

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146909.9 - phase: 0
(229 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
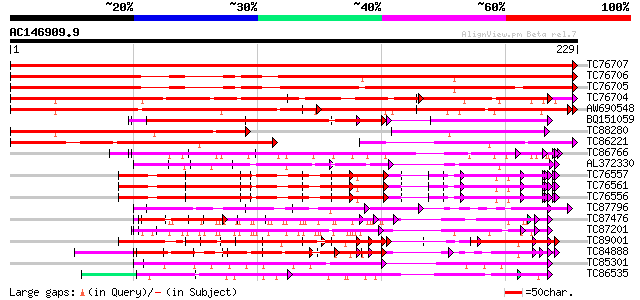
Score E
Sequences producing significant alignments: (bits) Value
TC76707 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulat... 516 e-147
TC76706 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulat... 375 e-105
TC76705 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulat... 327 2e-90
TC76704 homologue to SP|Q09134|GRPA_MEDFA Abscisic acid and envi... 247 2e-66
AW690548 similar to SP|Q09134|GRPA Abscisic acid and environment... 171 2e-43
BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melano... 117 5e-27
TC88280 weakly similar to GP|7024449|dbj|BAA92155.1 glycine-rich... 115 1e-26
TC86221 similar to PIR|T09608|T09608 environmental stress-induce... 110 4e-25
TC86766 weakly similar to PIR|T06291|T06291 extensin homolog T9E... 108 2e-24
AL372330 similar to GP|21322752|dbj cold shock protein-1 {Tritic... 104 3e-23
TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 99 1e-21
TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 99 1e-21
TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 99 1e-21
TC87796 weakly similar to SP|P03211|EBN1_EBV EBNA-1 nuclear prot... 90 8e-19
TC87476 homologue to PIR|T11622|T11622 extensin class 1 precurso... 89 1e-18
TC87201 similar to PIR|F86388|F86388 hypothetical protein AAG292... 87 7e-18
TC89001 similar to GP|5726567|gb|AAD48471.1| glycine-rich RNA-bi... 86 9e-18
TC84888 similar to GP|21322752|dbj|BAB78536. cold shock protein-... 86 1e-17
TC85301 similar to GP|8096255|dbj|BAA96106.1 TrPRP2 {Trifolium r... 82 2e-16
TC86535 similar to GP|9758076|dbj|BAB08520.1 contains similarity... 80 5e-16
>TC76707 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulated protein
CORA. [Alfalfa] {Medicago sativa}, partial (87%)
Length = 780
Score = 516 bits (1328), Expect = e-147
Identities = 228/229 (99%), Positives = 229/229 (99%)
Frame = +1
Query: 1 MDSKKAILMLGLLAMALISSVMSARDLTETSTDTKKEVVEKTNEVNDAKYGGYGGGYNHG 60
+DSKKAILMLGLLAMALISSVMSARDLTETSTDTKKEVVEKTNEVNDAKYGGYGGGYNHG
Sbjct: 7 VDSKKAILMLGLLAMALISSVMSARDLTETSTDTKKEVVEKTNEVNDAKYGGYGGGYNHG 186
Query: 61 GGGYNGGGYNHGGGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGYNGGGYNHGGGGYNHGG 120
GGGYNGGGYNHGGGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGYNGGGYNHGGGGYNHGG
Sbjct: 187 GGGYNGGGYNHGGGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGYNGGGYNHGGGGYNHGG 366
Query: 121 GGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGGYN 180
GGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGGYN
Sbjct: 367 GGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGGYN 546
Query: 181 HGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGGGGHGGHGAEQTEDKTQN 229
HGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGGGGHGGHGAEQTEDKTQN
Sbjct: 547 HGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGGGGHGGHGAEQTEDKTQN 693
>TC76706 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulated protein
CORA. [Alfalfa] {Medicago sativa}, partial (94%)
Length = 1034
Score = 375 bits (964), Expect = e-105
Identities = 188/243 (77%), Positives = 193/243 (79%), Gaps = 14/243 (5%)
Frame = +1
Query: 1 MDSKKAILMLGLLAMALISSVMSARDLTETSTDTKKEVVEKTNEVNDAKYGGYGGGYNHG 60
MDSKKAILMLGLLAMALISSVMSAR+LTETSTD KKEVVEKTNEVNDAKYGGY
Sbjct: 37 MDSKKAILMLGLLAMALISSVMSARELTETSTDAKKEVVEKTNEVNDAKYGGY------- 195
Query: 61 GGGYNGGGYNHGGGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGYNGGGYNHGGGGYNHGG 120
N GGYNH GGGYN GGGYNHGGG Y++GGGGY++GG
Sbjct: 196 ----NHGGYNH---------------GGGYN--GGGYNHGGG------YHNGGGGYHNGG 294
Query: 121 GGYNHGGGGYNGGGGHGGHGG-------GGYNGGGGHGGHGAAESVAVQTEEKTNEVNDA 173
GGYNHGGGGYNGGGGHGGHGG GGYNGGGGHGGHGAAESVAVQTEEKTNEVNDA
Sbjct: 295 GGYNHGGGGYNGGGGHGGHGGYNGGGGHGGYNGGGGHGGHGAAESVAVQTEEKTNEVNDA 474
Query: 174 KYGGG-------YNHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGGGGHGGHGAEQTEDK 226
KYGGG YNHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGGGGHGGHGAEQTEDK
Sbjct: 475 KYGGGSYNHGGSYNHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGGGGHGGHGAEQTEDK 654
Query: 227 TQN 229
TQN
Sbjct: 655 TQN 663
Score = 28.1 bits (61), Expect = 2.9
Identities = 28/130 (21%), Positives = 48/130 (36%), Gaps = 21/130 (16%)
Frame = -2
Query: 111 HGGGGYNHGGGGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGGHGAAESVAVQTEEKTNEV 170
H G + H GG Y G Y G + HG ++ H H + A Q ++ +
Sbjct: 586 HRGCNHLHRGGNYLLHGCNYLHHGCNFHHGCNYHHHIWHHLLH*SFPQFARQQIQQHRDH 407
Query: 171 NDAKYGGGYN---------------------HGGGSYNHGGGSYHHGGGGYNHGGGGHGG 209
+D + ++ H G ++ H G++HH G ++ G H
Sbjct: 406 HDLLHHCNHHDLLHHCNHRDLRGLHHHCNHLHHGCNHLHRCGNHHHRCGNHHRGYNLHHC 227
Query: 210 HGGGGHGGHG 219
+ G+ HG
Sbjct: 226 NHHRGYNHHG 197
>TC76705 similar to SP|Q07202|CORA_MEDSA Cold and drought-regulated protein
CORA. [Alfalfa] {Medicago sativa}, partial (80%)
Length = 854
Score = 327 bits (838), Expect = 2e-90
Identities = 171/232 (73%), Positives = 177/232 (75%), Gaps = 3/232 (1%)
Frame = +2
Query: 1 MDSKKAILMLGLLAMALISSVMSARDLTETSTDTKKEVVEKTNEVNDAKYGGYGGGYNHG 60
MDSKKAILMLGLLAMALISSVMSARDLTETSTD KKEVVEKTNEVNDAKYGGY
Sbjct: 47 MDSKKAILMLGLLAMALISSVMSARDLTETSTDAKKEVVEKTNEVNDAKYGGY------- 205
Query: 61 GGGYNGGGYNHGGGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGYNGGGYNHGGGGYNHGG 120
N GGYNH GGGYN GGGYNHGGG H GGGY++GG
Sbjct: 206 ----NHGGYNH---------------GGGYN--GGGYNHGGG-------YHNGGGYHNGG 301
Query: 121 GGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGG-- 178
GGY++GGGGYNGGGGHGGHGG YN GGGHGG GAAESVAVQTEEKTNEVNDA+ GGG
Sbjct: 302 GGYHNGGGGYNGGGGHGGHGG--YNRGGGHGGRGAAESVAVQTEEKTNEVNDARNGGGGG 475
Query: 179 -YNHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGGGGHGGHGAEQTEDKTQN 229
+N GG SYNHG GSYHHG GGYNHGG GGHGGGGHG HGAEQTEDKTQN
Sbjct: 476 SFNKGGASYNHGRGSYHHG*GGYNHGG---GGHGGGGHGSHGAEQTEDKTQN 622
>TC76704 homologue to SP|Q09134|GRPA_MEDFA Abscisic acid and environmental
stress inducible protein. [Sickle medic] {Medicago
falcata}, partial (81%)
Length = 771
Score = 247 bits (631), Expect = 2e-66
Identities = 131/170 (77%), Positives = 139/170 (81%), Gaps = 3/170 (1%)
Frame = +2
Query: 1 MDSKKAILMLGLLAMAL-ISSVMSARDLTETSTDTKKEVVEKTNEVNDAKYGG--YGGGY 57
MDSKKAIL+LGLLAM L ISS +SARDLTET++D KKEVVEKTNEVNDAKYGG GGGY
Sbjct: 38 MDSKKAILILGLLAMVLLISSEVSARDLTETTSDAKKEVVEKTNEVNDAKYGGGYNGGGY 217
Query: 58 NHGGGGYNGGGYNHGGGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGYNGGGYNHGGGGYN 117
NHGGG YNGGGYNHGGGGYNNGGG YNHGGGGYNN GGGYNHGGGGYNGGGYNHGGGGYN
Sbjct: 218 NHGGG-YNGGGYNHGGGGYNNGGG-YNHGGGGYNN-GGGYNHGGGGYNGGGYNHGGGGYN 388
Query: 118 HGGGGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGGHGAAESVAVQTEEKT 167
GGGYNHGGGGYN GG GG Y+ G H AE VA+Q ++ T
Sbjct: 389 --GGGYNHGGGGYNHGG-----GGCQYHCHGRCCSH--AEFVAMQAKDNT 511
Score = 108 bits (269), Expect = 2e-24
Identities = 69/144 (47%), Positives = 74/144 (50%), Gaps = 27/144 (18%)
Frame = +2
Query: 113 GGGYNHGGGGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGGHGAAESVAVQTEEKTNEVND 172
GGGYN GGGYNHGGG YNGGG + HGGGGYN G
Sbjct: 191 GGGYN--GGGYNHGGG-YNGGGYN--HGGGGYNNG------------------------- 280
Query: 173 AKYGGGYNHGGGSYNHGGGSYHHG-----GGGYNHGGGGHGG----HGGGG--HGGHG-- 219
GGYNHGGG YN+GGG Y+HG GGGYNHGGGG+ G HGGGG HGG G
Sbjct: 281 ----GGYNHGGGGYNNGGG-YNHGGGGYNGGGYNHGGGGYNGGGYNHGGGGYNHGGGGCQ 445
Query: 220 --------------AEQTEDKTQN 229
A Q +D TQN
Sbjct: 446 YHCHGRCCSHAEFVAMQAKDNTQN 517
Score = 88.2 bits (217), Expect = 2e-18
Identities = 42/64 (65%), Positives = 45/64 (69%), Gaps = 9/64 (14%)
Frame = +2
Query: 165 EKTNEVNDAKYGGGYNHG---------GGSYNHGGGSYHHGGGGYNHGGGGHGGHGGGGH 215
EKTNEVNDAKYGGGYN G GG YNHGGG Y++ GGGYNHGGGG+ GG H
Sbjct: 158 EKTNEVNDAKYGGGYNGGGYNHGGGYNGGGYNHGGGGYNN-GGGYNHGGGGYNNGGGYNH 334
Query: 216 GGHG 219
GG G
Sbjct: 335 GGGG 346
>AW690548 similar to SP|Q09134|GRPA Abscisic acid and environmental stress
inducible protein. [Sickle medic] {Medicago falcata},
partial (68%)
Length = 599
Score = 171 bits (434), Expect = 2e-43
Identities = 95/159 (59%), Positives = 110/159 (68%), Gaps = 33/159 (20%)
Frame = +1
Query: 1 MDSKKAILMLGLLAMAL-ISSVMSARDLTETSTDTKKEVVEKTNEVNDAKYGGYGGGYNH 59
MDSKKAIL+LGLLAM L ISS +SARDLTETS+D KKEVVEKTNEVNDAKYGG GG Y++
Sbjct: 22 MDSKKAILILGLLAMVLLISSEVSARDLTETSSDAKKEVVEKTNEVNDAKYGG-GGNYHN 198
Query: 60 GGGGYNGGGYNHGGGGYNNGGGGYN-----------------------------HGGGGY 90
GGG Y GGG +HGGGG ++GGGG + +GGGGY
Sbjct: 199 GGGHYYGGGSHHGGGGSHHGGGGCHYYCHGHCCSYAEFVAVQTEEKTNEVNDAKYGGGGY 378
Query: 91 NNGGGGYNHGGGGYNGGGYNHGGGGYNHGGGG---YNHG 126
+NGGG Y++GGG Y GGG +HGGGG +HGGGG Y HG
Sbjct: 379 HNGGGNYHNGGGHYYGGG-SHGGGGSHHGGGGCHYYCHG 492
Score = 129 bits (325), Expect = 7e-31
Identities = 72/124 (58%), Positives = 83/124 (66%), Gaps = 13/124 (10%)
Frame = +1
Query: 119 GGGGYNHGGGGYNGGGGHGGHGGGGYNGGGGH---GGH--GAAESVAVQTEEKTNEVNDA 173
GGG Y++GGG Y GGG H G GG + GGG H GH AE VAVQTEEKTNEVNDA
Sbjct: 178 GGGNYHNGGGHYYGGGSHHGGGGSHHGGGGCHYYCHGHCCSYAEFVAVQTEEKTNEVNDA 357
Query: 174 KY-GGGYNHGGGSYNHGGGSYH----HGGGGYNHGGGGHGGHGGG---GHGGHGAEQTED 225
KY GGGY++GGG+Y++GGG Y+ HGGGG +HGGGG + G H A Q ED
Sbjct: 358 KYGGGGYHNGGGNYHNGGGHYYGGGSHGGGGSHHGGGGCHYYCHGHCCSHAEFVAVQAED 537
Query: 226 KTQN 229
KTQN
Sbjct: 538 KTQN 549
Score = 72.4 bits (176), Expect = 1e-13
Identities = 40/67 (59%), Positives = 46/67 (67%), Gaps = 4/67 (5%)
Frame = +1
Query: 165 EKTNEVNDAKYGGGYN-HGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGGG---GHGGHGA 220
EKTNEVNDAKYGGG N H GG + +GGGS HHGGGG +HGGGG + G + A
Sbjct: 142 EKTNEVNDAKYGGGGNYHNGGGHYYGGGS-HHGGGGSHHGGGGCHYYCHGHCCSYAEFVA 318
Query: 221 EQTEDKT 227
QTE+KT
Sbjct: 319 VQTEEKT 339
>BQ151059 similar to GP|7291525|gb|A CG9888-PA {Drosophila melanogaster},
partial (26%)
Length = 308
Score = 117 bits (292), Expect = 5e-27
Identities = 60/97 (61%), Positives = 60/97 (61%)
Frame = +2
Query: 56 GYNHGGGGYNGGGYNHGGGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGYNGGGYNHGGGG 115
G N GGGG GGG GGGG GGGG GGGG GGGG GGGG GGG GGGG
Sbjct: 23 GENPGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 202
Query: 116 YNHGGGGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGG 152
GGGG GGGG GGGG GG GGGG GGGG GG
Sbjct: 203 GGGGGGGGGGGGGG--GGGGGGGGGGGGGGGGGGGGG 307
Score = 114 bits (284), Expect = 4e-26
Identities = 59/99 (59%), Positives = 59/99 (59%)
Frame = +3
Query: 56 GYNHGGGGYNGGGYNHGGGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGYNGGGYNHGGGG 115
G GGGG GGG GGGG GGGG GGGG GGGG GGGG GGG GGGG
Sbjct: 24 GKTPGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 203
Query: 116 YNHGGGGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGGHG 154
GGGG GGG GGGG GG GGGG GGGG GG G
Sbjct: 204 GGGGGGG----GGGGGGGGGGGGGGGGGGGGGGGGGGGG 308
Score = 113 bits (283), Expect = 5e-26
Identities = 61/105 (58%), Positives = 62/105 (58%)
Frame = +1
Query: 50 YGGYGGGYNHGGGGYNGGGYNHGGGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGYNGGGY 109
Y + G GGGG GGG GGGG GGGG GGGG GGGG GGGG GGG
Sbjct: 10 YPPFRGKPRGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 189
Query: 110 NHGGGGYNHGGGGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGGHG 154
GGGG GGGG GGGG GGGG GG GGGG GGGG GG G
Sbjct: 190 GGGGGG---GGGG---GGGGGGGGGGGGGGGGGGGGGGGGGGGGG 306
Score = 110 bits (274), Expect = 6e-25
Identities = 56/94 (59%), Positives = 56/94 (59%)
Frame = +3
Query: 49 KYGGYGGGYNHGGGGYNGGGYNHGGGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGYNGGG 108
K G GGG GGGG GGG GGGG GGGG GGGG GGGG GGGG GGG
Sbjct: 27 KTPGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 206
Query: 109 YNHGGGGYNHGGGGYNHGGGGYNGGGGHGGHGGG 142
GGGG GGGG GGGG GGGG GG GGG
Sbjct: 207 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 308
Score = 107 bits (268), Expect = 3e-24
Identities = 64/124 (51%), Positives = 64/124 (51%)
Frame = +2
Query: 96 GYNHGGGGYNGGGYNHGGGGYNHGGGGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGGHGA 155
G N GGGG GGG GGGG GGGG GGGG GGGG GG GGGG GGGG GG G
Sbjct: 23 GENPGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG- 199
Query: 156 AESVAVQTEEKTNEVNDAKYGGGYNHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGGGGH 215
GGG GGG GGG GGGG GGGG GG GGGG
Sbjct: 200 --------------------GGGGGGGGG----GGG----GGGGGGGGGGGGGGGGGGGG 295
Query: 216 GGHG 219
GG G
Sbjct: 296 GGGG 307
Score = 68.6 bits (166), Expect = 2e-12
Identities = 40/89 (44%), Positives = 41/89 (45%)
Frame = +2
Query: 131 NGGGGHGGHGGGGYNGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGGYNHGGGSYNHGG 190
+G GG GGGG GGGG GG G GGG GGG GG
Sbjct: 20 SGENPGGGGGGGGGGGGGGGGGGGGG-------------------GGGGGGGGGGGGGGG 142
Query: 191 GSYHHGGGGYNHGGGGHGGHGGGGHGGHG 219
G GGGG GGGG GG GGGG GG G
Sbjct: 143 GGGGGGGGGGGGGGGGGGGGGGGGGGGGG 229
Score = 48.5 bits (114), Expect = 2e-06
Identities = 25/49 (51%), Positives = 26/49 (53%)
Frame = +1
Query: 171 NDAKYGGGYNHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGGGGHGGHG 219
N + G GGG GGG GGGG GGGG GG GGGG GG G
Sbjct: 7 NYPPFRGKPRGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 153
>TC88280 weakly similar to GP|7024449|dbj|BAA92155.1 glycine-rich protein
{Citrus unshiu}, partial (56%)
Length = 603
Score = 115 bits (288), Expect = 1e-26
Identities = 63/99 (63%), Positives = 73/99 (73%), Gaps = 2/99 (2%)
Frame = +1
Query: 1 MDSKKAILMLGLLAMAL-ISSVMSARDLTETSTDTKKEVVEKTNEVNDAKYGGYGGGY-N 58
MDSKKAIL+LGLLA+ L ISS +SARDLTE S++T+KE ++TNE+NDAKYG YGGGY
Sbjct: 37 MDSKKAILILGLLAIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYPG 216
Query: 59 HGGGGYNGGGYNHGGGGYNNGGGGYNHGGGGYNNGGGGY 97
GGGG GGGY GGY GG N+GGG N GG Y
Sbjct: 217 RGGGGNYGGGYPRNRGGYPGNRGG-NYGGGYPGNRGGNY 330
Score = 55.5 bits (132), Expect = 2e-08
Identities = 28/67 (41%), Positives = 40/67 (58%), Gaps = 3/67 (4%)
Frame = +1
Query: 155 AAESVAVQTEEKTNEVNDAKYG---GGYNHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHG 211
A+ + + +++TNE+NDAKYG GGY GG N+GGG + GG + GG +GG
Sbjct: 127 ASSNTQKEADDQTNELNDAKYGSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGY 306
Query: 212 GGGHGGH 218
G GG+
Sbjct: 307 PGNRGGN 327
Score = 26.6 bits (57), Expect = 8.5
Identities = 12/29 (41%), Positives = 16/29 (54%), Gaps = 5/29 (17%)
Frame = -3
Query: 180 NHGGGSYNHGG-----GSYHHGGGGYNHG 203
+H +Y+HG SYHHG G +HG
Sbjct: 343 SHSDNNYHHGFQDNHHHSYHHGFQGNHHG 257
>TC86221 similar to PIR|T09608|T09608 environmental stress-induced protein -
alfalfa (fragment), partial (25%)
Length = 655
Score = 110 bits (275), Expect = 4e-25
Identities = 63/109 (57%), Positives = 73/109 (66%), Gaps = 1/109 (0%)
Frame = +1
Query: 1 MDSKKAILMLGLLAMALISSVMSARDLTETSTDTKKEVVEKTNEVNDAKYGGYGGGYNHG 60
M SKKAILMLGLLAM LISSV+S ETSTD + VE+T ++N K GGY GG N+G
Sbjct: 43 MVSKKAILMLGLLAMVLISSVIS-----ETSTDPNWDSVEQT-DLNYGKDGGYVGGSNNG 204
Query: 61 GGGYNGGGYNHGGGG-YNNGGGGYNHGGGGYNNGGGGYNHGGGGYNGGG 108
G NG N+G G + GG YNHGG GYN GGGG+NHG G+ G G
Sbjct: 205 GDSNNGDDSNNGDGDVHEKGGHVYNHGGEGYNGGGGGHNHGRRGHGGHG 351
Score = 73.6 bits (179), Expect = 6e-14
Identities = 40/89 (44%), Positives = 48/89 (52%), Gaps = 1/89 (1%)
Frame = +1
Query: 142 GGYNGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGGYNHGGGSYNHGGGS-YHHGGGGY 200
GGY GG +GG D+ G N+G G + GG Y+HGG GY
Sbjct: 175 GGYVGGSNNGG-------------------DSNNGDDSNNGDGDVHEKGGHVYNHGGEGY 297
Query: 201 NHGGGGHGGHGGGGHGGHGAEQTEDKTQN 229
N GGGGH HG GHGGHGA +TED+T+N
Sbjct: 298 NGGGGGHN-HGRRGHGGHGAVETEDETRN 381
>TC86766 weakly similar to PIR|T06291|T06291 extensin homolog T9E8.80 -
Arabidopsis thaliana, partial (18%)
Length = 1260
Score = 108 bits (269), Expect = 2e-24
Identities = 83/190 (43%), Positives = 86/190 (44%), Gaps = 21/190 (11%)
Frame = -1
Query: 49 KYGGYGGGYNHGGGG--YNGGG------YNHGGGGYNNGGGG----YNHGGGGYNNGGG- 95
K GG G Y GGGG Y GGG GGG GGGG Y GGG +GGG
Sbjct: 552 KTGGIGPSYTGGGGGDEYTGGGGEGYAYTGDGGGEVYTGGGGEG*AYTGDGGGDGDGGG* 373
Query: 96 -GYNHGGGGYNGGGYNHGGGGYNHGGGGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGGHG 154
G G G GGG GGGG GGGG GGG +GGGG GGG GGGG G
Sbjct: 372 *GV*CGAGDDGGGGSTQGGGG---GGGGSTQGGG--DGGGGE*TGGGGECTGGGGGG--- 217
Query: 155 AAESVAVQTEEKTNEVNDAKYGGGYNHGGGSYNHGGGSYH-----HGGGGYNHGGGGHGG 209
T V + GGG GGG GGG GGGG N G GG GG
Sbjct: 216 -------DKYGYTGGVGE*GGGGGEXGGGGGE*AGGGGGEL***GTGGGGENTGAGGGGG 58
Query: 210 HG--GGGHGG 217
G GGG GG
Sbjct: 57 GGEFGGGGGG 28
Score = 95.5 bits (236), Expect = 1e-20
Identities = 72/169 (42%), Positives = 78/169 (45%), Gaps = 12/169 (7%)
Frame = -1
Query: 50 YGGYGGGYNHGGGGYNGGGYNHGGGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGYNGG-- 107
Y G GGG + GGG G Y GGG +GGG G ++GGGG GGGG GG
Sbjct: 471 YTGDGGGEVYTGGGGEG*AYTGDGGGDGDGGG**GV*CGAGDDGGGGSTQGGGGGGGGST 292
Query: 108 -GYNHGGGGYNHGGGGYNHGGGG------YNGGGGHGGHGGGGYNGGGGH-GGHGAAESV 159
G GGGG GGGG GGGG Y GG G G GGG GGGG G G E
Sbjct: 291 QGGGDGGGGE*TGGGGECTGGGGGGDKYGYTGGVGE*GGGGGEXGGGGGE*AGGGGGEL* 112
Query: 160 AVQTEEKTNEVNDAKYGGGYNHGGGSYNHGGGSYHHGGGG--YNHGGGG 206
T GGG N G G GGG + GGGG +GG G
Sbjct: 111 **GTG-----------GGGENTGAGG-GGGGGEFGGGGGGDLTQYGGVG 1
Score = 94.4 bits (233), Expect = 3e-20
Identities = 73/189 (38%), Positives = 81/189 (42%), Gaps = 8/189 (4%)
Frame = -1
Query: 41 KTNEVNDAKYGGYGGGYNHGGGGYNGGGYNHGGGGYNNGGGGYNHGGGGYNNGGGGYNHG 100
++++ N K GG G Y G G Y GGGG GGG G Y GGG +
Sbjct: 612 RSSQKNQ*KGGGGGDAYEIPKTGGIGPSYTGGGGGDEYTGGGGE--GYAYTGDGGGEVYT 439
Query: 101 GGGYNGGGYNHGGGGYNHGGG--------GYNHGGGGYNGGGGHGGHGGGGYNGGGGHGG 152
GGG G Y GGG GGG G + GGG GGGG GGGG GGG GG
Sbjct: 438 GGGGEG*AYTGDGGGDGDGGG**GV*CGAGDDGGGGSTQGGGG----GGGGSTQGGGDGG 271
Query: 153 HGAAESVAVQTEEKTNEVNDAKYGGGYNHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGG 212
G E T + GGG GG Y + GG GGGG G GG GG
Sbjct: 270 GG----------E*TGGGGECTGGGG---GGDKYGYTGGVGE*GGGG-----GEXGGGGG 145
Query: 213 GGHGGHGAE 221
GG G E
Sbjct: 144 E*AGGGGGE 118
Score = 68.9 bits (167), Expect = 1e-12
Identities = 64/163 (39%), Positives = 70/163 (42%), Gaps = 13/163 (7%)
Frame = -1
Query: 73 GGGYNNGGGGYN---HGGGGYNNGGGGYNHGGGGYNGGGYNHGGGGYNHGGGGYNHGGGG 129
GGG GG Y GG G + GGG GG Y GGG G GY + G G GG
Sbjct: 585 GGG---GGDAYEIPKTGGIGPSYTGGG---GGDEYTGGG----GEGYAYTGDG---GGEV 445
Query: 130 YNGGGGHG----GHGGGGYNGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGGYNHGGGS 185
Y GGGG G G GGG +GGG G G G + GGGS
Sbjct: 444 YTGGGGEG*AYTGDGGGDGDGGG**G---------------------V*CGAGDDGGGGS 328
Query: 186 YNHGGGSYHHGGGGYNHGGGGHGG---HGGGGH---GGHGAEQ 222
GGG GGGG GGG GG GGGG GG G ++
Sbjct: 327 TQGGGG----GGGGSTQGGGDGGGGE*TGGGGECTGGGGGGDK 211
Score = 59.7 bits (143), Expect = 9e-10
Identities = 56/143 (39%), Positives = 60/143 (41%), Gaps = 19/143 (13%)
Frame = -1
Query: 100 GGGGYNGGGYN---HGGGGYNHGGGGYNHGGGGYNGGGGHG----GHGGGG-YNGGGGHG 151
GGGG G Y GG G ++ GGG GG Y GGGG G G GGG Y GGGG G
Sbjct: 585 GGGG--GDAYEIPKTGGIGPSYTGGG---GGDEYTGGGGEGYAYTGDGGGEVYTGGGGEG 421
Query: 152 GHGAAESVAVQTEEKTNEVNDAKYGGGYNHGGG--------SYNHGGGSYHHGGGGYNHG 203
+ GGG GGG + GGGS GGGG G
Sbjct: 420 *AYTGD------------------GGGDGDGGG**GV*CGAGDDGGGGSTQGGGGG---G 304
Query: 204 GGGHGGHGGGGHG---GHGAEQT 223
GG G G GG G G G E T
Sbjct: 303 GGSTQGGGDGGGGE*TGGGGECT 235
>AL372330 similar to GP|21322752|dbj cold shock protein-1 {Triticum
aestivum}, partial (33%)
Length = 435
Score = 104 bits (259), Expect = 3e-23
Identities = 59/109 (54%), Positives = 65/109 (59%), Gaps = 4/109 (3%)
Frame = +2
Query: 51 GGYGGGYNHGGGGYNGGGY--NHGGGGYNNGG-GGYNHGGGGYNNGGGGYNHGGGGYNGG 107
GGYGG + G GG + Y + GGGY G GGY GGY GGY GGGGY GG
Sbjct: 23 GGYGGSSSGGYGGSSSSSYPASSSGGGYGGGSSGGYGGSSGGYGGSSGGY--GGGGYGGG 196
Query: 108 GYN-HGGGGYNHGGGGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGGHGA 155
+GGGGY G GG +GGGGY GGGG+GG GGGGY GG G GA
Sbjct: 197 ASGGYGGGGYGGGSGG-GYGGGGY-GGGGYGGGGGGGY-GGDKMGALGA 334
Score = 90.9 bits (224), Expect = 4e-19
Identities = 62/147 (42%), Positives = 68/147 (46%)
Frame = +2
Query: 74 GGYNNGGGGYNHGGGGYNNGGGGYNHGGGGYNGGGYNHGGGGYNHGGGGYNHGGGGYNGG 133
GGY GG + G GG ++ + GGGY GG GGY GGY GGY GG
Sbjct: 23 GGY---GGSSSGGYGGSSSSSYPASSSGGGYGGGS----SGGYGGSSGGYGGSSGGY-GG 178
Query: 134 GGHGGHGGGGYNGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGGYNHGGGSYNHGGGSY 193
GG+GG GGY GGG YGGG GG Y GGG Y
Sbjct: 179 GGYGGGASGGYGGGG--------------------------YGGG---SGGGY--GGGGY 265
Query: 194 HHGGGGYNHGGGGHGGHGGGGHGGHGA 220
GGGGY GGGG GG+GG G GA
Sbjct: 266 --GGGGY--GGGGGGGYGGDKMGALGA 334
Score = 90.1 bits (222), Expect = 6e-19
Identities = 64/158 (40%), Positives = 74/158 (46%)
Frame = +2
Query: 65 NGGGYNHGGGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGYNGGGYNHGGGGYNHGGGGYN 124
+ GGY GG + G GG + ++ GGGY GG + GGY GGY GGY
Sbjct: 17 SSGGY---GGSSSGGYGGSSSSSYPASSSGGGY----GGGSSGGYGGSSGGYGGSSGGY- 172
Query: 125 HGGGGYNGGGGHGGHGGGGYNGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGGYNHGGG 184
GGGGY GGG GG+GGGGY GG G GGY GGG
Sbjct: 173 -GGGGY-GGGASGGYGGGGYGGGSG---------------------------GGY--GGG 259
Query: 185 SYNHGGGSYHHGGGGYNHGGGGHGGHGGGGHGGHGAEQ 222
Y GGG Y GGG GGGG+GG G G + +Q
Sbjct: 260 GY--GGGGY---GGG---GGGGYGGDKMGALGANLQKQ 349
Score = 85.9 bits (211), Expect = 1e-17
Identities = 54/133 (40%), Positives = 66/133 (49%), Gaps = 1/133 (0%)
Frame = +2
Query: 91 NNGGGGYNHGGGGYNGGGYNHGGGGYNHGGGGYNHGGGGYNGGGGHGGHGGGGYNGGGGH 150
++ GGY GG + GGY GG + + GGGY GGG GG+GG GG
Sbjct: 11 SSSSGGY----GGSSSGGY----GGSSSSSYPASSSGGGY-GGGSSGGYGGSSGGYGGSS 163
Query: 151 GGHGAAESVAVQTEEKTNEVNDAKYGGGYNHGGGSYNHGGGSYHHG-GGGYNHGGGGHGG 209
GG+G GGGY GG S +GGG Y G GGGY GG G GG
Sbjct: 164 GGYG---------------------GGGYG-GGASGGYGGGGYGGGSGGGYGGGGYGGGG 277
Query: 210 HGGGGHGGHGAEQ 222
+GGGG GG+G ++
Sbjct: 278 YGGGGGGGYGGDK 316
Score = 85.1 bits (209), Expect = 2e-17
Identities = 43/81 (53%), Positives = 46/81 (56%)
Frame = +2
Query: 51 GGYGGGYNHGGGGYNGGGYNHGGGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGYNGGGYN 110
GG GGY GGY G +GGGGY G G +GGGGY G GG +GGGGY GGGY
Sbjct: 110 GGSSGGYGGSSGGYGGSSGGYGGGGYGGGASG-GYGGGGYGGGSGG-GYGGGGYGGGGYG 283
Query: 111 HGGGGYNHGGGGYNHGGGGYN 131
GGGG G GG G G N
Sbjct: 284 GGGGG---GYGGDKMGALGAN 337
>TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 912
Score = 99.0 bits (245), Expect = 1e-21
Identities = 55/95 (57%), Positives = 60/95 (62%)
Frame = +1
Query: 45 VNDAKYGGYGGGYNHGGGGYNGGGYNHGGGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGY 104
VN A+ G GGG GGGG GGGY GGGGY GY GGGGY GGG GGGGY
Sbjct: 478 VNQAQSRGSGGG---GGGGRGGGGYGGGGGGYGGERRGYG-GGGGY---GGG---GGGGY 627
Query: 105 NGGGYNHGGGGYNHGGGGYNHGGGGYNGGGGHGGH 139
G GGGGY+ GGGG +GGGGY+ GGG GG+
Sbjct: 628 --GERRGGGGGYSRGGGGGGYGGGGYSRGGGDGGY 726
Score = 98.2 bits (243), Expect = 2e-21
Identities = 49/82 (59%), Positives = 53/82 (63%)
Frame = +1
Query: 72 GGGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGYNGGGYNHGGGGYNHGGGGYNHGGGGYN 131
GGGG GGGGY GGGGY GY GGGGY GGG GGGY G GGGGY+
Sbjct: 508 GGGGGGRGGGGYGGGGGGYGGERRGYG-GGGGYGGGG----GGGYGERRG----GGGGYS 660
Query: 132 GGGGHGGHGGGGYNGGGGHGGH 153
GGG GG+GGGGY+ GGG GG+
Sbjct: 661 RGGGGGGYGGGGYSRGGGDGGY 726
Score = 84.7 bits (208), Expect = 3e-17
Identities = 51/111 (45%), Positives = 56/111 (49%)
Frame = +1
Query: 98 NHGGGGYNGGGYNHGGGGYNHGGGGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGGHGAAE 157
+ G GG GGG GGGGY GGGGY GY GGGG+GG GGGGY G GG
Sbjct: 493 SRGSGGGGGGG--RGGGGYGGGGGGYGGERRGYGGGGGYGGGGGGGY--GERRGG----- 645
Query: 158 SVAVQTEEKTNEVNDAKYGGGYNHGGGSYNHGGGSYHHGGGGYNHGGGGHG 208
GGGY+ GGG GGG +GGGGY+ GGG G
Sbjct: 646 ------------------GGGYSRGGG----GGG---YGGGGYSRGGGDGG 723
Score = 79.0 bits (193), Expect = 1e-15
Identities = 45/100 (45%), Positives = 49/100 (49%)
Frame = +1
Query: 119 GGGGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGG 178
GGGG GGGGY GGGG G GY GGGG+GG G GGG
Sbjct: 508 GGGGGGRGGGGYGGGGGGYGGERRGYGGGGGYGGGG---------------------GGG 624
Query: 179 YNHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGGGGHGGH 218
Y G GGG Y GGGG +GGGG+ GGG GG+
Sbjct: 625 YGERRG----GGGGYSRGGGGGGYGGGGYS--RGGGDGGY 726
Score = 78.6 bits (192), Expect = 2e-15
Identities = 47/96 (48%), Positives = 51/96 (52%), Gaps = 5/96 (5%)
Frame = +1
Query: 94 GGGYNHGGGGYNGGGYNHGGGGYNHGGGGYNHGGGGYNGGGGHGGH-----GGGGYNGGG 148
GGG GGGG GGGY GGGGY GY GGGGY GGGG GG+ GGGGY+ GG
Sbjct: 505 GGG---GGGGRGGGGYGGGGGGYGGERRGYG-GGGGYGGGGG-GGYGERRGGGGGYSRGG 669
Query: 149 GHGGHGAAESVAVQTEEKTNEVNDAKYGGGYNHGGG 184
G GG+G GGGY+ GGG
Sbjct: 670 GGGGYG---------------------GGGYSRGGG 714
Score = 75.9 bits (185), Expect = 1e-14
Identities = 44/94 (46%), Positives = 51/94 (53%), Gaps = 3/94 (3%)
Frame = +1
Query: 131 NGGGGHGGHGGGGYNGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGGYNHGGGSYNHGG 190
+GGGG GG GGGGY GGGG G+G YGGG +GGG GG
Sbjct: 502 SGGGGGGGRGGGGYGGGGG--GYGGERR---------------GYGGGGGYGGG----GG 618
Query: 191 GSYHH---GGGGYNHGGGGHGGHGGGGHGGHGAE 221
G Y GGGGY+ GGGG GG+GGGG+ G +
Sbjct: 619 GGYGERRGGGGGYSRGGGG-GGYGGGGYSRGGGD 717
Score = 52.8 bits (125), Expect = 1e-07
Identities = 28/53 (52%), Positives = 32/53 (59%), Gaps = 3/53 (5%)
Frame = +1
Query: 170 VNDAKYGGGYNHGGGSYNHGGGSYHHGGGGY---NHGGGGHGGHGGGGHGGHG 219
VN A+ G + GGG GGG Y GGGGY G GG GG+GGGG GG+G
Sbjct: 478 VNQAQSRG--SGGGGGGGRGGGGYGGGGGGYGGERRGYGGGGGYGGGGGGGYG 630
Score = 43.9 bits (102), Expect = 5e-05
Identities = 24/47 (51%), Positives = 26/47 (55%)
Frame = +1
Query: 176 GGGYNHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGGGGHGGHGAEQ 222
GGG GGG Y GGG Y GG G GG GG+ GGG GG E+
Sbjct: 511 GGGGGRGGGGYGGGGGGY----GGERRGYGGGGGY-GGGGGGGYGER 636
Score = 40.0 bits (92), Expect = 7e-04
Identities = 22/59 (37%), Positives = 29/59 (48%)
Frame = +1
Query: 159 VAVQTEEKTNEVNDAKYGGGYNHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGGGGHGG 217
V E+ N+ +A G + + N GGGG GGGG+GG GGGG+GG
Sbjct: 397 VTFANEKSMNDAIEAMNGQDLDGRNITVNQAQSRGSGGGGGGGRGGGGYGG-GGGGYGG 570
>TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 1054
Score = 99.0 bits (245), Expect = 1e-21
Identities = 55/95 (57%), Positives = 60/95 (62%)
Frame = +1
Query: 45 VNDAKYGGYGGGYNHGGGGYNGGGYNHGGGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGY 104
VN A+ G GGG GGGG GGGY GGGGY GY GGGGY GGG GGGGY
Sbjct: 574 VNQAQSRGSGGG---GGGGRGGGGYGGGGGGYGGERRGYG-GGGGY---GGG---GGGGY 723
Query: 105 NGGGYNHGGGGYNHGGGGYNHGGGGYNGGGGHGGH 139
G GGGGY+ GGGG +GGGGY+ GGG GG+
Sbjct: 724 --GERRGGGGGYSRGGGGGGYGGGGYSRGGGDGGY 822
Score = 98.2 bits (243), Expect = 2e-21
Identities = 49/82 (59%), Positives = 53/82 (63%)
Frame = +1
Query: 72 GGGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGYNGGGYNHGGGGYNHGGGGYNHGGGGYN 131
GGGG GGGGY GGGGY GY GGGGY GGG GGGY G GGGGY+
Sbjct: 604 GGGGGGRGGGGYGGGGGGYGGERRGYG-GGGGYGGGG----GGGYGERRG----GGGGYS 756
Query: 132 GGGGHGGHGGGGYNGGGGHGGH 153
GGG GG+GGGGY+ GGG GG+
Sbjct: 757 RGGGGGGYGGGGYSRGGGDGGY 822
Score = 84.7 bits (208), Expect = 3e-17
Identities = 51/111 (45%), Positives = 56/111 (49%)
Frame = +1
Query: 98 NHGGGGYNGGGYNHGGGGYNHGGGGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGGHGAAE 157
+ G GG GGG GGGGY GGGGY GY GGGG+GG GGGGY G GG
Sbjct: 589 SRGSGGGGGGG--RGGGGYGGGGGGYGGERRGYGGGGGYGGGGGGGY--GERRGG----- 741
Query: 158 SVAVQTEEKTNEVNDAKYGGGYNHGGGSYNHGGGSYHHGGGGYNHGGGGHG 208
GGGY+ GGG GGG +GGGGY+ GGG G
Sbjct: 742 ------------------GGGYSRGGG----GGG---YGGGGYSRGGGDGG 819
Score = 79.0 bits (193), Expect = 1e-15
Identities = 45/100 (45%), Positives = 49/100 (49%)
Frame = +1
Query: 119 GGGGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGG 178
GGGG GGGGY GGGG G GY GGGG+GG G GGG
Sbjct: 604 GGGGGGRGGGGYGGGGGGYGGERRGYGGGGGYGGGG---------------------GGG 720
Query: 179 YNHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGGGGHGGH 218
Y G GGG Y GGGG +GGGG+ GGG GG+
Sbjct: 721 YGERRG----GGGGYSRGGGGGGYGGGGYS--RGGGDGGY 822
Score = 78.6 bits (192), Expect = 2e-15
Identities = 47/96 (48%), Positives = 51/96 (52%), Gaps = 5/96 (5%)
Frame = +1
Query: 94 GGGYNHGGGGYNGGGYNHGGGGYNHGGGGYNHGGGGYNGGGGHGGH-----GGGGYNGGG 148
GGG GGGG GGGY GGGGY GY GGGGY GGGG GG+ GGGGY+ GG
Sbjct: 601 GGG---GGGGRGGGGYGGGGGGYGGERRGYG-GGGGYGGGGG-GGYGERRGGGGGYSRGG 765
Query: 149 GHGGHGAAESVAVQTEEKTNEVNDAKYGGGYNHGGG 184
G GG+G GGGY+ GGG
Sbjct: 766 GGGGYG---------------------GGGYSRGGG 810
Score = 75.9 bits (185), Expect = 1e-14
Identities = 44/94 (46%), Positives = 51/94 (53%), Gaps = 3/94 (3%)
Frame = +1
Query: 131 NGGGGHGGHGGGGYNGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGGYNHGGGSYNHGG 190
+GGGG GG GGGGY GGGG G+G YGGG +GGG GG
Sbjct: 598 SGGGGGGGRGGGGYGGGGG--GYGGERR---------------GYGGGGGYGGG----GG 714
Query: 191 GSYHH---GGGGYNHGGGGHGGHGGGGHGGHGAE 221
G Y GGGGY+ GGGG GG+GGGG+ G +
Sbjct: 715 GGYGERRGGGGGYSRGGGG-GGYGGGGYSRGGGD 813
Score = 52.8 bits (125), Expect = 1e-07
Identities = 28/53 (52%), Positives = 32/53 (59%), Gaps = 3/53 (5%)
Frame = +1
Query: 170 VNDAKYGGGYNHGGGSYNHGGGSYHHGGGGY---NHGGGGHGGHGGGGHGGHG 219
VN A+ G + GGG GGG Y GGGGY G GG GG+GGGG GG+G
Sbjct: 574 VNQAQSRG--SGGGGGGGRGGGGYGGGGGGYGGERRGYGGGGGYGGGGGGGYG 726
Score = 43.9 bits (102), Expect = 5e-05
Identities = 24/47 (51%), Positives = 26/47 (55%)
Frame = +1
Query: 176 GGGYNHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGGGGHGGHGAEQ 222
GGG GGG Y GGG Y GG G GG GG+ GGG GG E+
Sbjct: 607 GGGGGRGGGGYGGGGGGY----GGERRGYGGGGGY-GGGGGGGYGER 732
Score = 40.0 bits (92), Expect = 7e-04
Identities = 22/59 (37%), Positives = 29/59 (48%)
Frame = +1
Query: 159 VAVQTEEKTNEVNDAKYGGGYNHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGGGGHGG 217
V E+ N+ +A G + + N GGGG GGGG+GG GGGG+GG
Sbjct: 493 VTFANEKSMNDAIEAMNGQDLDGRNITVNQAQSRGSGGGGGGGRGGGGYGG-GGGGYGG 666
>TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 754
Score = 99.0 bits (245), Expect = 1e-21
Identities = 55/95 (57%), Positives = 60/95 (62%)
Frame = +2
Query: 45 VNDAKYGGYGGGYNHGGGGYNGGGYNHGGGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGY 104
VN A+ G GGG GGGG GGGY GGGGY GY GGGGY GGG GGGGY
Sbjct: 320 VNQAQSRGSGGG---GGGGRGGGGYGGGGGGYGGERRGYG-GGGGY---GGG---GGGGY 469
Query: 105 NGGGYNHGGGGYNHGGGGYNHGGGGYNGGGGHGGH 139
G GGGGY+ GGGG +GGGGY+ GGG GG+
Sbjct: 470 --GERRGGGGGYSRGGGGGGYGGGGYSRGGGDGGY 568
Score = 98.2 bits (243), Expect = 2e-21
Identities = 49/82 (59%), Positives = 53/82 (63%)
Frame = +2
Query: 72 GGGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGYNGGGYNHGGGGYNHGGGGYNHGGGGYN 131
GGGG GGGGY GGGGY GY GGGGY GGG GGGY G GGGGY+
Sbjct: 350 GGGGGGRGGGGYGGGGGGYGGERRGYG-GGGGYGGGG----GGGYGERRG----GGGGYS 502
Query: 132 GGGGHGGHGGGGYNGGGGHGGH 153
GGG GG+GGGGY+ GGG GG+
Sbjct: 503 RGGGGGGYGGGGYSRGGGDGGY 568
Score = 84.7 bits (208), Expect = 3e-17
Identities = 51/111 (45%), Positives = 56/111 (49%)
Frame = +2
Query: 98 NHGGGGYNGGGYNHGGGGYNHGGGGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGGHGAAE 157
+ G GG GGG GGGGY GGGGY GY GGGG+GG GGGGY G GG
Sbjct: 335 SRGSGGGGGGG--RGGGGYGGGGGGYGGERRGYGGGGGYGGGGGGGY--GERRGG----- 487
Query: 158 SVAVQTEEKTNEVNDAKYGGGYNHGGGSYNHGGGSYHHGGGGYNHGGGGHG 208
GGGY+ GGG GGG +GGGGY+ GGG G
Sbjct: 488 ------------------GGGYSRGGG----GGG---YGGGGYSRGGGDGG 565
Score = 79.0 bits (193), Expect = 1e-15
Identities = 45/100 (45%), Positives = 49/100 (49%)
Frame = +2
Query: 119 GGGGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGG 178
GGGG GGGGY GGGG G GY GGGG+GG G GGG
Sbjct: 350 GGGGGGRGGGGYGGGGGGYGGERRGYGGGGGYGGGG---------------------GGG 466
Query: 179 YNHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGGGGHGGH 218
Y G GGG Y GGGG +GGGG+ GGG GG+
Sbjct: 467 YGERRG----GGGGYSRGGGGGGYGGGGYS--RGGGDGGY 568
Score = 78.6 bits (192), Expect = 2e-15
Identities = 47/96 (48%), Positives = 51/96 (52%), Gaps = 5/96 (5%)
Frame = +2
Query: 94 GGGYNHGGGGYNGGGYNHGGGGYNHGGGGYNHGGGGYNGGGGHGGH-----GGGGYNGGG 148
GGG GGGG GGGY GGGGY GY GGGGY GGGG GG+ GGGGY+ GG
Sbjct: 347 GGG---GGGGRGGGGYGGGGGGYGGERRGYG-GGGGYGGGGG-GGYGERRGGGGGYSRGG 511
Query: 149 GHGGHGAAESVAVQTEEKTNEVNDAKYGGGYNHGGG 184
G GG+G GGGY+ GGG
Sbjct: 512 GGGGYG---------------------GGGYSRGGG 556
Score = 75.9 bits (185), Expect = 1e-14
Identities = 44/94 (46%), Positives = 51/94 (53%), Gaps = 3/94 (3%)
Frame = +2
Query: 131 NGGGGHGGHGGGGYNGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGGYNHGGGSYNHGG 190
+GGGG GG GGGGY GGGG G+G YGGG +GGG GG
Sbjct: 344 SGGGGGGGRGGGGYGGGGG--GYGGERR---------------GYGGGGGYGGG----GG 460
Query: 191 GSYHH---GGGGYNHGGGGHGGHGGGGHGGHGAE 221
G Y GGGGY+ GGGG GG+GGGG+ G +
Sbjct: 461 GGYGERRGGGGGYSRGGGG-GGYGGGGYSRGGGD 559
Score = 52.8 bits (125), Expect = 1e-07
Identities = 28/53 (52%), Positives = 32/53 (59%), Gaps = 3/53 (5%)
Frame = +2
Query: 170 VNDAKYGGGYNHGGGSYNHGGGSYHHGGGGY---NHGGGGHGGHGGGGHGGHG 219
VN A+ G + GGG GGG Y GGGGY G GG GG+GGGG GG+G
Sbjct: 320 VNQAQSRG--SGGGGGGGRGGGGYGGGGGGYGGERRGYGGGGGYGGGGGGGYG 472
Score = 43.9 bits (102), Expect = 5e-05
Identities = 24/47 (51%), Positives = 26/47 (55%)
Frame = +2
Query: 176 GGGYNHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGGGGHGGHGAEQ 222
GGG GGG Y GGG Y GG G GG GG+ GGG GG E+
Sbjct: 353 GGGGGRGGGGYGGGGGGY----GGERRGYGGGGGY-GGGGGGGYGER 478
Score = 40.0 bits (92), Expect = 7e-04
Identities = 22/59 (37%), Positives = 29/59 (48%)
Frame = +2
Query: 159 VAVQTEEKTNEVNDAKYGGGYNHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGGGGHGG 217
V E+ N+ +A G + + N GGGG GGGG+GG GGGG+GG
Sbjct: 239 VTFANEKSMNDAIEAMNGQDLDGRNITVNQAQSRGSGGGGGGGRGGGGYGG-GGGGYGG 412
>TC87796 weakly similar to SP|P03211|EBN1_EBV EBNA-1 nuclear protein.
[strain B95-8 Human herpesvirus 4] {Epstein-barr
virus}, partial (23%)
Length = 431
Score = 89.7 bits (221), Expect = 8e-19
Identities = 56/112 (50%), Positives = 63/112 (56%)
Frame = +1
Query: 56 GYNHGGGGYNGGGYNHGGGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGYNGGGYNHGGGG 115
G +HGGG GGG GGGG G GG + G GY +GGG GGGY GG GGGG
Sbjct: 1 GRDHGGGYGGGGGSGGGGGG---GAGGAH--GVGYGSGGG----TGGGYGGGSPGGGGGG 153
Query: 116 YNHGGGGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGGHGAAESVAVQTEEKT 167
GGGG GGGG +GG GG GGG GGGHGG+ A+Q E+T
Sbjct: 154 GGSGGGG---GGGGAHGGAYGGGIGGG---EGGGHGGYEP*GFNAIQCYERT 291
Score = 83.6 bits (205), Expect = 6e-17
Identities = 50/96 (52%), Positives = 54/96 (56%), Gaps = 1/96 (1%)
Frame = +1
Query: 51 GGYGGGYNHGGGGYNGGGYNHGGGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGYNGGGYN 110
GGYGGG GGGG G G HG G Y +GGG GGGY G G GGGG GG
Sbjct: 16 GGYGGGGGSGGGGGGGAGGAHGVG-YGSGGGT----GGGYGGGSPGGGGGGGGSGGG--- 171
Query: 111 HGGGGYNHGGGGYNHGGG-GYNGGGGHGGHGGGGYN 145
GGGG HGG +GGG G GGGHGG+ G+N
Sbjct: 172 -GGGGGAHGGA---YGGGIGGGEGGGHGGYEP*GFN 267
Score = 81.3 bits (199), Expect = 3e-16
Identities = 58/132 (43%), Positives = 67/132 (49%)
Frame = +1
Query: 96 GYNHGGGGYNGGGYNHGGGGYNHGGGGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGGHGA 155
G +HGGG Y GGG + GGGG GG G HG G +GGG GG+GGG GGGG GG G
Sbjct: 1 GRDHGGG-YGGGGGSGGGGG---GGAGGAHGVGYGSGGGTGGGYGGGSPGGGGGGGGSGG 168
Query: 156 AESVAVQTEEKTNEVNDAKYGGGYNHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGGGGH 215
GGG GGG++ GG+Y GGG GGG GGHGG
Sbjct: 169 --------------------GGG---GGGAH---GGAY---GGGI--GGGEGGGHGGYEP 255
Query: 216 GGHGAEQTEDKT 227
G A Q ++T
Sbjct: 256 *GFNAIQCYERT 291
Score = 81.3 bits (199), Expect = 3e-16
Identities = 49/105 (46%), Positives = 50/105 (46%)
Frame = +1
Query: 115 GYNHGGGGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGGHGAAESVAVQTEEKTNEVNDAK 174
G +HGGG GG G GGGG GG G GY GGG GG
Sbjct: 1 GRDHGGGYGGGGGSGGGGGGGAGGAHGVGYGSGGGTGGG--------------------- 117
Query: 175 YGGGYNHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGGGGHGGHG 219
YGGG GGG GGGS GGGG HGG GG GGG GGHG
Sbjct: 118 YGGGSPGGGGG---GGGSGGGGGGGGAHGGAYGGGIGGGEGGGHG 243
>TC87476 homologue to PIR|T11622|T11622 extensin class 1 precursor - cowpea,
partial (48%)
Length = 939
Score = 89.0 bits (219), Expect = 1e-18
Identities = 60/114 (52%), Positives = 61/114 (52%), Gaps = 18/114 (15%)
Frame = -1
Query: 54 GGGYNHGGGG----YNGGGYNHGGGGY--NNGGGGYNHGGGGY--NNGGGGYNHGGGG-- 103
GGG GGGG Y GGG GGGG GGGG GGGG GGGG GGGG
Sbjct: 342 GGGEGDGGGGDL*TYGGGGDGDGGGGDL*TYGGGGEGDGGGGDL*I*GGGGEGVGGGGDL 163
Query: 104 --YNGGGYNHGGGGY--NHGGGGYNHGGGG--YNGGGGHGGHGGGG--YNGGGG 149
Y GGG GGGG +GGGG GGGG Y GGG G GGGG Y GGG
Sbjct: 162 *TYGGGGDGEGGGGLL*TYGGGGDGDGGGGDLYTYGGGGEGDGGGGDLYIYGGG 1
Score = 81.6 bits (200), Expect = 2e-16
Identities = 60/135 (44%), Positives = 64/135 (46%), Gaps = 13/135 (9%)
Frame = -1
Query: 93 GGGGYNHGGGG----YNGGGYNHGGGGY--NHGGGGYNHGGGG---YNGGGGHGGHGGGG 143
GGGG GGGG Y GGG GGGG +GGGG GGGG GGGG G GGG
Sbjct: 345 GGGGEGDGGGGDL*TYGGGGDGDGGGGDL*TYGGGGEGDGGGGDL*I*GGGGEGVGGGGD 166
Query: 144 ---YNGGG-GHGGHGAAESVAVQTEEKTNEVNDAKYGGGYNHGGGSYNHGGGSYHHGGGG 199
Y GGG G GG G + YGGG + GG GG Y +GGGG
Sbjct: 165 L*TYGGGGDGEGGGGLL*T----------------YGGGGDGDGG----GGDLYTYGGGG 46
Query: 200 YNHGGGGHGGHGGGG 214
GGGG GGG
Sbjct: 45 EGDGGGGDLYIYGGG 1
Score = 80.1 bits (196), Expect = 6e-16
Identities = 53/109 (48%), Positives = 55/109 (49%), Gaps = 15/109 (13%)
Frame = -1
Query: 64 YNGGGYNHGGGGY--NNGGGGYNHGGGGY--NNGGGGYNHGGGGY----NGGGYNHGGGG 115
Y GGG GGGG GGGG GGGG GGGG GGGG GGG GGGG
Sbjct: 348 YGGGGEGDGGGGDL*TYGGGGDGDGGGGDL*TYGGGGEGDGGGGDL*I*GGGGEGVGGGG 169
Query: 116 Y--NHGGGGYNHGGGG----YNGGGGHGGHGGGGYN-GGGGHGGHGAAE 157
+GGGG GGGG Y GGG G GG Y GGGG G G +
Sbjct: 168 DL*TYGGGGDGEGGGGLL*TYGGGGDGDGGGGDLYTYGGGGEGDGGGGD 22
Score = 79.7 bits (195), Expect = 8e-16
Identities = 52/109 (47%), Positives = 59/109 (53%), Gaps = 12/109 (11%)
Frame = -1
Query: 53 YGGGYNHGGGGYNGGGYNHGGGGYNNGGGGY--NHGGGGYNNGGGGY----NHGGGGYNG 106
YGGG GGG G +GGGG +GGGG +GGGG +GGGG GG G G
Sbjct: 348 YGGGGEGDGGG--GDL*TYGGGGDGDGGGGDL*TYGGGGEGDGGGGDL*I*GGGGEGVGG 175
Query: 107 GGY--NHGGGGYNHGGGGY--NHGGGGYNGGGGHG--GHGGGGYNGGGG 149
GG +GGGG GGGG +GGGG GGG +GGGG GGG
Sbjct: 174 GGDL*TYGGGGDGEGGGGLL*TYGGGGDGDGGGGDLYTYGGGGEGDGGG 28
Score = 77.0 bits (188), Expect = 5e-15
Identities = 53/107 (49%), Positives = 54/107 (49%), Gaps = 14/107 (13%)
Frame = -3
Query: 51 GGYGGGYNHGGGGY----NGGGYNHGGGGY--NNGGGGYNHGGGGYN--NGGGGYNHGGG 102
G YGGG GGGG GGG GGGG GGGG GGGG GGGG GGG
Sbjct: 688 G*YGGGDGDGGGGDL*I*GGGGEGEGGGGDL*PYGGGGDGEGGGGDL**YGGGGEAEGGG 509
Query: 103 GY----NGGGYNHGGGGY--NHGGGGYNHGGGGYNGGGGHGGHGGGG 143
G GGG GGGG +GGGG GGGG G GG G GG
Sbjct: 508 GDL*T*GGGGEGDGGGGDL*TYGGGGEGEGGGGDL*TYGGGGEGDGG 368
Score = 68.6 bits (166), Expect = 2e-12
Identities = 54/127 (42%), Positives = 59/127 (45%), Gaps = 11/127 (8%)
Frame = -1
Query: 104 YNGGGYNHGGGGY--NHGGGGYNHGGGG---YNGGGGHGGHGGGGYN--GGGGHGGHGAA 156
Y GGG GGGG +GGGG GGGG GGGG G GGG GGGG G G
Sbjct: 348 YGGGGEGDGGGGDL*TYGGGGDGDGGGGDL*TYGGGGEGDGGGGDL*I*GGGGEGVGGGG 169
Query: 157 ESVAVQTEEKTNEVNDAKYGGGYNHGGGSYNHGGGSYHHGGGGYNHGGGG----HGGHGG 212
+ YGGG + GG GG +GGGG GGGG +GG GG
Sbjct: 168 DL*T--------------YGGGGDGEGG----GGLL*TYGGGGDGDGGGGDLYTYGG-GG 46
Query: 213 GGHGGHG 219
G GG G
Sbjct: 45 EGDGGGG 25
Score = 65.1 bits (157), Expect = 2e-11
Identities = 46/102 (45%), Positives = 51/102 (49%), Gaps = 17/102 (16%)
Frame = -3
Query: 73 GGGYNNGGGGYNH--GGGGYNNGGGG----YNHGGGGYNGGGYN--HGGGGYNHGGGGY- 123
GGG +GGGG GGGG GGGG Y GG G GGG +GGGG GGGG
Sbjct: 679 GGGDGDGGGGDL*I*GGGGEGEGGGGDL*PYGGGGDGEGGGGDL**YGGGGEAEGGGGDL 500
Query: 124 ---NHGGGGYNGGGGHGGHGGGGYNGGGG-----HGGHGAAE 157
GG G GGG +GGGG GGG +GG G +
Sbjct: 499 *T*GGGGEGDGGGGDL*TYGGGGEGEGGGGDL*TYGGGGEGD 374
Score = 62.0 bits (149), Expect = 2e-10
Identities = 55/137 (40%), Positives = 58/137 (42%), Gaps = 10/137 (7%)
Frame = -3
Query: 93 GGGGYNHGGGGYNGGGYNHGGGGYNH-GGGGYNHGGGG----YNGGGGHGGHGGGG---- 143
GGGG G Y GG + GGG GGGG GGGG Y GGG G GGGG
Sbjct: 715 GGGGEL*R*G*YGGGDGDGGGGDL*I*GGGGEGEGGGGDL*PYGGGGD--GEGGGGDL** 542
Query: 144 YNGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGGYNHGGGSYNHGGGSYHHGGGGYNHG 203
Y GGG G G D GG G G GG +GGGG G
Sbjct: 541 YGGGGEAEGGG----------------GDL*T*GGGGEGDGG---GGDL*TYGGGGEGEG 419
Query: 204 GGGH-GGHGGGGHGGHG 219
GGG +GGGG G G
Sbjct: 418 GGGDL*TYGGGGEGDGG 368
Score = 59.3 bits (142), Expect = 1e-09
Identities = 55/146 (37%), Positives = 59/146 (39%), Gaps = 13/146 (8%)
Frame = -3
Query: 79 GGGGYNHGGGGYNNGGGGYNHGGGG---YNGGGYNHGGGGYN---HGGGGYNHGGGGY-- 130
GGGG G Y GGG GGGG GGG GGG + +GGGG GGGG
Sbjct: 715 GGGGEL*R*G*Y---GGGDGDGGGGDL*I*GGGGEGEGGGGDL*PYGGGGDGEGGGGDL* 545
Query: 131 -NGGGGHGGHGGGGY--NGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGGYNHGGGSYN 187
GGGG GGG GGGG G G GG
Sbjct: 544 *YGGGGEAEGGGGDL*T*GGGGEGDGG---------------------------GGDL*T 446
Query: 188 HGGGSYHHGGGGY--NHGGGGHGGHG 211
+GGG GGGG +GGGG G G
Sbjct: 445 YGGGGEGEGGGGDL*TYGGGGEGDGG 368
Score = 40.4 bits (93), Expect = 6e-04
Identities = 21/38 (55%), Positives = 25/38 (65%), Gaps = 2/38 (5%)
Frame = -1
Query: 53 YGGGYNHGGGGYNGGGYNHGGGGYNNGGGG--YNHGGG 88
YGGG + GGG G Y +GGGG +GGGG Y +GGG
Sbjct: 108 YGGGGDGDGGG--GDLYTYGGGGEGDGGGGDLYIYGGG 1
Score = 35.4 bits (80), Expect = 0.018
Identities = 30/88 (34%), Positives = 36/88 (40%), Gaps = 18/88 (20%)
Frame = -1
Query: 150 HGGHGAAESVAVQTEEKTNE-VNDAKYGGGYNH----------GGGSYNHGGGSYHHGGG 198
H G E V + + E V + + G Y H GG +GGG GGG
Sbjct: 495 HRVEGVKEMVGEEICKHMEEGVKEKEEEGTYRHMEVVVKGMVVGGDL*IYGGGGEGDGGG 316
Query: 199 GY--NHGGGGHGGHGGG-----GHGGHG 219
G +GGGG G GGG G GG G
Sbjct: 315 GDL*TYGGGGDGDGGGGDL*TYGGGGEG 232
>TC87201 similar to PIR|F86388|F86388 hypothetical protein AAG29221.1
[imported] - Arabidopsis thaliana, partial (41%)
Length = 991
Score = 86.7 bits (213), Expect = 7e-18
Identities = 80/198 (40%), Positives = 90/198 (45%), Gaps = 39/198 (19%)
Frame = -3
Query: 50 YGG----YGGG-----YNHGGGGYNGGGYN---HGGGG----YNNGGGGY-NHGGGG--- 89
YGG YGGG Y GG G GGG + GGGG Y GGG +GGGG
Sbjct: 572 YGGDS*TYGGGIGL**YGGGGDGEGGGGDS*T*GGGGGDL*TYCGGGGDS*TYGGGGGDL 393
Query: 90 --YNNGGGG-YNHGGGGYNGGGYN----HGGGGYNHGGGGYN---HGGGGYNGGGGH--- 136
Y GGG Y +GGGG GG +GGG Y +GGGG + +GGGG +GGGG
Sbjct: 392 *TYGGGGGDLYTYGGGGDGDGGGGDL*TYGGGVYTYGGGGGDL*IYGGGG-DGGGGDG*T 216
Query: 137 -GGHGGGGYNGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGGYNHGGGSYNHGGGS--- 192
GG GG Y GGG G YGGG + GGG GG+
Sbjct: 215 *GGGGGDLYTYGGGGGDL*I-------------------YGGGGDGGGGDG*T*GGARVV 93
Query: 193 --YHHGGGGYNHGGGGHG 208
GGGG GGGG G
Sbjct: 92 ICIRIGGGG--DGGGGDG 45
Score = 85.5 bits (210), Expect = 2e-17
Identities = 62/133 (46%), Positives = 73/133 (54%), Gaps = 32/133 (24%)
Frame = -3
Query: 51 GGYGGGYNHGGGG-----YNGGG---YNHGGGGYNNGGGGY--NHGGGGYNNGGGGYN-- 98
GG G +GGGG Y GGG Y +GGGG +GGGG +GGG Y GGGG +
Sbjct: 440 GGGGDS*TYGGGGGDL*TYGGGGGDLYTYGGGGDGDGGGGDL*TYGGGVYTYGGGGGDL* 261
Query: 99 -HGGGGYNGGGYN---HGGGG--YNHGGGGYN---HGGGGYNGGGGHG---GHG------ 140
+GGGG GGG GGGG Y +GGGG + +GGGG +GGGG G G
Sbjct: 260 IYGGGGDGGGGDG*T*GGGGGDLYTYGGGGGDL*IYGGGG-DGGGGDG*T*GGARVVICI 84
Query: 141 --GGGYNGGGGHG 151
GGG +GGGG G
Sbjct: 83 RIGGGGDGGGGDG 45
Score = 84.0 bits (206), Expect = 4e-17
Identities = 75/183 (40%), Positives = 82/183 (43%), Gaps = 21/183 (11%)
Frame = -3
Query: 53 YGGGYNHGGGG-----YNGGGYNHGGGGYN---NGGGG----YNHGGGG---YNNGGGGY 97
YGG GGG Y GGG GGGG + GGGG Y GGG Y GGG
Sbjct: 572 YGGDS*TYGGGIGL**YGGGGDGEGGGGDS*T*GGGGGDL*TYCGGGGDS*TYGGGGGDL 393
Query: 98 -NHGGGGYNGGGYNHGGGGYNHGGGGY--NHGGGGYNGGGGHGG---HGGGGYNGGGGHG 151
+GGGG G Y +GGGG GGGG +GGG Y GGG G +GGGG +GGGG G
Sbjct: 392 *TYGGGG--GDLYTYGGGGDGDGGGGDL*TYGGGVYTYGGGGGDL*IYGGGG-DGGGGDG 222
Query: 152 GHGAAESVAVQTEEKTNEVNDAKYGGGYNHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHG 211
GGG GG Y +GGG GG GGG G G
Sbjct: 221 *---------------------T*GGG---GGDLYTYGGG-----GGDL*IYGGGGDGGG 129
Query: 212 GGG 214
G G
Sbjct: 128 GDG 120
Score = 71.2 bits (173), Expect = 3e-13
Identities = 77/204 (37%), Positives = 86/204 (41%), Gaps = 44/204 (21%)
Frame = -3
Query: 60 GGGG--YNGGGYNHGGGGYNNGGGGYNHGGGGYN---NGGGGYNHGGGG----YNGGGYN 110
GGGG Y G Y GG GG G GG + GG +GGG Y GGG
Sbjct: 683 GGGGELYW*GLYGVGGDL*T*GGDL*T*GAGGGDL*IYGGDS*TYGGGIGL**YGGGGDG 504
Query: 111 HGGGGYN---HGGGG----YNHGGGG---YNGGGG----HGGHGGGGYNGGGGHGGHGAA 156
GGGG + GGGG Y GGG Y GGGG +GG GG Y GGG G G
Sbjct: 503 EGGGGDS*T*GGGGGDL*TYCGGGGDS*TYGGGGGDL*TYGGGGGDLYTYGGGGDGDGGG 324
Query: 157 ESVAVQTEEKTNEVNDAKYGGG-YNHGGGSYNHGGGSYHHGGGGYNHGGG-----GHGG- 209
+ YGGG Y +GGG GG +GGGG + GGG G GG
Sbjct: 323 GDL*T-------------YGGGVYTYGGG----GGDL*IYGGGG-DGGGGDG*T*GGGGG 198
Query: 210 --------------HGGGGHGGHG 219
+GGGG GG G
Sbjct: 197 DLYTYGGGGGDL*IYGGGGDGGGG 126
>TC89001 similar to GP|5726567|gb|AAD48471.1| glycine-rich RNA-binding
protein {Glycine max}, partial (90%)
Length = 488
Score = 86.3 bits (212), Expect = 9e-18
Identities = 44/71 (61%), Positives = 47/71 (65%)
Frame = +3
Query: 72 GGGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGYNGGGYNHGGGGYNHGGGGYNHGGGGYN 131
GGGG GGGGY GGGGY GGGGY GGGG GGY+ GGG +GGG G GY
Sbjct: 297 GGGGRGGGGGGYG-GGGGYGGGGGGYG-GGGGRRDGGYSRSGGGGGYGGG----GDRGYG 458
Query: 132 GGGGHGGHGGG 142
GGGG GG+GGG
Sbjct: 459 GGGG-GGYGGG 488
Score = 85.5 bits (210), Expect = 2e-17
Identities = 44/72 (61%), Positives = 47/72 (65%), Gaps = 2/72 (2%)
Frame = +3
Query: 78 NGGGGYNHGGGGYNNGGGGYNHGGGGYNGGGYNHGGGGYNHGGGGYNHGGGGYNGGG--G 135
+GGGG GGGGY GGGGY GGGGY GGG GGY+ G GGGGY GGG G
Sbjct: 294 SGGGGRGGGGGGYG-GGGGYGGGGGGYGGGG-GRRDGGYSRSG-----GGGGYGGGGDRG 452
Query: 136 HGGHGGGGYNGG 147
+GG GGGGY GG
Sbjct: 453 YGGGGGGGYGGG 488
Score = 83.6 bits (205), Expect = 6e-17
Identities = 41/67 (61%), Positives = 44/67 (65%)
Frame = +3
Query: 86 GGGGYNNGGGGYNHGGGGYNGGGYNHGGGGYNHGGGGYNHGGGGYNGGGGHGGHGGGGYN 145
GGGG GGGGY GGGGY GGG +GGGG GG GGGG GGGG G+GGG
Sbjct: 297 GGGGRGGGGGGYG-GGGGYGGGGGGYGGGGGRRDGGYSRSGGGGGYGGGGDRGYGGG--- 464
Query: 146 GGGGHGG 152
GGGG+GG
Sbjct: 465 GGGGYGG 485
Score = 81.6 bits (200), Expect = 2e-16
Identities = 48/84 (57%), Positives = 52/84 (61%)
Frame = +3
Query: 45 VNDAKYGGYGGGYNHGGGGYNGGGYNHGGGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGY 104
VN+A+ G GGG GGGG GY GGGGY GGGGY GGGG +GG + GGGGY
Sbjct: 270 VNEAQSRGSGGGGRGGGGG----GYG-GGGGYGGGGGGYG-GGGGRRDGGYSRSGGGGGY 431
Query: 105 NGGGYNHGGGGYNHGGGGYNHGGG 128
GGG GGG GGGGY GGG
Sbjct: 432 GGGGDRGYGGG---GGGGY--GGG 488
Score = 77.4 bits (189), Expect = 4e-15
Identities = 44/95 (46%), Positives = 50/95 (52%)
Frame = +3
Query: 119 GGGGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGG 178
GGGG GGGGY GGGG+GG GGGGY GGGG + GG
Sbjct: 297 GGGGRGGGGGGYGGGGGYGG-GGGGYGGGGG------------------------RRDGG 401
Query: 179 YNHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGGG 213
Y+ GG GGG + GGG +GGGG GG+GGG
Sbjct: 402 YSRSGG----GGG--YGGGGDRGYGGGGGGGYGGG 488
Score = 75.9 bits (185), Expect = 1e-14
Identities = 38/64 (59%), Positives = 42/64 (65%), Gaps = 1/64 (1%)
Frame = +3
Query: 92 NGGGGYNHGGGGYNGGG-YNHGGGGYNHGGGGYNHGGGGYNGGGGHGGHGGGGYNGGGGH 150
+GGGG GGGGY GGG Y GGGGY GGG + GGY+ GG GG+GGGG G GG
Sbjct: 294 SGGGGRGGGGGGYGGGGGYGGGGGGYGGGGGRRD---GGYSRSGGGGGYGGGGDRGYGGG 464
Query: 151 GGHG 154
GG G
Sbjct: 465 GGGG 476
Score = 75.5 bits (184), Expect = 2e-14
Identities = 44/89 (49%), Positives = 48/89 (53%)
Frame = +3
Query: 131 NGGGGHGGHGGGGYNGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGGYNHGGGSYNHGG 190
+GGGG GG GGGGY GGGG+GG G GGY GGG GG
Sbjct: 294 SGGGGRGG-GGGGYGGGGGYGGGG----------------------GGYG-GGGGRRDGG 401
Query: 191 GSYHHGGGGYNHGGGGHGGHGGGGHGGHG 219
S GGGGY GGGG G+GGGG GG+G
Sbjct: 402 YSRSGGGGGY--GGGGDRGYGGGGGGGYG 482
Score = 63.9 bits (154), Expect = 5e-11
Identities = 41/89 (46%), Positives = 44/89 (49%)
Frame = +3
Query: 103 GYNGGGYNHGGGGYNHGGGGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGGHGAAESVAVQ 162
G GGG GGGGY GGG +GGGG G GG GG GGY+ GG GG+G
Sbjct: 291 GSGGGGRGGGGGGYGGGGG---YGGGG-GGYGGGGGRRDGGYSRSGGGGGYGGG------ 440
Query: 163 TEEKTNEVNDAKYGGGYNHGGGSYNHGGG 191
D YGGG GGG Y GGG
Sbjct: 441 --------GDRGYGGG---GGGGY--GGG 488
Score = 54.7 bits (130), Expect = 3e-08
Identities = 29/62 (46%), Positives = 31/62 (49%), Gaps = 10/62 (16%)
Frame = +3
Query: 168 NEVNDAKYGGGYNHGGGSYNHGGGSYHHGGGGYNHGG----------GGHGGHGGGGHGG 217
NE GGG GGG GGG Y GGGGY GG GG GG+GGGG G
Sbjct: 273 NEAQSRGSGGGGRGGGGGGYGGGGGYGGGGGGYGGGGGRRDGGYSRSGGGGGYGGGGDRG 452
Query: 218 HG 219
+G
Sbjct: 453 YG 458
Score = 42.0 bits (97), Expect = 2e-04
Identities = 18/36 (50%), Positives = 22/36 (61%)
Frame = +3
Query: 187 NHGGGSYHHGGGGYNHGGGGHGGHGGGGHGGHGAEQ 222
+ G G GGGG +GGGG G GGGG+GG G +
Sbjct: 285 SRGSGGGGRGGGGGGYGGGGGYGGGGGGYGGGGGRR 392
>TC84888 similar to GP|21322752|dbj|BAB78536. cold shock protein-1 {Triticum
aestivum}, partial (58%)
Length = 612
Score = 85.9 bits (211), Expect = 1e-17
Identities = 49/82 (59%), Positives = 51/82 (61%)
Frame = +2
Query: 52 GYGGGYNHGGGGYNGGGYNHGGGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGYNGGGYNH 111
GYGGG GGGGY GGGY+ G GG GGG Y GGG+ G GG GGGGY GGY
Sbjct: 299 GYGGGGGRGGGGY-GGGYDQGYGG---GGGSY---GGGFGGGRGG-GRGGGGYXQGGYG- 451
Query: 112 GGGGYNHGGGGYNHGGGGYNGG 133
GGGGY GG G G GGY GG
Sbjct: 452 GGGGYXQGGYG---GQGGYGGG 508
Score = 80.1 bits (196), Expect = 6e-16
Identities = 51/118 (43%), Positives = 59/118 (49%), Gaps = 2/118 (1%)
Frame = +2
Query: 27 LTETSTDTKKEVVEKTNEVNDAKYGGYGGGYNHGGGGYNGGGYNHGGGGYNNGGGGYNHG 86
+ E D + + ++ T G +GGGG GGGGY GGGY+ G
Sbjct: 206 IVEAGDDGRTKAIDVTGPNGAPPQGAPRQEMGYGGGG------GRGGGGY---GGGYDQG 358
Query: 87 GGGYNNGGGGYNHGGGGYNGGGYNHGGGGYNHGGGGYNHGGGGY--NGGGGHGGHGGG 142
GG GGG Y G GG GGG GGGGY GG G GGGGY G GG GG+GGG
Sbjct: 359 YGG---GGGSYGGGFGGGRGGG--RGGGGYXQGGYG---GGGGYXQGGYGGQGGYGGG 508
Score = 78.6 bits (192), Expect = 2e-15
Identities = 48/86 (55%), Positives = 51/86 (58%), Gaps = 1/86 (1%)
Frame = +2
Query: 63 GYNGGGYNHGGGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGYNGGGYNHGGGGYNHGGGG 122
GY GGG GGGGY GGGY+ G GG GGG Y GGG+ GG GGG GGGG
Sbjct: 299 GYGGGG-GRGGGGY---GGGYDQGYGG---GGGSY---GGGFGGG---RGGG---RGGGG 430
Query: 123 YNHGGGGYNGGGGHGGHGG-GGYNGG 147
Y GG G GG GG+GG GGY GG
Sbjct: 431 YXQGGYGGGGGYXQGGYGGQGGYGGG 508
Score = 74.7 bits (182), Expect = 3e-14
Identities = 44/80 (55%), Positives = 48/80 (60%), Gaps = 2/80 (2%)
Frame = +2
Query: 75 GYNNGGGGYNHGGGGYNNGGGGYNHGGGGYNGGGYNHGGGGYNHGGGGYNHGGGGY--NG 132
GY GGG GGGGY GGGY+ G GG GG Y GGG+ G GG GGGGY G
Sbjct: 299 GYGGGGG---RGGGGY---GGGYDQGYGG-GGGSY---GGGFGGGRGG-GRGGGGYXQGG 445
Query: 133 GGGHGGHGGGGYNGGGGHGG 152
GG GG+ GGY G GG+GG
Sbjct: 446 YGGGGGYXQGGYGGQGGYGG 505
Score = 73.2 bits (178), Expect = 8e-14
Identities = 43/73 (58%), Positives = 45/73 (60%)
Frame = +2
Query: 51 GGYGGGYNHGGGGYNGGGYNHGGGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGYNGGGYN 110
GGYGGGY+ G GG GG Y GGG+ G GG GGGGY GG G GGGGY GGY
Sbjct: 329 GGYGGGYDQGYGG-GGGSY---GGGFGGGRGG-GRGGGGYXQGGYG---GGGGYXQGGYG 484
Query: 111 HGGGGYNHGGGGY 123
G GGY GGGY
Sbjct: 485 -GQGGY---GGGY 511
Score = 72.0 bits (175), Expect = 2e-13
Identities = 44/95 (46%), Positives = 49/95 (51%), Gaps = 2/95 (2%)
Frame = +2
Query: 87 GGGYNNGGGGYNHGGGGYNGGGYNHGGGGYNHGGGGYNHGGGGYNGGGGH--GGHGGGGY 144
GGG GGGGY GGGY+ GY GGG Y GGG+ G GG GGGG+ GG+GGGG
Sbjct: 305 GGGGGRGGGGY---GGGYD-QGYGGGGGSY---GGGFGGGRGGGRGGGGYXQGGYGGGGG 463
Query: 145 NGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGGY 179
GG+GG G YGGGY
Sbjct: 464 YXQGGYGGQGG-------------------YGGGY 511
Score = 70.5 bits (171), Expect = 5e-13
Identities = 52/126 (41%), Positives = 55/126 (43%), Gaps = 1/126 (0%)
Frame = +2
Query: 89 GYNNGGGGYNHGGGGYNGGGYNHGGGGYNHGGGGYNHGGGGYNGGGGHGGHGGGGYNGGG 148
GY GGG GGGGY GGGY+ G GG GGG Y GGG+ GG G GG GGGGY GG
Sbjct: 299 GYGGGGG---RGGGGY-GGGYDQGYGG---GGGSY---GGGFGGGRG-GGRGGGGYXQGG 445
Query: 149 GHGGHGAAESVAVQTEEKTNEVNDAKYGGGYNHGGGSYNHGGGSYHHGGGGYNHGG-GGH 207
YGGG GGY GG GG
Sbjct: 446 --------------------------YGGG-------------------GGYXQGGYGGQ 490
Query: 208 GGHGGG 213
GG+GGG
Sbjct: 491 GGYGGG 508
Score = 67.4 bits (163), Expect = 4e-12
Identities = 47/114 (41%), Positives = 51/114 (44%)
Frame = +2
Query: 103 GYNGGGYNHGGGGYNHGGGGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGGHGAAESVAVQ 162
GY GGG GGGGY GGGY+ G GG GGG +GG GGG GG G
Sbjct: 299 GYGGGG-GRGGGGY---GGGYDQGYGG--GGGSYGGGFGGGRGGGRG------------- 421
Query: 163 TEEKTNEVNDAKYGGGYNHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGGGGHG 216
GGGY GG + GGGGY GG +GG GG G G
Sbjct: 422 -------------GGGYXQGG----------YGGGGGYXQGG--YGGQGGYGGG 508
Score = 66.6 bits (161), Expect = 7e-12
Identities = 42/96 (43%), Positives = 47/96 (48%), Gaps = 1/96 (1%)
Frame = +2
Query: 125 HGGGGYNGGGGHGGHGGGGYNGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGGYNHGGG 184
+GGGG GGGG+GG GY GGGG YGGG+ GGG
Sbjct: 302 YGGGGGRGGGGYGGGYDQGYGGGGG------------------------SYGGGF--GGG 403
Query: 185 SYNHGGGSYHHGGGGYNHGG-GGHGGHGGGGHGGHG 219
GGG GGGGY GG GG GG+ GG+GG G
Sbjct: 404 ---RGGG---RGGGGYXQGGYGGGGGYXQGGYGGQG 493
Score = 51.2 bits (121), Expect = 3e-07
Identities = 33/80 (41%), Positives = 34/80 (42%)
Frame = +2
Query: 143 GYNGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGGYNHGGGSYNHGGGSYHHGGGGYNH 202
GY GGGG GG G YGGGY+ G Y GGGSY
Sbjct: 299 GYGGGGGRGGGG--------------------YGGGYDQG---YGGGGGSY--------- 382
Query: 203 GGGGHGGHGGGGHGGHGAEQ 222
GGG GG GGG GG G Q
Sbjct: 383 -GGGFGGGRGGGRGGGGYXQ 439
Score = 29.3 bits (64), Expect = 1.3
Identities = 13/27 (48%), Positives = 15/27 (55%)
Frame = +2
Query: 191 GSYHHGGGGYNHGGGGHGGHGGGGHGG 217
G+ G G GG GG GGGG+GG
Sbjct: 263 GAPPQGAPRQEMGYGGGGGRGGGGYGG 343
>TC85301 similar to GP|8096255|dbj|BAA96106.1 TrPRP2 {Trifolium repens},
partial (92%)
Length = 1318
Score = 82.0 bits (201), Expect = 2e-16
Identities = 71/208 (34%), Positives = 101/208 (48%), Gaps = 43/208 (20%)
Frame = -2
Query: 55 GGYNHGG--------GGYNGGGYNHGG---GGYNNGG---GGYNHGG---GGYNNGG--- 94
GG+++GG GG GG+++GG GG++ GG GG++ GG GGY+ GG
Sbjct: 948 GGFSNGGL*TGGSSTGGI*NGGFSNGGL*KGGFSTGGL*MGGFSKGGLYTGGYSIGGL*I 769
Query: 95 GGYNHGG--------GGYNGGGYNHGG---GGYNHGG---GGYNHGGGGYNGGGGHGGHG 140
GG+++GG GG GG++ GG GG++ GG GG++ GG Y GG GG
Sbjct: 768 GGFSNGGL*TGGFSTGGL*TGGFSTGGLYTGGFSTGGL*IGGFSTGGL-YTGGFSTGGL* 592
Query: 141 GGGYNGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGGYNHGG---GSYNHGG---GSYH 194
GG++ GG + G Y GG++ GG G ++ GG G +
Sbjct: 591 TGGFSTGGLYTG--------------------GLYTGGFSTGGL*TGGFSTGGLYTGGFS 472
Query: 195 HGG---GGYNHGGGGHGGHGGGGHGGHG 219
GG GG++ GG GG GG G
Sbjct: 471 TGGL*TGGFSTGGLYTGGFSTGGFSTGG 388
Score = 68.9 bits (167), Expect = 1e-12
Identities = 52/126 (41%), Positives = 71/126 (56%), Gaps = 24/126 (19%)
Frame = -2
Query: 51 GGYGGGYNHGG---GGYNGGGYNHGG---GGYNNGG---GGYNHGG---GGYNNGG---G 95
G Y GG++ GG GG++ GG GG GG++ GG GG++ GG GG++ GG G
Sbjct: 495 GLYTGGFSTGGL*TGGFSTGGLYTGGFSTGGFSTGGL*AGGFSTGGLYTGGFSTGGL*IG 316
Query: 96 GYNHGG---GGYNGGGYNHGG---GGYNHGG---GGYNHGGGGYNGGGGHGGHGGGGYNG 146
G++ GG GG++ GG GG GG GG GG++ GG Y GG GG GG++
Sbjct: 315 GFSTGGLYTGGFSTGGL*TGGFSTGGLYTGGL*TGGFSTGGL-YTGGFSTGGL*TGGFST 139
Query: 147 GGGHGG 152
GG + G
Sbjct: 138 GGLYTG 121
>TC86535 similar to GP|9758076|dbj|BAB08520.1 contains similarity to RNA
binding protein~gene_id:MNF13.1 {Arabidopsis thaliana},
partial (48%)
Length = 1555
Score = 80.5 bits (197), Expect = 5e-16
Identities = 63/195 (32%), Positives = 78/195 (39%), Gaps = 17/195 (8%)
Frame = +1
Query: 30 TSTDTKKEVVEKTNEVNDAKYGGYGGGYNHGGGGYNGGGYNHGGGGYNNGGGGYNHGGGG 89
T + KK +K N + ++G GGY G Y+ GGG+ GGGY+ G
Sbjct: 739 TQVEIKKAEPKKANAPPPSSKRYNDSRSSYGSGGY-GDAYDGFGGGFGGVGGGYSRSGSA 915
Query: 90 YNNGGGGYNHGGGGYNG-GGYNHGGGGYNHGGGGYNHGG------------GGYNGGG-- 134
Y GG Y G + G GGY GG G + G GY G G
Sbjct: 916 YGGRGGPYGGYGSEFAGYGGYAGAMGGPYRGDPSLAYAGRYGGSFRSSYDLSGYGGPGES 1095
Query: 135 --GHGGHGGGGYNGGGGHGGHGAAESVAVQTEEKTNEVNDAKYGGGYNHGGGSYNHGGGS 192
+GG G G GGGG G GA +S DA GGGY GG + G S
Sbjct: 1096YGAYGGAGAGAAAGGGGSSGAGAYQSGY-----------DASLGGGY--GGAA---SGAS 1227
Query: 193 YHHGGGGYNHGGGGH 207
++ GGY G H
Sbjct: 1228FYGSRGGYGGAGRYH 1272
Score = 68.2 bits (165), Expect = 3e-12
Identities = 56/168 (33%), Positives = 71/168 (41%), Gaps = 24/168 (14%)
Frame = +1
Query: 76 YNNGGGGYNHGGGG--YNNGGGGYNHGGGGYNGGGYNHGGGGYNHGGGGYNHGG-GGYNG 132
YN+ Y GG G Y+ GGG+ GGGY+ G +GG G +GG G G GGY G
Sbjct: 805 YNDSRSSYGSGGYGDAYDGFGGGFGGVGGGYSRSGSAYGGRGGPYGGYGSEFAGYGGYAG 984
Query: 133 GGG----------HGGHGGG---------GYNG-GGGHGGHGAAESVAVQTEEKTNEVND 172
G + G GG GY G G +G +G A +
Sbjct: 985 AMGGPYRGDPSLAYAGRYGGSFRSSYDLSGYGGPGESYGAYGGAGA-------------G 1125
Query: 173 AKYGGGYNHGGGSYNHGGGSYHHGGGGYNHGGGGHGGHGG-GGHGGHG 219
A GGG + G G+Y G + GGGY G +G GG+GG G
Sbjct: 1126AAAGGGGSSGAGAYQSGYDA--SLGGGYGGAASGASFYGSRGGYGGAG 1263
Score = 45.8 bits (107), Expect = 1e-05
Identities = 30/71 (42%), Positives = 35/71 (49%), Gaps = 8/71 (11%)
Frame = +1
Query: 52 GYGG-GYNHGGGGYNGGGYNHGGGGYNNGGGGYNHG-----GGGYNNGGGGYNHGG--GG 103
GYGG G ++G G G G GGGG ++G G Y G GGGY G + G GG
Sbjct: 1072 GYGGPGESYGAYGGAGAGAAAGGGG-SSGAGAYQSGYDASLGGGYGGAASGASFYGSRGG 1248
Query: 104 YNGGGYNHGGG 114
Y G G H G
Sbjct: 1249 YGGAGRYHPYG 1281
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.307 0.143 0.456
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,578,924
Number of Sequences: 36976
Number of extensions: 208832
Number of successful extensions: 24908
Number of sequences better than 10.0: 597
Number of HSP's better than 10.0 without gapping: 1484
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 9070
length of query: 229
length of database: 9,014,727
effective HSP length: 93
effective length of query: 136
effective length of database: 5,575,959
effective search space: 758330424
effective search space used: 758330424
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.9 bits)
S2: 56 (26.2 bits)
Medicago: description of AC146909.9


