

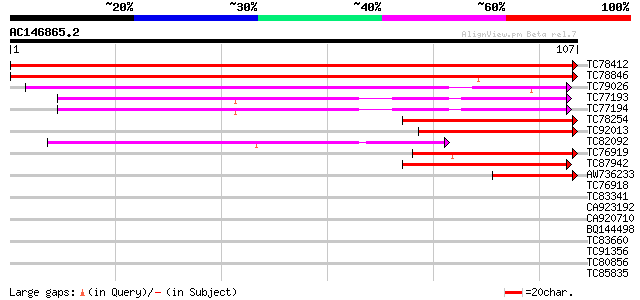
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146865.2 - phase: 0
(107 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC78412 homologue to GP|17978995|gb|AAL47458.1 AT3g07590/MLP3_4 ... 217 6e-58
TC78846 homologue to PIR|B85036|B85036 small nuclear riboprotein... 212 2e-56
TC79026 homologue to GP|21555384|gb|AAM63846.1 small nuclear rib... 57 1e-09
TC77193 homologue to GP|21593495|gb|AAM65462.1 glycine rich prot... 54 7e-09
TC77194 homologue to GP|21593495|gb|AAM65462.1 glycine rich prot... 54 7e-09
TC78254 similar to GP|15795167|dbj|BAB03155. gene_id:MQC3.30~unk... 44 1e-05
TC92013 homologue to GP|11595557|emb|CAC18142. related to c-modu... 40 1e-04
TC82092 similar to PIR|H86164|H86164 hypothetical protein AAC721... 40 1e-04
TC76919 similar to GP|15724324|gb|AAL06555.1 At1g76010/T4O12_22 ... 40 2e-04
TC87942 similar to GP|15795167|dbj|BAB03155. gene_id:MQC3.30~unk... 39 4e-04
AW736233 GP|21105451|gb small nuclear ribonucleoprotein D1 {Dani... 39 4e-04
TC76918 similar to GP|2231329|gb|AAB62000.1| bactinecin 11 {Ovis... 36 0.002
TC83341 weakly similar to GP|8978076|dbj|BAA98104.1 gene_id:K14A... 33 0.017
CA923192 similar to GP|20196906|gb| expressed protein {Arabidops... 33 0.017
CA920710 similar to PIR|S64314|S643 probable membrane protein YG... 33 0.017
BQ144498 similar to GP|6330472|dbj KIAA1205 protein {Homo sapien... 32 0.038
TC83660 GP|12835827|dbj|BAB23379. data source:MGD source key:MG... 32 0.050
TC91356 similar to GP|20151585|gb|AAM11152.1 LD24077p {Drosophil... 32 0.050
TC80856 similar to GP|15706274|emb|CAC69997. HMG I/Y like protei... 32 0.050
TC85835 similar to PIR|T12180|T12180 probable transcription fact... 32 0.050
>TC78412 homologue to GP|17978995|gb|AAL47458.1 AT3g07590/MLP3_4
{Arabidopsis thaliana}, complete
Length = 740
Score = 217 bits (552), Expect = 6e-58
Identities = 107/107 (100%), Positives = 107/107 (100%)
Frame = +1
Query: 1 MKLNNETVSIELKNGTIVHGTITGVDISMNTHLKTVKLTLKGKNPVTLDHLSVRGNNIRY 60
MKLNNETVSIELKNGTIVHGTITGVDISMNTHLKTVKLTLKGKNPVTLDHLSVRGNNIRY
Sbjct: 127 MKLNNETVSIELKNGTIVHGTITGVDISMNTHLKTVKLTLKGKNPVTLDHLSVRGNNIRY 306
Query: 61 YILPDSLNLETLLVEEAPRVKPKKPTAGKPLGRGRGRGRGRGRGRGR 107
YILPDSLNLETLLVEEAPRVKPKKPTAGKPLGRGRGRGRGRGRGRGR
Sbjct: 307 YILPDSLNLETLLVEEAPRVKPKKPTAGKPLGRGRGRGRGRGRGRGR 447
>TC78846 homologue to PIR|B85036|B85036 small nuclear riboprotein Sm-D1-like
protein [imported] - Arabidopsis thaliana, partial (98%)
Length = 642
Score = 212 bits (540), Expect = 2e-56
Identities = 107/108 (99%), Positives = 107/108 (99%), Gaps = 1/108 (0%)
Frame = +1
Query: 1 MKLNNETVSIELKNGTIVHGTITGVDISMNTHLKTVKLTLKGKNPVTLDHLSVRGNNIRY 60
MKLNNETVSIELKNGTIVHGTITGVDISMNTHLKTVKLTLKGKNPVTLDHLSVRGNNIRY
Sbjct: 130 MKLNNETVSIELKNGTIVHGTITGVDISMNTHLKTVKLTLKGKNPVTLDHLSVRGNNIRY 309
Query: 61 YILPDSLNLETLLVEEAPRVKPKKPTA-GKPLGRGRGRGRGRGRGRGR 107
YILPDSLNLETLLVEEAPRVKPKKPTA GKPLGRGRGRGRGRGRGRGR
Sbjct: 310 YILPDSLNLETLLVEEAPRVKPKKPTAGGKPLGRGRGRGRGRGRGRGR 453
>TC79026 homologue to GP|21555384|gb|AAM63846.1 small nuclear
ribonucleoprotein putative {Arabidopsis thaliana},
partial (85%)
Length = 753
Score = 56.6 bits (135), Expect = 1e-09
Identities = 34/114 (29%), Positives = 55/114 (47%), Gaps = 11/114 (9%)
Frame = +2
Query: 4 NNETVSIELKNGTIVHGTITGVDISMNTHLKTVKLTLKGKNPVTLDHLSVRGNNIRYYIL 63
+ V++ELK+G + G + + + N L+++ T K L+H+ +RG+ +R+ ++
Sbjct: 122 SGHVVTVELKSGELYRGNMIECEDNWNCQLESITYTAKDGKTSQLEHVFIRGSKVRFMVI 301
Query: 64 PDSLNLETLLVEEAPRVKPKKPTAGKPLGRGRGR-----------GRGRGRGRG 106
PD L + R+K K G LG GRGR GRG GRG
Sbjct: 302 PDMLKNAPMFKRLDARIKGK----GASLGVGRGRAVAMRAKAQAAGRGAPPGRG 451
>TC77193 homologue to GP|21593495|gb|AAM65462.1 glycine rich protein-like
{Arabidopsis thaliana}, partial (85%)
Length = 989
Score = 54.3 bits (129), Expect = 7e-09
Identities = 36/98 (36%), Positives = 49/98 (49%), Gaps = 1/98 (1%)
Frame = +2
Query: 10 IELKNGTIVHGTITGVDISMNTHLKTVKLTLK-GKNPVTLDHLSVRGNNIRYYILPDSLN 68
+ELKNG +G + D MN HL+ V T K G + +RGN I+Y +PD
Sbjct: 155 VELKNGETYNGHLVNCDTWMNIHLREVICTSKDGDRFWRMPECYIRGNTIKYLRVPDE-- 328
Query: 69 LETLLVEEAPRVKPKKPTAGKPLGRGRGRGRGRGRGRG 106
++++ K + T KP G GRGRGRGR G
Sbjct: 329 ----VIDKVQEEKSR--TDRKPPGVGRGRGRGREDAAG 424
>TC77194 homologue to GP|21593495|gb|AAM65462.1 glycine rich protein-like
{Arabidopsis thaliana}, partial (96%)
Length = 700
Score = 54.3 bits (129), Expect = 7e-09
Identities = 36/98 (36%), Positives = 49/98 (49%), Gaps = 1/98 (1%)
Frame = +3
Query: 10 IELKNGTIVHGTITGVDISMNTHLKTVKLTLK-GKNPVTLDHLSVRGNNIRYYILPDSLN 68
+ELKNG +G + D MN HL+ V T K G + +RGN I+Y +PD
Sbjct: 210 VELKNGETYNGHLVNCDTWMNIHLREVICTSKDGDRFWRMPECYIRGNTIKYLRVPDE-- 383
Query: 69 LETLLVEEAPRVKPKKPTAGKPLGRGRGRGRGRGRGRG 106
++++ K + T KP G GRGRGRGR G
Sbjct: 384 ----VIDKVQEEKSR--TDRKPPGVGRGRGRGREDAAG 479
>TC78254 similar to GP|15795167|dbj|BAB03155. gene_id:MQC3.30~unknown
protein {Arabidopsis thaliana}, partial (51%)
Length = 1464
Score = 43.9 bits (102), Expect = 1e-05
Identities = 22/33 (66%), Positives = 23/33 (69%)
Frame = +1
Query: 75 EEAPRVKPKKPTAGKPLGRGRGRGRGRGRGRGR 107
EEA R K + P GRGRGRGRGRGRGRGR
Sbjct: 628 EEAKRGKMLELGHTSPTGRGRGRGRGRGRGRGR 726
>TC92013 homologue to GP|11595557|emb|CAC18142. related to c-module-binding
factor {Neurospora crassa}, partial (2%)
Length = 1437
Score = 40.0 bits (92), Expect = 1e-04
Identities = 18/30 (60%), Positives = 22/30 (73%)
Frame = -2
Query: 78 PRVKPKKPTAGKPLGRGRGRGRGRGRGRGR 107
P ++ + G+ GRGRGRGRGRGRGRGR
Sbjct: 281 PSMRGRGGGRGRGGGRGRGRGRGRGRGRGR 192
Score = 36.6 bits (83), Expect = 0.002
Identities = 15/15 (100%), Positives = 15/15 (100%)
Frame = -2
Query: 92 GRGRGRGRGRGRGRG 106
GRGRGRGRGRGRGRG
Sbjct: 233 GRGRGRGRGRGRGRG 189
Score = 33.9 bits (76), Expect = 0.010
Identities = 14/17 (82%), Positives = 15/17 (87%)
Frame = -2
Query: 88 GKPLGRGRGRGRGRGRG 104
G+ GRGRGRGRGRGRG
Sbjct: 239 GRGRGRGRGRGRGRGRG 189
Score = 32.3 bits (72), Expect = 0.029
Identities = 15/23 (65%), Positives = 15/23 (65%)
Frame = -2
Query: 85 PTAGKPLGRGRGRGRGRGRGRGR 107
P P RGRG GRGRG GRGR
Sbjct: 296 PAMRGPSMRGRGGGRGRGGGRGR 228
>TC82092 similar to PIR|H86164|H86164 hypothetical protein AAC72111.1
[imported] - Arabidopsis thaliana, partial (80%)
Length = 311
Score = 40.0 bits (92), Expect = 1e-04
Identities = 24/78 (30%), Positives = 44/78 (55%), Gaps = 2/78 (2%)
Frame = +1
Query: 8 VSIELKNGTIVHGTITGVDISMNTHLKTVKLTLKGKNP--VTLDHLSVRGNNIRYYILPD 65
V++ELKN + GT+ VD +N L+ ++ + + P +++ + +RG+ +RY LP
Sbjct: 70 VTVELKNDLAIRGTLHSVDQYLNIKLENTRVVDQDRYPHMLSVRNCFIRGSVVRYVQLPP 249
Query: 66 SLNLETLLVEEAPRVKPK 83
+ LV +A R KP+
Sbjct: 250 E-GGDIELVHDAQRKKPR 300
>TC76919 similar to GP|15724324|gb|AAL06555.1 At1g76010/T4O12_22
{Arabidopsis thaliana}, partial (42%)
Length = 1207
Score = 39.7 bits (91), Expect = 2e-04
Identities = 21/35 (60%), Positives = 23/35 (65%), Gaps = 4/35 (11%)
Frame = +3
Query: 77 APRVKP----KKPTAGKPLGRGRGRGRGRGRGRGR 107
A +VKP + G P RGRGRGRGRGRGRGR
Sbjct: 510 ADQVKPLNEYEDEGEGSPRMRGRGRGRGRGRGRGR 614
Score = 36.6 bits (83), Expect = 0.002
Identities = 15/15 (100%), Positives = 15/15 (100%)
Frame = +3
Query: 92 GRGRGRGRGRGRGRG 106
GRGRGRGRGRGRGRG
Sbjct: 573 GRGRGRGRGRGRGRG 617
Score = 32.3 bits (72), Expect = 0.029
Identities = 14/18 (77%), Positives = 14/18 (77%)
Frame = +3
Query: 90 PLGRGRGRGRGRGRGRGR 107
P RGRGR RGR RGRGR
Sbjct: 780 PAPRGRGRERGRARGRGR 833
Score = 31.2 bits (69), Expect = 0.065
Identities = 16/32 (50%), Positives = 20/32 (62%)
Frame = +3
Query: 75 EEAPRVKPKKPTAGKPLGRGRGRGRGRGRGRG 106
E +PR++ G+ GRGRGRGRGRG G
Sbjct: 552 EGSPRMR------GRGRGRGRGRGRGRGMYNG 629
Score = 30.4 bits (67), Expect = 0.11
Identities = 14/23 (60%), Positives = 15/23 (64%)
Frame = +3
Query: 85 PTAGKPLGRGRGRGRGRGRGRGR 107
P A + GR RGR RGRGR GR
Sbjct: 777 PPAPRGRGRERGRARGRGRDAGR 845
Score = 27.7 bits (60), Expect = 0.72
Identities = 12/19 (63%), Positives = 13/19 (68%)
Frame = +3
Query: 88 GKPLGRGRGRGRGRGRGRG 106
G+ RGR RGRGR GRG
Sbjct: 792 GRGRERGRARGRGRDAGRG 848
>TC87942 similar to GP|15795167|dbj|BAB03155. gene_id:MQC3.30~unknown
protein {Arabidopsis thaliana}, partial (29%)
Length = 1222
Score = 38.5 bits (88), Expect = 4e-04
Identities = 20/32 (62%), Positives = 21/32 (65%)
Frame = +1
Query: 75 EEAPRVKPKKPTAGKPLGRGRGRGRGRGRGRG 106
EEA R K + GRGRGRGRGRGRGRG
Sbjct: 412 EEAKRSKMLELGHTGSTGRGRGRGRGRGRGRG 507
>AW736233 GP|21105451|gb small nuclear ribonucleoprotein D1 {Danio rerio},
partial (14%)
Length = 510
Score = 38.5 bits (88), Expect = 4e-04
Identities = 16/16 (100%), Positives = 16/16 (100%)
Frame = +1
Query: 92 GRGRGRGRGRGRGRGR 107
GRGRGRGRGRGRGRGR
Sbjct: 229 GRGRGRGRGRGRGRGR 276
Score = 33.9 bits (76), Expect = 0.010
Identities = 16/28 (57%), Positives = 19/28 (67%)
Frame = +1
Query: 76 EAPRVKPKKPTAGKPLGRGRGRGRGRGR 103
E+ V + AG+ GRGRGRGRGRGR
Sbjct: 193 ESQEVGTEYYNAGRGRGRGRGRGRGRGR 276
>TC76918 similar to GP|2231329|gb|AAB62000.1| bactinecin 11 {Ovis aries},
partial (13%)
Length = 576
Score = 36.2 bits (82), Expect = 0.002
Identities = 15/18 (83%), Positives = 16/18 (88%)
Frame = +1
Query: 90 PLGRGRGRGRGRGRGRGR 107
P RG+GRGRGRGRGRGR
Sbjct: 205 PAPRGQGRGRGRGRGRGR 258
Score = 35.8 bits (81), Expect = 0.003
Identities = 16/23 (69%), Positives = 17/23 (73%)
Frame = +1
Query: 85 PTAGKPLGRGRGRGRGRGRGRGR 107
P A + GRGRGRGRGRGR GR
Sbjct: 202 PPAPRGQGRGRGRGRGRGRDAGR 270
Score = 34.3 bits (77), Expect = 0.008
Identities = 14/14 (100%), Positives = 14/14 (100%)
Frame = +1
Query: 93 RGRGRGRGRGRGRG 106
RGRGRGRGRGRGRG
Sbjct: 1 RGRGRGRGRGRGRG 42
Score = 32.3 bits (72), Expect = 0.029
Identities = 13/13 (100%), Positives = 13/13 (100%)
Frame = +1
Query: 92 GRGRGRGRGRGRG 104
GRGRGRGRGRGRG
Sbjct: 4 GRGRGRGRGRGRG 42
Score = 32.0 bits (71), Expect = 0.038
Identities = 13/13 (100%), Positives = 13/13 (100%)
Frame = +1
Query: 95 RGRGRGRGRGRGR 107
RGRGRGRGRGRGR
Sbjct: 1 RGRGRGRGRGRGR 39
Score = 28.9 bits (63), Expect = 0.32
Identities = 12/15 (80%), Positives = 12/15 (80%)
Frame = +1
Query: 92 GRGRGRGRGRGRGRG 106
GRGRGRGRGRG G
Sbjct: 10 GRGRGRGRGRGMYNG 54
Score = 26.6 bits (57), Expect = 1.6
Identities = 16/38 (42%), Positives = 17/38 (44%), Gaps = 18/38 (47%)
Frame = +1
Query: 88 GKPLGRGRGRGRGR------------------GRGRGR 107
G+ GRGRGRGRG GRGRGR
Sbjct: 4 GRGRGRGRGRGRGMYNGGMEYGDGYDGGRGYGGRGRGR 117
>TC83341 weakly similar to GP|8978076|dbj|BAA98104.1 gene_id:K14A3.4~unknown
protein {Arabidopsis thaliana}, partial (80%)
Length = 1253
Score = 33.1 bits (74), Expect = 0.017
Identities = 14/15 (93%), Positives = 14/15 (93%)
Frame = +3
Query: 93 RGRGRGRGRGRGRGR 107
RGRGRGRGRGRGR R
Sbjct: 588 RGRGRGRGRGRGRRR 632
Score = 31.6 bits (70), Expect = 0.050
Identities = 14/16 (87%), Positives = 14/16 (87%)
Frame = +3
Query: 92 GRGRGRGRGRGRGRGR 107
GRGRGRGRGRGR R R
Sbjct: 591 GRGRGRGRGRGRRR*R 638
Score = 28.1 bits (61), Expect = 0.55
Identities = 13/20 (65%), Positives = 15/20 (75%)
Frame = +3
Query: 86 TAGKPLGRGRGRGRGRGRGR 105
T+ + GRGRGRGRGR R R
Sbjct: 579 TSRRGRGRGRGRGRGRRR*R 638
>CA923192 similar to GP|20196906|gb| expressed protein {Arabidopsis
thaliana}, partial (16%)
Length = 762
Score = 33.1 bits (74), Expect = 0.017
Identities = 14/20 (70%), Positives = 16/20 (80%)
Frame = -3
Query: 88 GKPLGRGRGRGRGRGRGRGR 107
G+ G+G GRGRG GRGRGR
Sbjct: 382 GRGQGQGWGRGRGSGRGRGR 323
Score = 32.3 bits (72), Expect = 0.029
Identities = 14/19 (73%), Positives = 15/19 (78%)
Frame = -3
Query: 88 GKPLGRGRGRGRGRGRGRG 106
G+ G GRGRG GRGRGRG
Sbjct: 376 GQGQGWGRGRGSGRGRGRG 320
Score = 30.4 bits (67), Expect = 0.11
Identities = 13/20 (65%), Positives = 15/20 (75%)
Frame = -3
Query: 88 GKPLGRGRGRGRGRGRGRGR 107
G GRG+G+G GRGRG GR
Sbjct: 394 GGARGRGQGQGWGRGRGSGR 335
Score = 28.9 bits (63), Expect = 0.32
Identities = 12/19 (63%), Positives = 15/19 (78%)
Frame = -3
Query: 88 GKPLGRGRGRGRGRGRGRG 106
G+ RGRG+G+G GRGRG
Sbjct: 400 GRGGARGRGQGQGWGRGRG 344
Score = 27.7 bits (60), Expect = 0.72
Identities = 12/21 (57%), Positives = 15/21 (71%)
Frame = -3
Query: 87 AGKPLGRGRGRGRGRGRGRGR 107
+G G RGRG+G+G GRGR
Sbjct: 409 SGGGRGGARGRGQGQGWGRGR 347
Score = 27.3 bits (59), Expect = 0.94
Identities = 11/19 (57%), Positives = 14/19 (72%)
Frame = -3
Query: 88 GKPLGRGRGRGRGRGRGRG 106
G+ G GRG RGRG+G+G
Sbjct: 418 GRVSGGGRGGARGRGQGQG 362
Score = 25.0 bits (53), Expect = 4.6
Identities = 11/16 (68%), Positives = 11/16 (68%)
Frame = -3
Query: 92 GRGRGRGRGRGRGRGR 107
GRGR G GRG RGR
Sbjct: 424 GRGRVSGGGRGGARGR 377
>CA920710 similar to PIR|S64314|S643 probable membrane protein YGR023w -
yeast (Saccharomyces cerevisiae), partial (7%)
Length = 774
Score = 33.1 bits (74), Expect = 0.017
Identities = 14/15 (93%), Positives = 14/15 (93%)
Frame = -1
Query: 93 RGRGRGRGRGRGRGR 107
RGRGRGRGRGRGR R
Sbjct: 384 RGRGRGRGRGRGRER 340
Score = 31.2 bits (69), Expect = 0.065
Identities = 13/14 (92%), Positives = 13/14 (92%)
Frame = -1
Query: 92 GRGRGRGRGRGRGR 105
GRGRGRGRGRGR R
Sbjct: 381 GRGRGRGRGRGRER 340
Score = 24.3 bits (51), Expect = 7.9
Identities = 10/14 (71%), Positives = 11/14 (78%)
Frame = -1
Query: 88 GKPLGRGRGRGRGR 101
G+ GRGRGRGR R
Sbjct: 381 GRGRGRGRGRGRER 340
>BQ144498 similar to GP|6330472|dbj KIAA1205 protein {Homo sapiens}, partial
(3%)
Length = 1284
Score = 32.0 bits (71), Expect = 0.038
Identities = 12/16 (75%), Positives = 14/16 (87%)
Frame = +1
Query: 92 GRGRGRGRGRGRGRGR 107
G+GRGRGRG G G+GR
Sbjct: 568 GKGRGRGRGEGSGKGR 615
Score = 29.6 bits (65), Expect = 0.19
Identities = 11/15 (73%), Positives = 13/15 (86%)
Frame = +1
Query: 93 RGRGRGRGRGRGRGR 107
RG+GRGRGRG G G+
Sbjct: 565 RGKGRGRGRGEGSGK 609
Score = 27.7 bits (60), Expect = 0.72
Identities = 12/20 (60%), Positives = 13/20 (65%)
Frame = +1
Query: 84 KPTAGKPLGRGRGRGRGRGR 103
K GK GRGRG G G+GR
Sbjct: 556 KGERGKGRGRGRGEGSGKGR 615
>TC83660 GP|12835827|dbj|BAB23379. data source:MGD source key:MGI:105110
evidence:ISS~putative~ribosomal protein S2 {Mus
musculus}, partial (5%)
Length = 500
Score = 31.6 bits (70), Expect = 0.050
Identities = 13/15 (86%), Positives = 13/15 (86%)
Frame = +2
Query: 90 PLGRGRGRGRGRGRG 104
P RGRGRGRGRGRG
Sbjct: 89 PAPRGRGRGRGRGRG 133
Score = 30.0 bits (66), Expect = 0.14
Identities = 12/12 (100%), Positives = 12/12 (100%)
Frame = +2
Query: 95 RGRGRGRGRGRG 106
RGRGRGRGRGRG
Sbjct: 98 RGRGRGRGRGRG 133
Score = 27.7 bits (60), Expect = 0.72
Identities = 11/11 (100%), Positives = 11/11 (100%)
Frame = +2
Query: 97 RGRGRGRGRGR 107
RGRGRGRGRGR
Sbjct: 98 RGRGRGRGRGR 130
Score = 26.2 bits (56), Expect = 2.1
Identities = 11/16 (68%), Positives = 12/16 (74%)
Frame = +2
Query: 85 PTAGKPLGRGRGRGRG 100
P A + GRGRGRGRG
Sbjct: 86 PPAPRGRGRGRGRGRG 133
>TC91356 similar to GP|20151585|gb|AAM11152.1 LD24077p {Drosophila
melanogaster}, partial (71%)
Length = 698
Score = 31.6 bits (70), Expect = 0.050
Identities = 15/23 (65%), Positives = 15/23 (65%)
Frame = +2
Query: 85 PTAGKPLGRGRGRGRGRGRGRGR 107
P G RG RGRGRGRGRGR
Sbjct: 98 PRGGGFGSRGGDRGRGRGRGRGR 166
Score = 29.3 bits (64), Expect = 0.25
Identities = 12/15 (80%), Positives = 13/15 (86%)
Frame = +2
Query: 93 RGRGRGRGRGRGRGR 107
RGRGRGRGRGR G+
Sbjct: 134 RGRGRGRGRGRRGGK 178
Score = 27.7 bits (60), Expect = 0.72
Identities = 12/18 (66%), Positives = 13/18 (71%)
Frame = +2
Query: 88 GKPLGRGRGRGRGRGRGR 105
G GRGRGRGRGR G+
Sbjct: 125 GGDRGRGRGRGRGRRGGK 178
>TC80856 similar to GP|15706274|emb|CAC69997. HMG I/Y like protein {Glycine
max}, partial (35%)
Length = 1552
Score = 31.6 bits (70), Expect = 0.050
Identities = 16/24 (66%), Positives = 17/24 (70%), Gaps = 4/24 (16%)
Frame = +2
Query: 88 GKPLGRGRG----RGRGRGRGRGR 107
G+ GRGRG RGRG GRGRGR
Sbjct: 1067 GRGRGRGRGGVAGRGRGGGRGRGR 1138
Score = 31.6 bits (70), Expect = 0.050
Identities = 16/24 (66%), Positives = 17/24 (70%), Gaps = 4/24 (16%)
Frame = +2
Query: 88 GKPLGRGRGR----GRGRGRGRGR 107
G+ GRGRGR GRGRG GRGR
Sbjct: 1061 GRGRGRGRGRGGVAGRGRGGGRGR 1132
Score = 30.8 bits (68), Expect = 0.085
Identities = 15/26 (57%), Positives = 15/26 (57%)
Frame = +2
Query: 82 PKKPTAGKPLGRGRGRGRGRGRGRGR 107
P A GRGRGRGRG GRGR
Sbjct: 1037 PVAAVAATGRGRGRGRGRGGVAGRGR 1114
Score = 28.9 bits (63), Expect = 0.32
Identities = 13/18 (72%), Positives = 13/18 (72%)
Frame = +2
Query: 88 GKPLGRGRGRGRGRGRGR 105
G GRGRG GRGRGR R
Sbjct: 1091 GGVAGRGRGGGRGRGRPR 1144
Score = 28.5 bits (62), Expect = 0.42
Identities = 12/14 (85%), Positives = 12/14 (85%)
Frame = +2
Query: 94 GRGRGRGRGRGRGR 107
GRGRG GRGRGR R
Sbjct: 1103 GRGRGGGRGRGRPR 1144
Score = 28.5 bits (62), Expect = 0.42
Identities = 15/32 (46%), Positives = 18/32 (55%), Gaps = 3/32 (9%)
Frame = +2
Query: 79 RVKPKK---PTAGKPLGRGRGRGRGRGRGRGR 107
R +P+K A P+ GRGRGRGRGR
Sbjct: 995 RRRPRKNVNAVAAVPVAAVAATGRGRGRGRGR 1090
>TC85835 similar to PIR|T12180|T12180 probable transcription factor - fava
bean, partial (79%)
Length = 1737
Score = 31.6 bits (70), Expect = 0.050
Identities = 19/43 (44%), Positives = 23/43 (53%)
Frame = +3
Query: 64 PDSLNLETLLVEEAPRVKPKKPTAGKPLGRGRGRGRGRGRGRG 106
P L + L +A R +P+ G GRG GRG GRGRG G
Sbjct: 363 PAQLPSKPLPPSQAVRESRNEPSRG---GRGGGRGFGRGRGDG 482
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.315 0.138 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,759,503
Number of Sequences: 36976
Number of extensions: 34640
Number of successful extensions: 785
Number of sequences better than 10.0: 119
Number of HSP's better than 10.0 without gapping: 309
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 529
length of query: 107
length of database: 9,014,727
effective HSP length: 83
effective length of query: 24
effective length of database: 5,945,719
effective search space: 142697256
effective search space used: 142697256
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 50 (23.9 bits)
Medicago: description of AC146865.2


