
BF637906 weakly similar to PIR|A96686|A966 probable oxidoreducta... 274 3e-84
TC78953 weakly similar to GP|22137080|gb|AAM91385.1 At1g34200/F2... 218 2e-82
TC78952 weakly similar to GP|21554042|gb|AAM63123.1 AX110P-like ... 294 4e-80
TC89917 similar to GP|21554042|gb|AAM63123.1 AX110P-like protein... 287 5e-78
TC88767 weakly similar to PIR|T14319|T14319 protein AX110P - car... 246 8e-66
BE942050 similar to GP|21554042|gb AX110P-like protein {Arabidop... 177 7e-45
BF639435 weakly similar to GP|21554042|gb AX110P-like protein {A... 152 1e-40
TC88768 weakly similar to PIR|T14319|T14319 protein AX110P - car... 150 7e-37
AW697019 weakly similar to GP|21554042|gb| AX110P-like protein {... 120 8e-28
TC90674 similar to GP|20260136|gb|AAM12966.1 unknown protein {Ar... 36 0.027
TC88463 similar to GP|18447763|gb|AAL67992.1 putative serine car... 33 0.13
CB893690 weakly similar to GP|9759025|dbj gene_id:K6A12.9~simila... 33 0.17
TC80448 similar to PIR|C96495|C96495 probable ligand-gated ion c... 28 4.3
BQ751141 similar to SP|P29551|EF3 Elongation factor 3 (EF-3). {P... 28 7.3
AL385508 similar to SP|Q09167|SFR5_ Splicing factor arginine/se... 27 9.5
CB891121 similar to GP|1621467|gb| laccase {Liriodendron tulipif... 27 9.5
>TC92166 weakly similar to GP|21554042|gb|AAM63123.1 AX110P-like protein
{Arabidopsis thaliana}, partial (71%)
Length = 973
Score = 345 bits (886), Expect = 1e-95
Identities = 176/328 (53%), Positives = 227/328 (68%)
Frame = +1
Query: 18 ARKLARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRIYGSYDEVVEDSGVDAVYI 77
ARKL+RAI L+ ATISA+GSR++SKA FA N+ P + +IYGSY+EV++D VDAVY+
Sbjct: 1 ARKLSRAILLSSTATISAVGSRTLSKATTFAKLNNFPPTAKIYGSYEEVLDDPNVDAVYV 180
Query: 78 PLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEANGVQFMDGCMWLHHPRT 137
PLPT+LHV+WAVAAA K KHVL+EKP A+ V E D I++ CE GVQFMDG MW+HHPRT
Sbjct: 181 PLPTTLHVKWAVAAAKKGKHVLLEKPVAVSVKEFDEIVKVCEECGVQFMDGTMWMHHPRT 360
Query: 138 SNMKHLLDLSNSDGIGPVQFIHSTSTMPTTQEFLENNIRVKPELDALGALGDLAWYCIGA 197
MK L+ + + G V+ IH+ T +FLEN+IRVKP+LDALG+LGD WYC+ A
Sbjct: 361 EVMKGFLE--DGEKFGKVKSIHTCFTFGADPDFLENDIRVKPDLDALGSLGDEGWYCVRA 534
Query: 198 SLWAKGYKLPTTVTALPDVTRNMAGVILSITTSLQWDQPNQTVANIHCSFHSHTSMDLSI 257
LWA Y+LP TV A + N AGV+LS SL W + V +CSF S+ +MD++
Sbjct: 535 ILWAYNYELPKTVLATREPVLNKAGVLLSCGASLYWQ--DGRVTTFYCSFLSNLTMDITA 708
Query: 258 SGSNGSMHLKDFIIPYKETSASFDFTFGAKFVDLHIGWNVRPEEVHVVNKLPQEALMVQE 317
G+ G++H+ DFIIPY+E ASF + F DL W +P + + N LPQEALMV E
Sbjct: 709 VGTKGTLHVHDFIIPYEEKKASFYAASESSFDDLVTKWGSQPSKHVIENDLPQEALMVTE 888
Query: 318 FAMLVASIRDSGSQPSIKWPEISRKTQL 345
F+ LVA I+ ++P KWP ISRKTQL
Sbjct: 889 FSRLVAEIKSKNAKPEKKWPTISRKTQL 972
>BF637906 weakly similar to PIR|A96686|A966 probable oxidoreductase F15E12.2
[imported] - Arabidopsis thaliana, partial (25%)
Length = 574
Score = 274 bits (700), Expect(2) = 3e-84
Identities = 134/134 (100%), Positives = 134/134 (100%)
Frame = +2
Query: 230 SLQWDQPNQTVANIHCSFHSHTSMDLSISGSNGSMHLKDFIIPYKETSASFDFTFGAKFV 289
SLQWDQPNQTVANIHCSFHSHTSMDLSISGSNGSMHLKDFIIPYKETSASFDFTFGAKFV
Sbjct: 83 SLQWDQPNQTVANIHCSFHSHTSMDLSISGSNGSMHLKDFIIPYKETSASFDFTFGAKFV 262
Query: 290 DLHIGWNVRPEEVHVVNKLPQEALMVQEFAMLVASIRDSGSQPSIKWPEISRKTQLVVDA 349
DLHIGWNVRPEEVHVVNKLPQEALMVQEFAMLVASIRDSGSQPSIKWPEISRKTQLVVDA
Sbjct: 263 DLHIGWNVRPEEVHVVNKLPQEALMVQEFAMLVASIRDSGSQPSIKWPEISRKTQLVVDA 442
Query: 350 VKKSLELGCKPVSL 363
VKKSLELGCKPVSL
Sbjct: 443 VKKSLELGCKPVSL 484
Score = 56.2 bits (134), Expect(2) = 3e-84
Identities = 27/27 (100%), Positives = 27/27 (100%)
Frame = +1
Query: 203 GYKLPTTVTALPDVTRNMAGVILSITT 229
GYKLPTTVTALPDVTRNMAGVILSITT
Sbjct: 1 GYKLPTTVTALPDVTRNMAGVILSITT 81
>TC78953 weakly similar to GP|22137080|gb|AAM91385.1 At1g34200/F23M19.12
{Arabidopsis thaliana}, partial (57%)
Length = 1138
Score = 218 bits (556), Expect(3) = 2e-82
Identities = 104/186 (55%), Positives = 131/186 (69%)
Frame = +3
Query: 40 SISKAAKFAVENSLPASVRIYGSYDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVL 99
S+ KA KFA N+ P+ ++YGSYD V++D VD VYIPLPTSLH+ WAV AA KKKH+L
Sbjct: 3 SLEKATKFAASNNFPSHAKVYGSYDAVLDDPDVDVVYIPLPTSLHLHWAVLAAQKKKHLL 182
Query: 100 VEKPAALDVAELDLILEACEANGVQFMDGCMWLHHPRTSNMKHLLDLSNSDGIGPVQFIH 159
++KP AL+V ELD ILEACE+NG+Q+MD MW+HHPRT M +S+ + G +Q +H
Sbjct: 183 LDKPVALNVGELDKILEACESNGLQYMDATMWMHHPRTEKMFQF--ISDPNLFGSLQLVH 356
Query: 160 STSTMPTTQEFLENNIRVKPELDALGALGDLAWYCIGASLWAKGYKLPTTVTALPDVTRN 219
+ T + FL N+IRVKP+LDALGALGD WYCI A LWA Y+LP T L N
Sbjct: 357 AAFTFGASPHFLANDIRVKPDLDALGALGDAGWYCIRAILWAANYELPKTAKVLHKPKYN 536
Query: 220 MAGVIL 225
AGV+L
Sbjct: 537 EAGVLL 554
Score = 92.4 bits (228), Expect(3) = 2e-82
Identities = 48/115 (41%), Positives = 68/115 (58%)
Frame = +1
Query: 222 GVILSITTSLQWDQPNQTVANIHCSFHSHTSMDLSISGSNGSMHLKDFIIPYKETSASFD 281
G S SL W+ + VA CSF + SMD++ G+ GS+ + D++IP +E A F
Sbjct: 544 GYFWSCEASLTWE--DNKVATFFCSFLADMSMDITAIGAKGSLRVHDYVIPNEENEALFR 717
Query: 282 FTFGAKFVDLHIGWNVRPEEVHVVNKLPQEALMVQEFAMLVASIRDSGSQPSIKW 336
+ + F++L GWN +P EV V N +PQEALMV+EFA LV I+ S+ KW
Sbjct: 718 TSSNSYFLELSAGWNPKPSEVVVKNDIPQEALMVKEFAHLVGGIKYGNSKVENKW 882
Score = 34.3 bits (77), Expect(3) = 2e-82
Identities = 15/25 (60%), Positives = 21/25 (84%)
Frame = +2
Query: 339 ISRKTQLVVDAVKKSLELGCKPVSL 363
ISRKTQ+V+DAVK S++ G +PV +
Sbjct: 890 ISRKTQVVIDAVKASIDNGLQPVPI 964
>TC78952 weakly similar to GP|21554042|gb|AAM63123.1 AX110P-like protein
{Arabidopsis thaliana}, partial (58%)
Length = 858
Score = 294 bits (752), Expect = 4e-80
Identities = 145/258 (56%), Positives = 186/258 (71%)
Frame = +2
Query: 3 EKDTVRFGILGCATIARKLARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRIYGS 62
E +RFGILGCA IARK++RAITL+PN+T+ AIGSRS+ KA K+A N+ P+ +IYGS
Sbjct: 95 ETKPIRFGILGCAEIARKVSRAITLSPNSTLYAIGSRSLEKATKYAASNNFPSHTKIYGS 274
Query: 63 YDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEANG 122
YD V++D VDAVY+PLPTSLH+ WAV AA KKKH+L+EKP AL+V ELD ILEACE+NG
Sbjct: 275 YDAVLDDPDVDAVYVPLPTSLHLHWAVLAAQKKKHLLLEKPVALNVGELDKILEACESNG 454
Query: 123 VQFMDGCMWLHHPRTSNMKHLLDLSNSDGIGPVQFIHSTSTMPTTQEFLENNIRVKPELD 182
+Q+MD MW+HHPRT M +S+ + G +Q +H+T + PT+ FL N+IRVKP+LD
Sbjct: 455 LQYMDATMWMHHPRTEKMFQF--ISDPNLFGSLQSVHATFSYPTSPYFLANDIRVKPDLD 628
Query: 183 ALGALGDLAWYCIGASLWAKGYKLPTTVTALPDVTRNMAGVILSITTSLQWDQPNQTVAN 242
+LG LGD WYCI A LWA Y+LP T AL N AGV+LS SL W+ + VA
Sbjct: 629 SLGTLGDTGWYCIRAILWAANYELPKTAKALHKPKYNEAGVLLSCEASLTWE--DNKVAT 802
Query: 243 IHCSFHSHTSMDLSISGS 260
CSF + SMD++ G+
Sbjct: 803 XFCSFLADMSMDITAIGA 856
>TC89917 similar to GP|21554042|gb|AAM63123.1 AX110P-like protein
{Arabidopsis thaliana}, partial (74%)
Length = 852
Score = 287 bits (734), Expect = 5e-78
Identities = 142/284 (50%), Positives = 192/284 (67%)
Frame = +3
Query: 8 RFGILGCATIARKLARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRIYGSYDEVV 67
+ G++GCA IARK++RAI L+PNATI A+ SR+ KAA FA N P + ++YG+Y+ ++
Sbjct: 3 QIGVVGCADIARKVSRAINLSPNATICAVASRNYDKAAAFAAANGYPLTAKVYGNYEALL 182
Query: 68 EDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEANGVQFMD 127
ED VDAVY+PLPTSLH++WAV AA KKH+L+EKP ALD+AE D I+ ACE NGVQFMD
Sbjct: 183 EDPDVDAVYMPLPTSLHLKWAVLAAQNKKHLLLEKPVALDIAEFDEIVGACEENGVQFMD 362
Query: 128 GCMWLHHPRTSNMKHLLDLSNSDGIGPVQFIHSTSTMPTTQEFLENNIRVKPELDALGAL 187
MW+H+PRT+ M ++ G ++ + + T E+L+ +IRVKP+LDA G+L
Sbjct: 363 NTMWVHNPRTAKMAQF--FNDPHRFGQLKSVRTCFTFAADSEYLKKDIRVKPDLDAHGSL 536
Query: 188 GDLAWYCIGASLWAKGYKLPTTVTALPDVTRNMAGVILSITTSLQWDQPNQTVANIHCSF 247
GD WYCI A L A Y+LP TV A + N GVIL SL W+ + VA HCSF
Sbjct: 537 GDAGWYCIRAILLAANYELPKTVIASREPVLNKDGVILDCGASLYWE--DGKVATFHCSF 710
Query: 248 HSHTSMDLSISGSNGSMHLKDFIIPYKETSASFDFTFGAKFVDL 291
++ +MD++ G+ G++H+ DFIIPY+E ASF F F DL
Sbjct: 711 LANLTMDITAIGTKGTLHVHDFIIPYEENEASFHFGTETSFDDL 842
>TC88767 weakly similar to PIR|T14319|T14319 protein AX110P - carrot,
partial (42%)
Length = 652
Score = 246 bits (629), Expect = 8e-66
Identities = 122/204 (59%), Positives = 149/204 (72%)
Frame = +2
Query: 1 MTEKDTVRFGILGCATIARKLARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRIY 60
M + VRFGILGCA I+ KL +AI+ NAT+ AIGSRS+ KA FA E LP VR+Y
Sbjct: 44 MANETVVRFGILGCAQISIKLCKAISKAQNATLIAIGSRSLEKATAFAAEQGLPEVVRVY 223
Query: 61 GSYDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEA 120
GSY+ V+ED+ VDAVYIPLPT+LHV WAV AA + KHVL+EKP A++V+ELD ILEACE
Sbjct: 224 GSYEAVLEDNEVDAVYIPLPTALHVTWAVKAAERGKHVLLEKPVAMNVSELDRILEACET 403
Query: 121 NGVQFMDGCMWLHHPRTSNMKHLLDLSNSDGIGPVQFIHSTSTMPTTQEFLENNIRVKPE 180
NGVQFMDG MWLHHPRT+ MK L + G +++IHS T EFL+N+IR+KP+
Sbjct: 404 NGVQFMDGSMWLHHPRTAKMKEA--LCDEQRFGQLKWIHSCMTYNPGPEFLKNSIRMKPD 577
Query: 181 LDALGALGDLAWYCIGASLWAKGY 204
LD LGALGD+ WY I LW+ Y
Sbjct: 578 LDGLGALGDIXWYSIQXILWSVNY 649
>BE942050 similar to GP|21554042|gb AX110P-like protein {Arabidopsis
thaliana}, partial (37%)
Length = 656
Score = 177 bits (448), Expect = 7e-45
Identities = 80/131 (61%), Positives = 104/131 (79%)
Frame = +2
Query: 10 GILGCATIARKLARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRIYGSYDEVVED 69
G++GCA IARK++RAI L+PNATI A+ SR+ KAA FA N P + ++YG+Y+ ++ED
Sbjct: 2 GVVGCADIARKVSRAINLSPNATICAVASRNYDKAAAFAAANGYPLTAKVYGNYEALLED 181
Query: 70 SGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEANGVQFMDGC 129
VDAVY+PLPTSLH++WAV AA KKH+L+EKP ALD+AE D I+ ACE NGVQFMD
Sbjct: 182 PDVDAVYMPLPTSLHLKWAVLAAQNKKHLLLEKPVALDIAEFDEIVGACEENGVQFMDNT 361
Query: 130 MWLHHPRTSNM 140
MW+H+PRT+ M
Sbjct: 362 MWVHNPRTAKM 394
>BF639435 weakly similar to GP|21554042|gb AX110P-like protein {Arabidopsis
thaliana}, partial (32%)
Length = 470
Score = 152 bits (383), Expect(2) = 1e-40
Identities = 73/128 (57%), Positives = 96/128 (74%)
Frame = +3
Query: 1 MTEKDTVRFGILGCATIARKLARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRIY 60
M++ + +RFGILGCA IARKL+R I L+ ATIS +GSR++SKA FA N+ + ++Y
Sbjct: 21 MSQTEPIRFGILGCADIARKLSRTILLSSTATISGVGSRTLSKATTFAKLNNFRPTAKVY 200
Query: 61 GSYDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLILEACEA 120
GSY+EV++D VDAVY+PLPT+LHV+WAVAAA K KHVL+EKP A+ V E D I++ CE
Sbjct: 201 GSYEEVLDDPNVDAVYLPLPTTLHVKWAVAAAKKGKHVLLEKPVAVSVKEFDEIVKVCEE 380
Query: 121 NGVQFMDG 128
GV G
Sbjct: 381 CGVHLWMG 404
Score = 32.7 bits (73), Expect(2) = 1e-40
Identities = 11/13 (84%), Positives = 12/13 (91%)
Frame = +2
Query: 125 FMDGCMWLHHPRT 137
FMDG MW+HHPRT
Sbjct: 392 FMDGTMWMHHPRT 430
>TC88768 weakly similar to PIR|T14319|T14319 protein AX110P - carrot,
partial (27%)
Length = 767
Score = 150 bits (379), Expect = 7e-37
Identities = 83/212 (39%), Positives = 124/212 (58%), Gaps = 3/212 (1%)
Frame = +2
Query: 155 VQFIHSTSTMPTTQEFLENNIRVKPELDALGALGDLAWYCIGASLWAKGYKLPTTVTALP 214
+++IHS T E L+N+IR+KP+LD LGALGD+ WY I A LW+ Y+LP +V A P
Sbjct: 41 LKWIHSAMTYNPVPESLKNSIRMKPDLDGLGALGDIGWYSIQAILWSVNYELPKSVLAFP 220
Query: 215 DVTRNMAGVILSITTSLQWDQPNQTVANIHCSFHSHTSMDLSISGSNGSMHLKDFIIPYK 274
T N VI+S +SL W+ A HCSF ++ + D++I G+ GS+ L D +P++
Sbjct: 221 KATLNEDNVIISCGSSLHWEDGKS--ATFHCSFLTYVNFDVTILGTKGSLRLHDLTLPFE 394
Query: 275 ETSASFDFTFGAK--FVDLHIG-WNVRPEEVHVVNKLPQEALMVQEFAMLVASIRDSGSQ 331
E+ F+ ++ + + G W R E V Q+ MV+EFA LV I +
Sbjct: 395 ESLGFGTFSESSEIDYAKIEQGRWCPRANEHVVETMFSQDVWMVKEFADLVGKIMRLEMK 574
Query: 332 PSIKWPEISRKTQLVVDAVKKSLELGCKPVSL 363
P W +SRKTQ+V+DAVK+S++ G + V +
Sbjct: 575 PEKTWEVVSRKTQIVLDAVKESIQRGYESVEI 670
>AW697019 weakly similar to GP|21554042|gb| AX110P-like protein {Arabidopsis
thaliana}, partial (22%)
Length = 759
Score = 120 bits (301), Expect = 8e-28
Identities = 56/94 (59%), Positives = 74/94 (78%)
Frame = +1
Query: 3 EKDTVRFGILGCATIARKLARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRIYGS 62
E +RFGILG A IARK++RAITL+PN+T+ AIGSRS+ KA K+A N+ P+ +IYGS
Sbjct: 88 ETKPIRFGILGWAEIARKVSRAITLSPNSTLYAIGSRSLEKATKYAASNNFPSHTKIYGS 267
Query: 63 YDEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKK 96
YD V++D VDAVY+PLPTSLH+ WA A +K++
Sbjct: 268 YDAVLDDPDVDAVYVPLPTSLHLHWAGACCSKEE 369
>TC90674 similar to GP|20260136|gb|AAM12966.1 unknown protein {Arabidopsis
thaliana}, partial (51%)
Length = 566
Score = 35.8 bits (81), Expect = 0.027
Identities = 18/52 (34%), Positives = 29/52 (55%)
Frame = +2
Query: 64 DEVVEDSGVDAVYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLIL 115
+++++DS + AV + L V ++ KHVL EKPAA + EL+ L
Sbjct: 185 NDIIQDSSITAVLVVLAGQYQVDISLKLLKAGKHVLQEKPAASGINELETAL 340
>TC88463 similar to GP|18447763|gb|AAL67992.1 putative serine
carboxypeptidase precursor {Gossypium hirsutum}, partial
(71%)
Length = 1452
Score = 33.5 bits (75), Expect = 0.13
Identities = 25/73 (34%), Positives = 33/73 (44%)
Frame = +3
Query: 10 GILGCATIARKLARAITLTPNATISAIGSRSISKAAKFAVENSLPASVRIYGSYDEVVED 69
G LG A+KL R + L P+ ++ S + S + VEN L I D VED
Sbjct: 129 GDLGLKLEAKKLVRDLNLFPDVNLNIASSAANSSSRGKIVENPLNFRNLISDDDDVSVED 308
Query: 70 SGVDAVYIPLPTS 82
G A Y P+ S
Sbjct: 309 FGHYAGYYPIQHS 347
>CB893690 weakly similar to GP|9759025|dbj gene_id:K6A12.9~similar to unknown
protein~sp|O15736 {Arabidopsis thaliana}, partial (59%)
Length = 916
Score = 33.1 bits (74), Expect = 0.17
Identities = 17/67 (25%), Positives = 35/67 (51%)
Frame = +3
Query: 232 QWDQPNQTVANIHCSFHSHTSMDLSISGSNGSMHLKDFIIPYKETSASFDFTFGAKFVDL 291
+W + V + SFHS TS+ ++ + G MH+K+ ++PY ++ + G + + L
Sbjct: 180 RWMKLFVEVKKVLNSFHSQTSLPIA---NTG*MHMKEVVLPYCSSTILVN*LLGDRIIQL 350
Query: 292 HIGWNVR 298
G ++
Sbjct: 351 KCGIQIQ 371
>TC80448 similar to PIR|C96495|C96495 probable ligand-gated ion channel
[imported] - Arabidopsis thaliana, partial (28%)
Length = 1200
Score = 28.5 bits (62), Expect = 4.3
Identities = 12/33 (36%), Positives = 22/33 (66%)
Frame = -3
Query: 224 ILSITTSLQWDQPNQTVANIHCSFHSHTSMDLS 256
+L +++S+ + Q + ++H SF H+SMDLS
Sbjct: 937 LLFVSSSVAFPVDLQVIFHLHSSFFFHSSMDLS 839
>BQ751141 similar to SP|P29551|EF3 Elongation factor 3 (EF-3). {Pneumocystis
carinii}, partial (15%)
Length = 569
Score = 27.7 bits (60), Expect = 7.3
Identities = 18/63 (28%), Positives = 25/63 (39%), Gaps = 3/63 (4%)
Frame = -1
Query: 75 VYIPLPTSLHVRWAVAAANKKKHVLVEKPAALDVAELDLI---LEACEANGVQFMDGCMW 131
V +PLP + VRW ++ +L P L+ L C G Q + G W
Sbjct: 524 VGLPLPHARSVRWPACQGRPRRLLLAASPLDRS*RAHQLLGP*LSRCPVQGSQGLRG--W 351
Query: 132 LHH 134
HH
Sbjct: 350 CHH 342
>AL385508 similar to SP|Q09167|SFR5_ Splicing factor arginine/serine-rich 5
(Pre-mRNA splicing factor SRP40), partial (7%)
Length = 465
Score = 27.3 bits (59), Expect = 9.5
Identities = 14/25 (56%), Positives = 16/25 (64%)
Frame = +2
Query: 164 MPTTQEFLENNIRVKPELDALGALG 188
MPT +EFLE I VK LD + LG
Sbjct: 269 MPTWKEFLEGIIAVKRLLDLVALLG 343
>CB891121 similar to GP|1621467|gb| laccase {Liriodendron tulipifera},
partial (25%)
Length = 706
Score = 27.3 bits (59), Expect = 9.5
Identities = 17/54 (31%), Positives = 24/54 (43%)
Frame = -3
Query: 205 KLPTTVTALPDVTRNMAGVILSITTSLQWDQPNQTVANIHCSFHSHTSMDLSIS 258
K P + V RN AGV +L++ N V +HC HTS L ++
Sbjct: 488 KDPAKFNLVDPVERNTAGVPAGGWVALRFLADNPGVWFMHCHLEVHTSWGLKMA 327
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.132 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,928,385
Number of Sequences: 36976
Number of extensions: 143100
Number of successful extensions: 819
Number of sequences better than 10.0: 35
Number of HSP's better than 10.0 without gapping: 808
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 810
length of query: 363
length of database: 9,014,727
effective HSP length: 97
effective length of query: 266
effective length of database: 5,428,055
effective search space: 1443862630
effective search space used: 1443862630
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC147472.2


