

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146854.1 + phase: 0 /pseudo
(1390 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
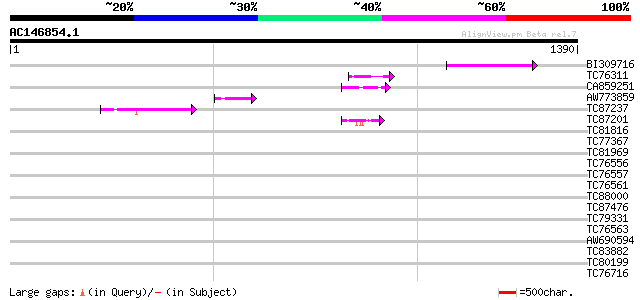
Score E
Sequences producing significant alignments: (bits) Value
BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F2... 72 2e-12
TC76311 homologue to GP|3204132|emb|CAA07235.1 extensin {Cicer a... 50 8e-06
CA859251 similar to GP|21305823|gb DNA polymerase I {Hz-1 insect... 48 2e-05
AW773859 47 4e-05
TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-inf... 45 2e-04
TC87201 similar to PIR|F86388|F86388 hypothetical protein AAG292... 43 7e-04
TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana taba... 41 0.004
TC77367 similar to GP|7269579|emb|CAB79581.1 GH3 like protein {A... 40 0.005
TC81969 similar to GP|6523547|emb|CAB62280.1 hydroxyproline-rich... 40 0.005
TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 40 0.008
TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 40 0.008
TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding pr... 40 0.008
TC88000 weakly similar to GP|13486661|dbj|BAB39898. contains EST... 39 0.011
TC87476 homologue to PIR|T11622|T11622 extensin class 1 precurso... 39 0.011
TC79331 similar to GP|10798760|dbj|BAB16432. WRKY transcription ... 39 0.014
TC76563 homologue to PIR|C29356|C29356 hydroxyproline-rich glyco... 39 0.014
AW690594 similar to GP|23498163|emb hypothetical protein {Plasmo... 39 0.018
TC83882 similar to GP|20197942|gb|AAD31580.2 putative farnesylat... 39 0.018
TC80199 weakly similar to GP|21593112|gb|AAM65061.1 unknown {Ara... 37 0.041
TC76716 similar to PIR|T10863|T10863 extensin precursor - kidney... 37 0.041
>BI309716 weakly similar to PIR|G96722|G9 hypothetical protein F20P5.25
[imported] - Arabidopsis thaliana, partial (10%)
Length = 744
Score = 71.6 bits (174), Expect = 2e-12
Identities = 76/224 (33%), Positives = 106/224 (46%)
Frame = +3
Query: 1071 KFANCPNLSMASSKLVDNGMKS*LDCSSTINIYKLLLMHPCLLRQLLIALLHFLFMLMI* 1130
++ + NL MASS+ VDNG+ S L+ S + IY LL+ P L ++ LLH +MLM
Sbjct: 18 RYVSFKNLYMASSRQVDNGILSYLNLSFPLVIYNPLLIFPFLQNLKILHLLHC*YMLMTL 197
Query: 1131 SLLEILSLKLISSRMFYTKPSKSKTLVF*NIF*VLKLHTPKMVSLFAKGSIVWICYKIQV 1190
LE++ LK +F KSKTLV +IF*VL+L K +I+ +I V
Sbjct: 198 F*LEMIYLKSNMLNVFLLIVLKSKTLVPYDIF*VLRLLEANKAFYLIKENILLNF*RIVV 377
Query: 1191 YLAQSRLPHLQIHPLSCTMTLHHSSLMSLLTEDWLGD*SISTQLGLTLHS*LNSSVNF*V 1250
L + L L I + T+ + ++M L ED + I L L N+ N
Sbjct: 378 ILL*NPLLLLMIFL*NSTILIRPFTMMKLNIEDS*ANLFI*LLLVLIYPLLFNN*ANLFR 557
Query: 1251 NQPTLITMLL*EFLNILKALQVVAFFFLELQDYIFKVTQMLIGQ 1294
N LL EF NI K + +L F V Q+LIGQ
Sbjct: 558 NLSKFTIRLLLEFYNI*KLPLPKDYSTQQLLI*NFLVLQILIGQ 689
>TC76311 homologue to GP|3204132|emb|CAA07235.1 extensin {Cicer arietinum},
partial (77%)
Length = 973
Score = 49.7 bits (117), Expect = 8e-06
Identities = 36/113 (31%), Positives = 54/113 (46%)
Frame = +3
Query: 831 LPLPLIFLILRILIFHNHHLLSHHLHSPLLHNLHHYLLMSLYLLELPSEIKLHLHIYMTT 890
LPLPL+ I+ I H+HH L HHL + H LHH+ L+LL + S + LHLH++
Sbjct: 9 LPLPLLHTII---IPHHHHRLLHHLPTTTNH-LHHH-PRHLHLLTITSHLHLHLHLH--- 164
Query: 891 FAMHLFLLLITVSVNIPFPILFHMLVYLTLILFLLCLLLLILNQNPMMRQSNM 943
H+L+ + ++L L LL+L NP+ Q +
Sbjct: 165 ----------------------HLLITIKVLLHHLPLLILPTTTNPLHHQRRL 257
>CA859251 similar to GP|21305823|gb DNA polymerase I {Hz-1 insect virus}
[Heliothis zea virus 1], partial (2%)
Length = 803
Score = 48.1 bits (113), Expect = 2e-05
Identities = 39/120 (32%), Positives = 64/120 (52%), Gaps = 1/120 (0%)
Frame = +3
Query: 814 LTIHHHPLLPLKIGNIFLP-LPLIFLILRILIFHNHHLLSHHLHSPLLHNLHHYLLMSLY 872
+ +HH LL LKI NI + L I ++ I+I NH++ LH L N HHY ++ +
Sbjct: 309 IQVHHLYLLYLKIINIIIIYLIKIIQVIVIIILINHNIQ*FMLH---LTN-HHYRMLKQH 476
Query: 873 LLELPSEIKLHLHIYMTTFAMHLFLLLITVSVNIPFPILFHMLVYLTLILFLLCLLLLIL 932
L L +K+ + + M + + +I + PIL +++ L LIL L+ LLL+I+
Sbjct: 477 HLLLTELVKVVIEVKTPPLTMGV*IKVIIIQ-----PILLLLILILILILILILLLLIII 641
>AW773859
Length = 538
Score = 47.4 bits (111), Expect = 4e-05
Identities = 39/103 (37%), Positives = 54/103 (51%)
Frame = -1
Query: 503 LPQVFVLQFL*TLVILFVLV*LFHVFLPMLFGTLD*DICLIKDCPKCIICILLFLLIIKL 562
LP +L+F L +LF +L FGTLD DI I++C I+ LL L I L
Sbjct: 295 LPP*HLLKFNLNLNLLF--------YLKKPFGTLDWDIYPIENCLAYIVIFLL*L*IKIL 140
Query: 563 LVTYVTLLSKENYLIIPANLLLILNLNFFTLIFGVL*LKHLFM 605
VTYV + ++YL + L +N FTLI+GV +LF+
Sbjct: 139 FVTYVIIPDIKSYLFSLVQIELPNVMNCFTLIYGVPFPPNLFI 11
>TC87237 similar to GP|160409|gb|AAA29651.1|| mature-parasite-infected
erythrocyte surface antigen {Plasmodium falciparum},
partial (2%)
Length = 2007
Score = 45.1 bits (105), Expect = 2e-04
Identities = 70/256 (27%), Positives = 125/256 (48%), Gaps = 22/256 (8%)
Frame = -1
Query: 224 IIL*LLLFLVKNLVYL*MHQM-LECPMAVVNLLRDHLNLRTIQDIALFAIGIITQWI-FV 281
IIL ++ L+ L+++*+ + L C + + L+ L T+ +A+ G I+ + F+
Sbjct: 888 IILGGVILLLLLLLFI*LRRRTLRCIILCLCLI-----LLTLISLAVLCNGRISGFSSFI 724
Query: 282 TRCMVIPMQINLMLLQMLL--LMKAFLIL------RKMVKDLPLSLKLVLLKS--NMVIL 331
T + + +++ L L L L++ FL L ++ + +L++L + N++I
Sbjct: 723 TIFLRLQLKLILFLFVKLFFCLVRTFL*LVIHYSTGGVISGIIFLPRLIVLFTILNLIIR 544
Query: 332 LLYFSN--HLWFLKHLPQI-LLAPI--ILLLLF-HPFPQVSILLS--LVLYILKIIIGLL 383
F N L+FL I LL P+ ILL++F H F +++ +L +L+I I
Sbjct: 543 *SIFLNLIMLFFLSSFCSIGLLCPLFTILLVIFSHLFLRIASVLVAFFILFIFTICFTCF 364
Query: 384 IQVLMSIFAPLCICFI--PFIKLNLCMLLYLMVLL*LFTMLALLFFHLLSTLPMSYIHLI 441
+L+++F F PFI L +LL L + LF +L LFF + + + + I
Sbjct: 363 FILLLTLFLGNFSSFFLSPFIFLQFILLLTLFFIFVLFILLLTLFFIFVLFILLLNLFSI 184
Query: 442 SELT*FLFTNFALFCL 457
L FL T F +F L
Sbjct: 183 FNLFIFLLTLFFIFVL 136
>TC87201 similar to PIR|F86388|F86388 hypothetical protein AAG29221.1
[imported] - Arabidopsis thaliana, partial (41%)
Length = 991
Score = 43.1 bits (100), Expect = 7e-04
Identities = 40/139 (28%), Positives = 60/139 (42%), Gaps = 32/139 (23%)
Frame = +2
Query: 813 YLTIHHHPLLPLKIGNIFLP----LPLIFLILRILIFHNHH-----LLSHHLHSP----- 858
+L +HHH LL + LP P + + +L+ H HH S HLH
Sbjct: 224 HLLLHHHHLLHISTSR-HLPHHMCTPHLHMSTSLLLHHLHHHPHHMYTSRHLHHHMSTSH 400
Query: 859 -----------LLHN-----LHHYLLMSLYLLELPSEIKLHLHIYMTTFAMHLFLLLITV 902
LLHN LHH+L+ +LL P L LH + +H+F+ L +
Sbjct: 401 RHLLHMFTNHLLLHNMSTNPLHHHLMFMSHLLH-PHHHHL-LHTTTNLYPLHMFMSLHHI 574
Query: 903 SVN--IPFPILFHMLVYLT 919
S+N P P+ ++L+ T
Sbjct: 575 SINPHHPLPMFINLLLTFT 631
>TC81816 similar to GP|8096269|dbj|BAA95789.1 KED {Nicotiana tabacum},
partial (13%)
Length = 663
Score = 40.8 bits (94), Expect = 0.004
Identities = 53/186 (28%), Positives = 82/186 (43%), Gaps = 1/186 (0%)
Frame = -2
Query: 341 FLKHLPQILLAPIILLLLFHPFPQVSILLS-LVLYILKIIIGLLIQVLMSIFAPLCICFI 399
FL L + ++L F F ++I L+ L +++ L +L+ +F + FI
Sbjct: 623 FLLLLFLVFFFLLLLFTFFRGFFYINIPYRYFTLFFLILLLLFLFLILILLFLTSILTFI 444
Query: 400 PFIKLNLCMLLYLMVLL*LFTMLALLFFHLLSTLPMSYIHLISELT*FLFTNFALFCLTK 459
L L +LL + L LFT+L +FF LL + ++ +T F FT LF L
Sbjct: 443 IVFILVLFLLLVFLRFL-LFTILVFVFFSLLLLFLFLLLLILLFITIFFFTLNILFLL-- 273
Query: 460 SIFCLINA*YRI*NPRR*LVWVIFVMVSIG*ILLHAIRSLLFLLPQVFVLQFL*TLVILF 519
F L+ V +I ++ I +LL I LLFL +F+ F L I F
Sbjct: 272 FFFLLL------------FVLLILTLIYISFVLLLLI--LLFLFLIIFLY*F--NLQIFF 141
Query: 520 VLV*LF 525
L +F
Sbjct: 140 YLNLIF 123
Score = 40.0 bits (92), Expect = 0.006
Identities = 45/165 (27%), Positives = 74/165 (44%), Gaps = 3/165 (1%)
Frame = -2
Query: 368 LLSLVLYILKIIIGLLIQVLMSIFAPLCICFIPFIKLNL---CMLLYLMVLL*LFTMLAL 424
LL +++IL + LL V + L F F +N+ L+ ++LL LF L L
Sbjct: 656 LLGSLVFILSRFLLLLFLVFFFLLL-LFTFFRGFFYINIPYRYFTLFFLILLLLFLFLIL 480
Query: 425 LFFHLLSTLPMSYIHLISELT*FLFTNFALFCLTKSIFCLINA*YRI*NPRR*LVWVIFV 484
+ L S L + ++ +F F LF + +F + + L+ +I +
Sbjct: 479 ILLFLTSILTFIIVFILVLFLLLVFLRFLLFTILVFVFFSLLLLFLF------LLLLILL 318
Query: 485 MVSIG*ILLHAIRSLLFLLPQVFVLQFL*TLVILFVLV*LFHVFL 529
++I L+ I LLF +FVL L + I FVL+ L +FL
Sbjct: 317 FITIFFFTLN-ILFLLFFFLLLFVLLILTLIYISFVLLLLILLFL 186
Score = 39.7 bits (91), Expect = 0.008
Identities = 49/179 (27%), Positives = 84/179 (46%), Gaps = 17/179 (9%)
Frame = -2
Query: 305 FLILRKMVKDLPLSLKLVLLKSNMVILLLYFSNHLWFLKHLPQILLAPIILLLLFHPFPQ 364
F +L +V L L L+ L ++LL F +++ ++P L+LL
Sbjct: 662 FSLLGSLVFILSRFLLLLFLVFFFLLLLFTFFRGFFYI-NIPYRYFTLFFLILLLLFLFL 486
Query: 365 VSILLSLVLYILKIIIGLLIQVLMSIFAP-LCICFIPFIKLNLCML-LYLMVLL*LF--- 419
+ ILL L + II+ +L+ L+ +F L + F+ +L +L L+L++L+ LF
Sbjct: 485 ILILLFLTSILTFIIVFILVLFLLLVFLRFLLFTILVFVFFSLLLLFLFLLLLILLFITI 306
Query: 420 -----TMLALLFFHL-------LSTLPMSYIHLISELT*FLFTNFALFCLTKSIFCLIN 466
+L LLFF L L+ + +S++ L+ L FLF L+ IF +N
Sbjct: 305 FFFTLNILFLLFFFLLLFVLLILTLIYISFVLLLLILL-FLFLIIFLY*FNLQIFFYLN 132
Score = 38.9 bits (89), Expect = 0.014
Identities = 39/149 (26%), Positives = 73/149 (48%), Gaps = 6/149 (4%)
Frame = -2
Query: 287 IPMQINLMLLQMLLLMKAFLILRKMVKDLPLSLKLVLLKSNMVILLLYFSNHLWF----- 341
IP + + +LLL+ FLIL ++ L L +++ ++ LLL F L F
Sbjct: 545 IPYRYFTLFFLILLLLFLFLIL--ILLFLTSILTFIIVFILVLFLLLVFLRFLLFTILVF 372
Query: 342 -LKHLPQILLAPIILLLLFHPFPQVSILLSLVLYILKIIIGLLIQVLMSIFAPLCICFIP 400
L + L ++L+LLF +++ + L I+ L +L+ + L + +I
Sbjct: 371 VFFSLLLLFLFLLLLILLF---------ITIFFFTLNILFLLFFFLLLFVLLILTLIYIS 219
Query: 401 FIKLNLCMLLYLMVLL*LFTMLALLFFHL 429
F+ L L +LL+L +++ L+ +FF+L
Sbjct: 218 FVLL-LLILLFLFLIIFLY*FNLQIFFYL 135
Score = 35.4 bits (80), Expect = 0.16
Identities = 51/192 (26%), Positives = 81/192 (41%)
Frame = -2
Query: 741 FYMIHCLIFILSKFLVAFVMLLHFKLIEQNYNLEPESVFS*VTSLDSKVQFFLTYIPEKS 800
F ++ L+FILS+FL+ ++ F L+ +F+ FF IP +
Sbjct: 662 FSLLGSLVFILSRFLLLLFLVFFFLLL----------LFTFFRG------FFYINIPYR- 534
Query: 801 LFQEMLFSMN*YYLTIHHHPLLPLKIGNIFLPLPLIFLILRILIFHNHHLLSHHLHSPLL 860
Y T+ LL L + I + L L ++ I++F L LL
Sbjct: 533 ------------YFTLFFLILLLLFLFLILILLFLTSILTFIIVFILVLFLLLVFLRFLL 390
Query: 861 HNLHHYLLMSLYLLELPSEIKLHLHIYMTTFAMHLFLLLITVSVNIPFPILFHMLVYLTL 920
+ ++ SL LL L + + L I + F +++ LL F +L +L+ LTL
Sbjct: 389 FTILVFVFFSLLLLFLFLLLLILLFITIFFFTLNILFLLF-------FFLLLFVLLILTL 231
Query: 921 ILFLLCLLLLIL 932
I LLLLIL
Sbjct: 230 IYISFVLLLLIL 195
Score = 29.6 bits (65), Expect = 8.5
Identities = 46/165 (27%), Positives = 78/165 (46%), Gaps = 12/165 (7%)
Frame = -2
Query: 498 SLLFLLPQVFVLQFL*TLVILFVLV*L---FHVFLPMLFGTLD*DICLIKDCPKCIICIL 554
SL+F+L + +L FL +L + F++ +P + TL I L+ +
Sbjct: 647 SLVFILSRFLLLLFLVFFFLLLLFTFFRGFFYINIPYRYFTLFFLILLL---------LF 495
Query: 555 LFLLIIKLLVTYVTLLSKENYLIIPANLLLILNLNFFTL---IFGVL*LKHLFMA----- 606
LFL++I L +T + L ++++ LL+ L FT+ +F L L LF+
Sbjct: 494 LFLILILLFLTSI-LTFIIVFILVLFLLLVFLRFLLFTILVFVFFSLLLLFLFLLLLILL 318
Query: 607 -ISIF*P*LMILVDSYGSFYLKPKLKFLHMLKISFT*SIIIIMLL 650
I+IF L IL + F L L L ++ ISF ++I++ L
Sbjct: 317 FITIFFFTLNILFLLF-FFLLLFVLLILTLIYISFVLLLLILLFL 186
>TC77367 similar to GP|7269579|emb|CAB79581.1 GH3 like protein {Arabidopsis
thaliana}, partial (93%)
Length = 2088
Score = 40.4 bits (93), Expect = 0.005
Identities = 42/148 (28%), Positives = 72/148 (48%), Gaps = 11/148 (7%)
Frame = +1
Query: 289 MQINLMLLQMLLLMKAFLILRKMVKDLPLSLKLVLLKSNMVILLLY--------FSNHLW 340
M + L+LL + LL + IL + + L L L L++L V+ +L +W
Sbjct: 655 MILELVLLTL*LLTQQ*EIL--L*RSLNLILNLLILSKLSVVKILGRG*LLGFGLILSMW 828
Query: 341 FLKHLPQ---ILLAPIILLLLFHPFPQVSILLSLVLYILKIIIGLLIQVLMSIFAPLCIC 397
L L Q I L II+ +++H L +L + ++ + + L + +++ L +
Sbjct: 829 MLL*LEQCHSIFLFWIIIAMVYH-------LFALCMLLVNVTLVLT*TLFVNLVKCLTLL 987
Query: 398 FIPFIKLNLCMLLYLMVLL*LFTMLALL 425
F+P LN C+ + +MV L LFT L LL
Sbjct: 988 FLPCATLNFCL*IEVMVSLILFTHLDLL 1071
>TC81969 similar to GP|6523547|emb|CAB62280.1 hydroxyproline-rich
glycoprotein DZ-HRGP {Volvox carteri f. nagariensis},
partial (11%)
Length = 556
Score = 40.4 bits (93), Expect = 0.005
Identities = 35/94 (37%), Positives = 55/94 (58%), Gaps = 2/94 (2%)
Frame = -3
Query: 293 LMLLQMLLLMKAFLILR--KMVKDLPLSLKLVLLKSNMVILLLYFSNHLWFLKHLPQILL 350
L+LL +LLL+ F ILR ++ + L L L L+LL ++L+L F N + LP L
Sbjct: 347 LLLLLLLLLLLRFPILRLLQICRFLLLLLNLLLLVLPRILLVLKFPNLHCYAFPLPS--L 174
Query: 351 APIILLLLFHPFPQVSILLSLVLYILKIIIGLLI 384
+ LLL HP+ + +LSL+L + +I+ LL+
Sbjct: 173 HRVHLLLDLHPY--LPKILSLILQLHQIVSHLLM 78
>TC76556 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 754
Score = 39.7 bits (91), Expect = 0.008
Identities = 22/67 (32%), Positives = 35/67 (51%), Gaps = 7/67 (10%)
Frame = -3
Query: 840 LRILIFHNHHLLSHHL-------HSPLLHNLHHYLLMSLYLLELPSEIKLHLHIYMTTFA 892
L + H+HH+L HHL H +LH +HH+LL+ ++ L ++ H+H +
Sbjct: 554 LHLCCIHHHHILHHHLLCCSHRLHHGVLH-IHHHLLLRIHHLHRNHDVLHHIHRHHLHNH 378
Query: 893 MHLFLLL 899
H LLL
Sbjct: 377 RHHGLLL 357
Score = 34.7 bits (78), Expect = 0.26
Identities = 22/60 (36%), Positives = 28/60 (46%), Gaps = 1/60 (1%)
Frame = -3
Query: 814 LTIHHHPLLPLKIGNIFLPLPLIFLILRILIFHNHHLLSHHLHSPLLHN-LHHYLLMSLY 872
L IHHH L+LRI H +H + HH+H LHN HH LL+ L+
Sbjct: 473 LHIHHH------------------LLLRIHHLHRNHDVLHHIHRHHLHNHRHHGLLLHLH 348
>TC76557 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 912
Score = 39.7 bits (91), Expect = 0.008
Identities = 22/67 (32%), Positives = 35/67 (51%), Gaps = 7/67 (10%)
Frame = -3
Query: 840 LRILIFHNHHLLSHHL-------HSPLLHNLHHYLLMSLYLLELPSEIKLHLHIYMTTFA 892
L + H+HH+L HHL H +LH +HH+LL+ ++ L ++ H+H +
Sbjct: 712 LHLCCIHHHHILHHHLLCCSHRLHHGVLH-IHHHLLLRIHHLHRNHDVLHHIHRHHLHNH 536
Query: 893 MHLFLLL 899
H LLL
Sbjct: 535 RHHGLLL 515
Score = 34.7 bits (78), Expect = 0.26
Identities = 22/60 (36%), Positives = 28/60 (46%), Gaps = 1/60 (1%)
Frame = -3
Query: 814 LTIHHHPLLPLKIGNIFLPLPLIFLILRILIFHNHHLLSHHLHSPLLHN-LHHYLLMSLY 872
L IHHH L+LRI H +H + HH+H LHN HH LL+ L+
Sbjct: 631 LHIHHH------------------LLLRIHHLHRNHDVLHHIHRHHLHNHRHHGLLLHLH 506
>TC76561 similar to PIR|S71779|S71779 glycine-rich RNA-binding protein GRP1
- wheat, partial (95%)
Length = 1054
Score = 39.7 bits (91), Expect = 0.008
Identities = 22/67 (32%), Positives = 35/67 (51%), Gaps = 7/67 (10%)
Frame = -1
Query: 840 LRILIFHNHHLLSHHL-------HSPLLHNLHHYLLMSLYLLELPSEIKLHLHIYMTTFA 892
L + H+HH+L HHL H +LH +HH+LL+ ++ L ++ H+H +
Sbjct: 808 LHLCCIHHHHILHHHLLCCSHRLHHGVLH-IHHHLLLRIHHLHRNHDVLHHIHRHHLHNH 632
Query: 893 MHLFLLL 899
H LLL
Sbjct: 631 RHHGLLL 611
Score = 34.7 bits (78), Expect = 0.26
Identities = 22/60 (36%), Positives = 28/60 (46%), Gaps = 1/60 (1%)
Frame = -1
Query: 814 LTIHHHPLLPLKIGNIFLPLPLIFLILRILIFHNHHLLSHHLHSPLLHN-LHHYLLMSLY 872
L IHHH L+LRI H +H + HH+H LHN HH LL+ L+
Sbjct: 727 LHIHHH------------------LLLRIHHLHRNHDVLHHIHRHHLHNHRHHGLLLHLH 602
>TC88000 weakly similar to GP|13486661|dbj|BAB39898. contains EST
AU081362(R10239)~unknown protein {Oryza sativa (japonica
cultivar-group)}, partial (20%)
Length = 1096
Score = 39.3 bits (90), Expect = 0.011
Identities = 46/167 (27%), Positives = 68/167 (40%), Gaps = 20/167 (11%)
Frame = +2
Query: 787 SKVQFFLTYIPEKSLFQE-MLFSMN*YYLTIHHHPLLPLKIGNIFLPLPLIFLILRIL-- 843
S F +P L E +L S+ L + H +L L+ IFL L L LI+
Sbjct: 200 SSADFLQKKVPYPILLLE*LLLSLLFGNLNLAHQNILSLR--KIFLLLLLHLLIIHAKKH 373
Query: 844 --------IFHNHH---------LLSHHLHSPLLHNLHHYLLMSLYLLELPSEIKLHLHI 886
IF H L HHLH PL+H LH L+ L L LP HL
Sbjct: 374 QLRKTQDPIFFLHFSQN*ILRKILCLHHLHHPLIHLLH---LIILLLHVLPQTHHHHLPK 544
Query: 887 YMTTFAMHLFLLLITVSVNIPFPILFHMLVYLTLILFLLCLLLLILN 933
+ + ++ + +S +L H+L + L+ + L LI++
Sbjct: 545 LLQEGGVF*VMVHLLISQEKKMKMLLHLLQHYALVFLVQLALRLIVD 685
>TC87476 homologue to PIR|T11622|T11622 extensin class 1 precursor - cowpea,
partial (48%)
Length = 939
Score = 39.3 bits (90), Expect = 0.011
Identities = 32/88 (36%), Positives = 43/88 (48%), Gaps = 3/88 (3%)
Frame = +1
Query: 814 LTIHHHPL---LPLKIGNIFLPLPLIFLILRILIFHNHHLLSHHLHSPLLHNLHHYLLMS 870
+T HHPL L + I PL+ L+L L+ +LL HH LLH LH +
Sbjct: 358 VTTDHHPLHHHLHMSIS------PLLLLLLHPLLHMFTNLLPHH----LLHPLHPMSISH 507
Query: 871 LYLLELPSEIKLHLHIYMTTFAMHLFLL 898
L+LL+L H+ TT +HL LL
Sbjct: 508 LHLLQLH-------HLLHTTTNLHLHLL 570
Score = 37.7 bits (86), Expect = 0.031
Identities = 27/87 (31%), Positives = 43/87 (49%), Gaps = 6/87 (6%)
Frame = +3
Query: 814 LTIHHHPLLP----LKIGNIFLPLPLIFLILRILIFHNHHLLSHHLH--SPLLHNLHHYL 867
L +HHHP LP + + L PL + +R+ H+H L HHLH + LL + H +L
Sbjct: 24 LLLHHHPPLPHHMYISHHHPHLHPPLHHMSIRV---HHHPLHHHHLHMSTSLLLHQHPHL 194
Query: 868 LMSLYLLELPSEIKLHLHIYMTTFAMH 894
L +++ + L H T+ +H
Sbjct: 195 LHPIFISRHHLHLPLLPHHMFTSHHLH 275
Score = 34.3 bits (77), Expect = 0.35
Identities = 25/79 (31%), Positives = 38/79 (47%), Gaps = 2/79 (2%)
Frame = +3
Query: 814 LTIHHHPLL--PLKIGNIFLPLPLIFLILRILIFHNHHLLSHHLHSPLLHNLHHYLLMSL 871
L +H HP L P+ I L LPL+ +H SHHLH H+LH ++ + L
Sbjct: 168 LLLHQHPHLLHPIFISRHHLHLPLL---------PHHMFTSHHLHRLHPHHLHTFINLLL 320
Query: 872 YLLELPSEIKLHLHIYMTT 890
+ L LH ++++T
Sbjct: 321 HHL-------LHHRLHIST 356
>TC79331 similar to GP|10798760|dbj|BAB16432. WRKY transcription factor
Nt-SubD48 {Nicotiana tabacum}, partial (51%)
Length = 1336
Score = 38.9 bits (89), Expect = 0.014
Identities = 36/124 (29%), Positives = 56/124 (45%)
Frame = +1
Query: 833 LPLIFLILRILIFHNHHLLSHHLHSPLLHNLHHYLLMSLYLLELPSEIKLHLHIYMTTFA 892
L L+ L+ L++HNH +HH H P NL + +L+L + H
Sbjct: 400 LELVMLVSEKLLYHNH---NHHHHKPYNFNLLNQ*FSTLHLCN-----RFH--------- 528
Query: 893 MHLFLLLITVSVNIPFPILFHMLVYLTLILFLLCLLLLILNQNPMMRQSNMTAGNKQCRL 952
LFLL TV + IL +L++ TL+L +L L + + T N++CRL
Sbjct: 529 -QLFLLPFTVL*SREM-ILQKLLIFHTLLLVILSFLHSL---------ATTTTNNRRCRL 675
Query: 953 NLML 956
+L
Sbjct: 676 RRVL 687
>TC76563 homologue to PIR|C29356|C29356 hydroxyproline-rich glycoprotein
(clone Hyp2.13) - kidney bean (fragment), partial (34%)
Length = 507
Score = 38.9 bits (89), Expect = 0.014
Identities = 35/108 (32%), Positives = 51/108 (46%), Gaps = 3/108 (2%)
Frame = +3
Query: 816 IHHHPLLPLKIGNIFLPLPLIFLILRILIFHNHHLLSHHLHSPLLHNLHHYLLMSLYLLE 875
+HHH L + ++ P L L + HHLL HH + L H LH LL +
Sbjct: 180 LHHHTTTNLLLLHLLFPKHLTTTNLPHPL---HHLLHHHTTTNLPHPLHR-LLHHHTTIN 347
Query: 876 LP-SEIKLHLHIYMTTFAMHLFLLLITVSVNIPFPILFHML--VYLTL 920
LP LH H+ +T+ HL LL + P+L H+L ++LT+
Sbjct: 348 LPHHRHPLHHHLTITSLLPHLHHLLHHHTTT--NPLLLHLLFPIHLTI 485
Score = 34.3 bits (77), Expect = 0.35
Identities = 37/119 (31%), Positives = 53/119 (44%), Gaps = 17/119 (14%)
Frame = +3
Query: 837 FLILRILIFHN--HHLLSHHLHSPLLHNLHHYLLMSLYLL-----------ELPSEIKLH 883
FL+L+I+IF HH + +L PL LHH+ +L LL LP +
Sbjct: 96 FLLLQIVIFTLLLHHHTTTNLPHPLHRLLHHHTTTNLLLLHLLFPKHLTTTNLPHPLHHL 275
Query: 884 LHIYMTTFAMHLF--LLLITVSVNIPF--PILFHMLVYLTLILFLLCLLLLILNQNPMM 938
LH + TT H LL ++N+P L H L +L+ L LL NP++
Sbjct: 276 LHHHTTTNLPHPLHRLLHHHTTINLPHHRHPLHHHLTITSLLPHLHHLLHHHTTTNPLL 452
>AW690594 similar to GP|23498163|emb hypothetical protein {Plasmodium
falciparum 3D7}, partial (10%)
Length = 633
Score = 38.5 bits (88), Expect = 0.018
Identities = 28/80 (35%), Positives = 38/80 (47%), Gaps = 9/80 (11%)
Frame = -1
Query: 816 IHHHPLLPLKIGNIFLPLPLIFLILRILIFHNHH---------LLSHHLHSPLLHNLHHY 866
+HHHP P PL+ L+L +L+ H+ H LL HH+H LL H +
Sbjct: 234 LHHHP-----------PPPLLLLLLLLLLRHHVHLLLLLLLLLLLPHHVH--LLLPRHVH 94
Query: 867 LLMSLYLLELPSEIKLHLHI 886
LL+ L L LP L L +
Sbjct: 93 LLLLLLHLRLPPHYVLLLSV 34
>TC83882 similar to GP|20197942|gb|AAD31580.2 putative farnesylated protein
{Arabidopsis thaliana}, partial (4%)
Length = 670
Score = 38.5 bits (88), Expect = 0.018
Identities = 40/131 (30%), Positives = 64/131 (48%), Gaps = 12/131 (9%)
Frame = -2
Query: 816 IHHHPLLPLKIGNIFLPLPLIFLILRILIFH----------NHHLLSHHLHSPLLHNLHH 865
+HH P K+ F + +F++L L++H N HL HH H PL LHH
Sbjct: 624 LHH----PWKLLLHFTTVTNLFVLLSTLLYHQPPLLLSFSHNLHLQLHH-HLPLELYLHH 460
Query: 866 Y-LLMSLYLLELPSEIKLHLHIYMTTFAMHLFLLLITVSVNIPFPILFHMLVYLTLIL-F 923
+ LL++L+ + + K + +T L + V P ++L +LTL++ F
Sbjct: 459 HPLLLNLFKIFNTQKPK---SLNFSTLLSKFQTLTLLVKTQNP-----NLLNFLTLLVNF 304
Query: 924 LLCLLLLILNQ 934
L LLLL+L+Q
Sbjct: 303 PLLLLLLLLHQ 271
>TC80199 weakly similar to GP|21593112|gb|AAM65061.1 unknown {Arabidopsis
thaliana}, partial (41%)
Length = 934
Score = 37.4 bits (85), Expect = 0.041
Identities = 37/140 (26%), Positives = 59/140 (41%), Gaps = 6/140 (4%)
Frame = +3
Query: 763 HFKLIEQNYNLEPESVFS*VTSLDSKVQFFL-TYIPEKSLFQEMLFSMN*YYLTIHHHPL 821
HF LI ++ + S S + + + +F + +Y LFQ ML++ + HH
Sbjct: 18 HFLLILTQFSHQFPSKQSWILKVKFR*EFSI*SYYLCVLLFQLMLWNQESLLRSAHH--- 188
Query: 822 LPLKIGNIFLPLPLIFLILRILIFHNHHLLSHHLHSPLLH-----NLHHYLLMSLYLLEL 876
+ L I + I H+H L HH H L H +HH+ L+L L
Sbjct: 189 --------VVTLQYIHHLQ*YTIHHHHQLCIHHHHHHLRHRRQ*YTIHHH---HLHLRNL 335
Query: 877 PSEIKLHLHIYMTTFAMHLF 896
P +I LH H + H++
Sbjct: 336 PRQIVLHHHHHHHHHHHHMY 395
>TC76716 similar to PIR|T10863|T10863 extensin precursor - kidney bean,
partial (48%)
Length = 1275
Score = 37.4 bits (85), Expect = 0.041
Identities = 25/81 (30%), Positives = 36/81 (43%), Gaps = 8/81 (9%)
Frame = +1
Query: 813 YLTIHHHP------LLPLKIGNIFLPLPLIFLILRILIFHNHHLLS--HHLHSPLLHNLH 864
+ +IHHH LPL + I L+L + I H HH+ + HHL H+ H
Sbjct: 163 HTSIHHHHHQFTSISLPLHLSTHHHQFTSISLLLHLSIHHLHHISTNLHHLLLISTHHHH 342
Query: 865 HYLLMSLYLLELPSEIKLHLH 885
H + +LL + LH H
Sbjct: 343 HQFTSTSHLLPQFTSTNLHHH 405
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.357 0.162 0.524
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 50,392,287
Number of Sequences: 36976
Number of extensions: 941695
Number of successful extensions: 21452
Number of sequences better than 10.0: 225
Number of HSP's better than 10.0 without gapping: 7536
Number of HSP's successfully gapped in prelim test: 1835
Number of HSP's that attempted gapping in prelim test: 11088
Number of HSP's gapped (non-prelim): 12888
length of query: 1390
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1282
effective length of database: 5,021,319
effective search space: 6437330958
effective search space used: 6437330958
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.7 bits)
S2: 65 (29.6 bits)
Medicago: description of AC146854.1


