

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147405.9 + phase: 0
(589 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
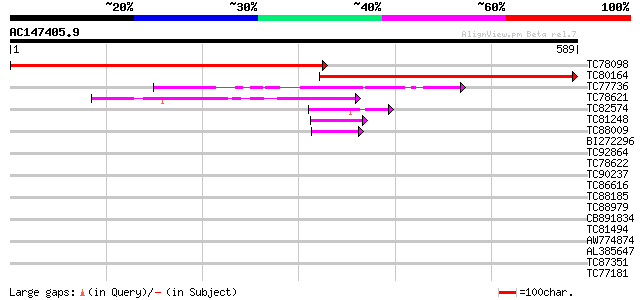
TC78098 similar to PIR|A84670|A84670 probable nucleotide-binding... 651 0.0
TC80164 similar to PIR|A84670|A84670 probable nucleotide-binding... 530 e-151
TC77736 similar to GP|12324651|gb|AAG52287.1 putative GTP-bindin... 132 3e-31
TC78621 similar to GP|6729012|gb|AAF27009.1| putative GTPase {Ar... 87 2e-17
TC82574 similar to GP|17473914|gb|AAL38371.1 GTP-binding protein... 46 4e-05
TC81248 similar to SP|O81004|YB87_ARATH Hypothetical GTP-binding... 44 1e-04
TC88009 similar to PIR|T48507|T48507 probable GTP-binding protei... 42 7e-04
BI272296 similar to GP|12322014|gb GTPase putative; 34281-30152... 41 0.001
TC92864 similar to GP|17644179|gb|AAL38787.1 unknown protein {Ar... 40 0.002
TC78622 weakly similar to GP|14587220|dbj|BAB61154. putative GTP... 39 0.004
TC90237 similar to GP|20466544|gb|AAM20589.1 unknown protein {Ar... 39 0.007
TC86616 similar to GP|7643796|gb|AAF65513.1| GTP-binding protein... 35 0.081
TC88185 homologue to PIR|T09368|T09368 GTP-binding protein F23K1... 33 0.24
TC88979 similar to PIR|H96601|H96601 hypothetical protein T6H22.... 33 0.40
CB891834 homologue to PIR|C96751|C96 probable GTP-binding protei... 31 1.2
TC81494 similar to GP|13173408|gb|AAK14395.1 response regulator ... 31 1.5
AW774874 similar to PIR|T46039|T4 hypothetical protein T16K5.80 ... 30 2.6
AL385647 30 3.4
TC87351 similar to GP|10176988|dbj|BAB10220. GTP-binding protein... 30 3.4
TC77181 similar to GP|510190|emb|CAA82196.1| chloroplast outer e... 30 3.4
>TC78098 similar to PIR|A84670|A84670 probable nucleotide-binding protein
[imported] - Arabidopsis thaliana, partial (40%)
Length = 1102
Score = 651 bits (1679), Expect = 0.0
Identities = 330/330 (100%), Positives = 330/330 (100%)
Frame = +3
Query: 1 MGKNQKTELGRALVKQHNQMIQQTKEKGKIYKKKFLESFTEVSDIDAIIEQADEDVDEQQ 60
MGKNQKTELGRALVKQHNQMIQQTKEKGKIYKKKFLESFTEVSDIDAIIEQADEDVDEQQ
Sbjct: 111 MGKNQKTELGRALVKQHNQMIQQTKEKGKIYKKKFLESFTEVSDIDAIIEQADEDVDEQQ 290
Query: 61 LLDIPLPPPTALINLDHASGSDFNGLTVEEMKKEQKIEEALHASSLRVPRRPFWSAEMSA 120
LLDIPLPPPTALINLDHASGSDFNGLTVEEMKKEQKIEEALHASSLRVPRRPFWSAEMSA
Sbjct: 291 LLDIPLPPPTALINLDHASGSDFNGLTVEEMKKEQKIEEALHASSLRVPRRPFWSAEMSA 470
Query: 121 DELHANETQHFLTWRRSLARLEENKKLVLTPFEKNLDIWRQLWRVVERSDLLVMVVDSRD 180
DELHANETQHFLTWRRSLARLEENKKLVLTPFEKNLDIWRQLWRVVERSDLLVMVVDSRD
Sbjct: 471 DELHANETQHFLTWRRSLARLEENKKLVLTPFEKNLDIWRQLWRVVERSDLLVMVVDSRD 650
Query: 181 PLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVREKWAEYFRAHDILFIFWSAKAAT 240
PLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVREKWAEYFRAHDILFIFWSAKAAT
Sbjct: 651 PLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVREKWAEYFRAHDILFIFWSAKAAT 830
Query: 241 AVLEGKKLGSSQADNMASADNPDTKIYGRDELLARLQSEAEAIVDMRRSSGSSKSSDDNA 300
AVLEGKKLGSSQADNMASADNPDTKIYGRDELLARLQSEAEAIVDMRRSSGSSKSSDDNA
Sbjct: 831 AVLEGKKLGSSQADNMASADNPDTKIYGRDELLARLQSEAEAIVDMRRSSGSSKSSDDNA 1010
Query: 301 SVSSSSSHVVVGFVGYPNVGKSSTINALVG 330
SVSSSSSHVVVGFVGYPNVGKSSTINALVG
Sbjct: 1011SVSSSSSHVVVGFVGYPNVGKSSTINALVG 1100
>TC80164 similar to PIR|A84670|A84670 probable nucleotide-binding protein
[imported] - Arabidopsis thaliana, partial (31%)
Length = 1215
Score = 530 bits (1364), Expect = e-151
Identities = 263/267 (98%), Positives = 263/267 (98%)
Frame = +3
Query: 323 STINALVGQKKTGVTSTPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLP 382
STINALVGQKKTGVTSTP KTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMIT GV P
Sbjct: 3 STINALVGQKKTGVTSTPRKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMIT*GVSP 182
Query: 383 IDRMTQHRECVQVVANRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRGQT 442
IDRM QHRECVQVVANRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRGQT
Sbjct: 183 IDRMNQHRECVQVVANRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRGQT 362
Query: 443 TSSGLPDETRASRQILKDYIDGKLPHYEMPPGLSTQELASEDSNEHDQVNPHVSDASDIE 502
TSSGLPDETRASRQILKDYIDGKLPHYEMPPGLSTQELASEDSNEHDQVNPHVSDASDIE
Sbjct: 363 TSSGLPDETRASRQILKDYIDGKLPHYEMPPGLSTQELASEDSNEHDQVNPHVSDASDIE 542
Query: 503 DSSVVETELAPKLEHVLDDLSSFDMANGLASNKVAPKKTKESQKHHRKPPRTKNRSWRAG 562
DSSVVETELAPKLEHVLDDLSSFDMANGLASNKVAPKKTKESQKHHRKPPRTKNRSWRAG
Sbjct: 543 DSSVVETELAPKLEHVLDDLSSFDMANGLASNKVAPKKTKESQKHHRKPPRTKNRSWRAG 722
Query: 563 NAGKDDTDGMPIARFHQKPVNSGPLKV 589
NAGKDDTDGMPIARFHQKPVNSGPLKV
Sbjct: 723 NAGKDDTDGMPIARFHQKPVNSGPLKV 803
>TC77736 similar to GP|12324651|gb|AAG52287.1 putative GTP-binding protein;
106556-109264 {Arabidopsis thaliana}, partial (76%)
Length = 2145
Score = 132 bits (333), Expect = 3e-31
Identities = 97/326 (29%), Positives = 156/326 (47%), Gaps = 2/326 (0%)
Frame = +3
Query: 150 TPFEKNLD--IWRQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVN 207
T FEK IW +L++V++ SD++V V+D+RDP RC LE + K+ HK+ +LL+N
Sbjct: 636 TMFEKGQSKRIWGELYKVIDSSDVVVQVIDARDPQGTRCYHLEKHLKKNCKHKHMVLLLN 815
Query: 208 KADLLPASVREKWAEYFRAHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIY 267
K DL+PA W+ K VL S + +A + + K +
Sbjct: 816 KCDLIPA-------------------WATKGWLRVL------SKEYPTLAFHASIN-KSF 917
Query: 268 GRDELLARLQSEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINA 327
G+ LL+ L+ A + S + VGFVGYPNVGKSS IN
Sbjct: 918 GKGSLLSVLRQFAR--------------------LKSDKQAISVGFVGYPNVGKSSVINT 1037
Query: 328 LVGQKKTGVTSTPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLPIDRMT 387
L + V PG+TK +Q + +++++ L DCPG+V+ + + S +++ GV+ + +
Sbjct: 1038LRTKNVCKVAPIPGETKVWQYITLTKRIFLIDCPGVVYHN-NDSETDVVLKGVVRVTNLK 1214
Query: 388 QHRECVQVVANRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRGQTTSSGL 447
+ + V RV + + Y I K + + ++ L C S G+ G
Sbjct: 1215DAADHIGEVLKRVKKEHLTRAYKI-----KEWVDE------NDFLLQLCKSSGKLLKGGE 1361
Query: 448 PDETRASRQILKDYIDGKLPHYEMPP 473
PD A++ IL D+ GK+P + PP
Sbjct: 1362PDLMTAAKMILHDWQRGKIPFFVPPP 1439
>TC78621 similar to GP|6729012|gb|AAF27009.1| putative GTPase {Arabidopsis
thaliana}, partial (56%)
Length = 1154
Score = 87.0 bits (214), Expect = 2e-17
Identities = 74/284 (26%), Positives = 123/284 (43%), Gaps = 5/284 (1%)
Frame = +3
Query: 86 LTVEEMKKEQKIEEALHASSLRVPRRPFWSAEMSADELHANETQHFLTWRRSLARLEENK 145
L + E ++E+ + E R R A + D+ ++N + EN
Sbjct: 234 LKLLEARRERALAELAKKKDERKERARKRKAGLPVDDDNSNSVE---------TASIENT 386
Query: 146 KLVLTPFEKNLD-----IWRQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHK 200
+ V T + N D L V+ SD+++ V+D+RDPL RC ++E ++ +K
Sbjct: 387 ESVPTVAKTNKDSSDRAFAGDLEAVIATSDVILEVLDARDPLGTRCVNIEKRVRDSGTYK 566
Query: 201 NTLLLVNKADLLPASVREKWAEYFRAHDILFIFWSAKAATAVLEGKKLGSSQADNMASAD 260
+LL+NK DL+P EKW +Y R F + K +T Q N+ +
Sbjct: 567 RHVLLLNKIDLVPRESVEKWLKYLREE---FPTVAFKCST---------QQQKSNLGRSK 710
Query: 261 NPDTKIYGRDELLARLQSEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVG 320
T S+ + D R+ K + A + + VG VG PNVG
Sbjct: 711 KIKT-------------SDTLQLSDWLRAETLLKLLKNYARSDKLKTSITVGLVGLPNVG 851
Query: 321 KSSTINALVGQKKTGVTSTPGKTKHFQTLIISEKLILCDCPGLV 364
SS IN+++ V+++ G T+ Q + + + + L DCPG+V
Sbjct: 852 NSSLINSVMRSHAVHVSASAGSTRSKQEIRLDKNVKLLDCPGVV 983
>TC82574 similar to GP|17473914|gb|AAL38371.1 GTP-binding protein-like
{Arabidopsis thaliana}, partial (54%)
Length = 915
Score = 46.2 bits (108), Expect = 4e-05
Identities = 29/91 (31%), Positives = 49/91 (52%), Gaps = 3/91 (3%)
Frame = +2
Query: 311 VGFVGYPNVGKSSTINALVGQKKTGVTSTPGKTKHFQTLIIS---EKLILCDCPGLVFPS 367
V +G PNVGKS+ N +VGQK + VT P T+H I S +++L D PG++
Sbjct: 350 VALLGKPNVGKSTLANQMVGQKLSIVTDKPQTTRHRVLCICSGSDYQMVLYDTPGVL--- 520
Query: 368 FSSSRYEMITCGVLPIDRMTQHRECVQVVAN 398
R+++ + + + T + +CV V+ +
Sbjct: 521 -QEQRHKLDSMMMQNVRSATVNADCVLVLVD 610
>TC81248 similar to SP|O81004|YB87_ARATH Hypothetical GTP-binding protein
At2g22870. [Mouse-ear cress] {Arabidopsis thaliana},
partial (78%)
Length = 1177
Score = 44.3 bits (103), Expect = 1e-04
Identities = 25/60 (41%), Positives = 36/60 (59%), Gaps = 1/60 (1%)
Frame = +3
Query: 313 FVGYPNVGKSSTINALVGQKKTGVTS-TPGKTKHFQTLIISEKLILCDCPGLVFPSFSSS 371
F+ NVGKSS IN+LV +K+ +TS PGKT+ ++++K L D PG F S +
Sbjct: 408 FLVVSNVGKSSLINSLVRKKELALTSKKPGKTQLINHFLVNKKWYLVDLPGYGFAKASEA 587
>TC88009 similar to PIR|T48507|T48507 probable GTP-binding protein -
Arabidopsis thaliana, partial (72%)
Length = 1399
Score = 42.0 bits (97), Expect = 7e-04
Identities = 23/55 (41%), Positives = 33/55 (59%), Gaps = 1/55 (1%)
Frame = +1
Query: 314 VGYPNVGKSSTINALVGQKKTGVTS-TPGKTKHFQTLIISEKLILCDCPGLVFPS 367
VG NVGKSS +N++V +KK +TS PGKT+ +++ L D PG + S
Sbjct: 640 VGRSNVGKSSLLNSIVRRKKLALTSKKPGKTQLINHFRVNDSWYLVDLPGYGYAS 804
>BI272296 similar to GP|12322014|gb GTPase putative; 34281-30152
{Arabidopsis thaliana}, partial (22%)
Length = 487
Score = 41.2 bits (95), Expect = 0.001
Identities = 22/55 (40%), Positives = 29/55 (52%)
Frame = +2
Query: 290 SGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVGQKKTGVTSTPGKTK 344
SG K + V + VG PNVGKSS +NALVG+ +T V+ G T+
Sbjct: 119 SGLXKVEEPQNLVEEEDYVPAISIVGRPNVGKSSILNALVGEDRTIVSPISGTTR 283
>TC92864 similar to GP|17644179|gb|AAL38787.1 unknown protein {Arabidopsis
thaliana}, partial (30%)
Length = 673
Score = 40.4 bits (93), Expect = 0.002
Identities = 21/85 (24%), Positives = 43/85 (49%)
Frame = +2
Query: 160 RQLWRVVERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVREK 219
++L ++ D+++ V D R PL P ++ + +K +L++N+ D++ + R
Sbjct: 344 KELKEQLKLMDVVIEVRDGRIPLSTSHPQMDLWLG----NKKRILVLNREDMISTADRNA 511
Query: 220 WAEYFRAHDILFIFWSAKAATAVLE 244
WAEYF H +F + + L+
Sbjct: 512 WAEYFTRHGTKVVFSNGQLGMGTLK 586
>TC78622 weakly similar to GP|14587220|dbj|BAB61154. putative GTPase {Oryza
sativa (japonica cultivar-group)}, partial (32%)
Length = 1025
Score = 39.3 bits (90), Expect = 0.004
Identities = 28/108 (25%), Positives = 48/108 (43%)
Frame = +1
Query: 411 ISLPKPKSYESQSRPPLASELLRTYCASRGQTTSSGLPDETRASRQILKDYIDGKLPHYE 470
ISL K S++S + L+ RG+ G+ D A+R +L D+ GK+P+Y
Sbjct: 46 ISLYKIPSFDS------VDDFLQIVATVRGKLKKGGIVDIDAAARIVLHDWNIGKIPYYT 207
Query: 471 MPPGLSTQELASEDSNEHDQVNPHVSDASDIEDSSVVETELAPKLEHV 518
MPP E + ++ + + E S + + +L+HV
Sbjct: 208 MPPVRDQGEPSEAKIVSQFSKEFNIDEVYNSETSFIGNLKPTVELDHV 351
>TC90237 similar to GP|20466544|gb|AAM20589.1 unknown protein {Arabidopsis
thaliana}, partial (79%)
Length = 1174
Score = 38.5 bits (88), Expect = 0.007
Identities = 52/214 (24%), Positives = 82/214 (38%), Gaps = 13/214 (6%)
Frame = +1
Query: 169 SDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVREKWAEYFRAHD 228
+DL++ V D+R P+ DL+ + K ++ +NK DL ++ KW YF +
Sbjct: 61 ADLVIEVRDARIPISSINSDLQPHLSL----KRRVVALNKKDLANPNIMHKWVNYFESCK 228
Query: 229 ILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDELLARLQSEAEAIVDMRR 288
I +A + ++V TK+ EL + EAI
Sbjct: 229 QDCIPINAHSKSSV---------------------TKLLELVELKLK-----EAI----- 315
Query: 289 SSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINAL-----------VGQKKTGVT 337
S ++V VG PNVGKS IN++ K+ V
Sbjct: 316 ---------------SKEPTLLVMVVGVPNVGKSCLINSIHQIAHSRFPVQEKMKRAAVG 450
Query: 338 STPGKTKHFQTLIISEK--LILCDCPGLVFPSFS 369
PG T+ I+ K + + D PG++ PS S
Sbjct: 451 PLPGVTQDIAGFKIANKPSIYVLDTPGVLVPSIS 552
>TC86616 similar to GP|7643796|gb|AAF65513.1| GTP-binding protein {Capsicum
annuum}, partial (57%)
Length = 886
Score = 35.0 bits (79), Expect = 0.081
Identities = 16/41 (39%), Positives = 25/41 (60%)
Frame = +2
Query: 288 RSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINAL 328
+S+ S ++ + + SSH+ +G VG PNVGKS+ N L
Sbjct: 116 KSAKSKEAPVERPILGRFSSHLKIGIVGLPNVGKSTLFNTL 238
>TC88185 homologue to PIR|T09368|T09368 GTP-binding protein F23K16.150 -
Arabidopsis thaliana, complete
Length = 1734
Score = 33.5 bits (75), Expect = 0.24
Identities = 22/68 (32%), Positives = 33/68 (48%), Gaps = 7/68 (10%)
Frame = +3
Query: 304 SSSSHVVVGFVGYPNVGKSSTINALVG-------QKKTGVTSTPGKTKHFQTLIISEKLI 356
+ S VG VG+P+VGKS+ +N L G + T +T PG + K+
Sbjct: 327 TKSGDARVGLVGFPSVGKSTLLNKLTGTFSEVASYEFTTLTCIPGVITY-----RGAKIQ 491
Query: 357 LCDCPGLV 364
L D PG++
Sbjct: 492 LLDLPGII 515
>TC88979 similar to PIR|H96601|H96601 hypothetical protein T6H22.14
[imported] - Arabidopsis thaliana, partial (89%)
Length = 1616
Score = 32.7 bits (73), Expect = 0.40
Identities = 17/41 (41%), Positives = 22/41 (53%)
Frame = +3
Query: 300 ASVSSSSSHVVVGFVGYPNVGKSSTINALVGQKKTGVTSTP 340
+S S S + G VG PNVGKS+ NA+V K + P
Sbjct: 177 SSSSKISMSLKAGIVGLPNVGKSTLFNAVVENGKAQAANFP 299
>CB891834 homologue to PIR|C96751|C96 probable GTP-binding protein F28P22.15
[imported] - Arabidopsis thaliana, partial (43%)
Length = 518
Score = 31.2 bits (69), Expect = 1.2
Identities = 20/64 (31%), Positives = 30/64 (46%), Gaps = 7/64 (10%)
Frame = +3
Query: 308 HVVVGFVGYPNVGKSSTINALVGQKK-------TGVTSTPGKTKHFQTLIISEKLILCDC 360
H V +G+P+VGKS+ + L G T +T PG + T K+ L D
Sbjct: 165 HGRVSLIGFPSVGKSTLLTLLTGTHSEAASYEFTTLTCIPGIIHYNDT-----KIQLLDL 329
Query: 361 PGLV 364
PG++
Sbjct: 330 PGII 341
>TC81494 similar to GP|13173408|gb|AAK14395.1 response regulator protein
{Dianthus caryophyllus}, partial (53%)
Length = 793
Score = 30.8 bits (68), Expect = 1.5
Identities = 23/63 (36%), Positives = 31/63 (48%)
Frame = +1
Query: 362 GLVFPSFSSSRYEMITCGVLPIDRMTQHRECVQVVANRVPRHVIEEIYNISLPKPKSYES 421
G FPSF SS+ M T V+ I +Q + V + V R VIE + IS K + +S
Sbjct: 4 GTRFPSFFSSKRVMETNSVVSIGVDSQEVHVLAVDDSLVDRKVIERLLKISACKVTAVDS 183
Query: 422 QSR 424
R
Sbjct: 184 GIR 192
>AW774874 similar to PIR|T46039|T4 hypothetical protein T16K5.80 -
Arabidopsis thaliana, partial (9%)
Length = 385
Score = 30.0 bits (66), Expect = 2.6
Identities = 21/85 (24%), Positives = 37/85 (42%)
Frame = +2
Query: 242 VLEGKKLGSSQADNMASADNPDTKIYGRDELLARLQSEAEAIVDMRRSSGSSKSSDDNAS 301
V+ + GS M+ A + ++ R LL R I ++RR+ ++
Sbjct: 131 VVSARGRGSGGQGFMSGAGETELQLQRR-RLLDRRNYLLTQIEEVRRTRAVQRAGRKRQG 307
Query: 302 VSSSSSHVVVGFVGYPNVGKSSTIN 326
SS+ + VGY N GKS+ ++
Sbjct: 308 GSSAQRLATIAVVGYTNAGKSTLVS 382
>AL385647
Length = 487
Score = 29.6 bits (65), Expect = 3.4
Identities = 13/31 (41%), Positives = 18/31 (57%)
Frame = +1
Query: 417 KSYESQSRPPLASELLRTYCASRGQTTSSGL 447
K ++ + R L S LLRT+C GQ SG+
Sbjct: 355 KQHQQEQRGLLYSTLLRTFCEQLGQVVQSGV 447
>TC87351 similar to GP|10176988|dbj|BAB10220. GTP-binding protein-like
{Arabidopsis thaliana}, partial (81%)
Length = 2266
Score = 29.6 bits (65), Expect = 3.4
Identities = 21/79 (26%), Positives = 38/79 (47%), Gaps = 2/79 (2%)
Frame = +3
Query: 268 GRDELLARLQSEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVV--GFVGYPNVGKSSTI 325
G +L LQ E + + S+ + D+++ + S + + VG PNVGKS+ +
Sbjct: 834 GMHDLFLSLQPVLEDYM-LNESAKENTCGDESSFPEADESKLPLQLAIVGRPNVGKSTLL 1010
Query: 326 NALVGQKKTGVTSTPGKTK 344
N L+ + + V G T+
Sbjct: 1011NTLLQEDRVLVGPEAGLTR 1067
Score = 29.3 bits (64), Expect = 4.5
Identities = 17/49 (34%), Positives = 27/49 (54%)
Frame = +3
Query: 292 SSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVGQKKTGVTSTP 340
+ K + D V S V+ +G PNVGKS+ N L+ +++ V +TP
Sbjct: 312 TKKKNLDFTKVEISRLPTVI-ILGRPNVGKSALFNRLIRRREALVYNTP 455
>TC77181 similar to GP|510190|emb|CAA82196.1| chloroplast outer envelope
protein 34 {Pisum sativum}, partial (96%)
Length = 1118
Score = 29.6 bits (65), Expect = 3.4
Identities = 16/56 (28%), Positives = 34/56 (60%), Gaps = 5/56 (8%)
Frame = +2
Query: 314 VGYPNVGKSSTINALVGQKKTGVTSTPGKTKHFQTLIISE-----KLILCDCPGLV 364
+G VGKSST+N+++G++ + +P +++ + +++S L + D PGL+
Sbjct: 299 MGKGGVGKSSTVNSIIGERVVAI--SPFQSEGPRPVMVSRARAGFTLNIIDTPGLI 460
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.313 0.130 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,510,719
Number of Sequences: 36976
Number of extensions: 239403
Number of successful extensions: 1187
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 1164
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1184
length of query: 589
length of database: 9,014,727
effective HSP length: 101
effective length of query: 488
effective length of database: 5,280,151
effective search space: 2576713688
effective search space used: 2576713688
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC147405.9


