

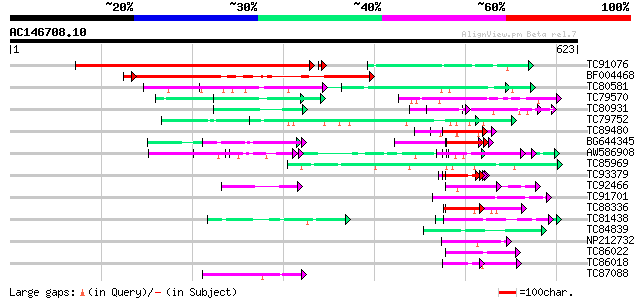
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146708.10 - phase: 0
(623 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC91076 similar to PIR|H96747|H96747 unknown protein T10D10.14 [... 524 e-149
BF004468 similar to PIR|H96747|H96 unknown protein T10D10.14 [im... 293 2e-82
TC80581 similar to GP|19683021|gb|AAL92644.1 hypothetical protei... 70 3e-12
TC79570 similar to PIR|F86427|F86427 auxin response factor 6 (AR... 65 1e-10
TC80931 similar to GP|14190783|gb|AAF65166.2 putative phloem tra... 59 1e-10
TC79752 similar to GP|11994670|dbj|BAB02898. gene_id:K7P8.17~unk... 62 5e-10
TC89480 similar to GP|20260684|gb|AAM13240.1 unknown protein {Ar... 59 6e-09
BG644345 weakly similar to GP|19697333|gb putative protein poten... 59 7e-09
AW586908 weakly similar to GP|2104814|emb| pleiotropic effects o... 59 7e-09
TC85969 similar to GP|21553908|gb|AAM62991.1 unknown {Arabidopsi... 57 2e-08
TC93379 similar to GP|19569994|gb|AAL92295.1 hypothetical protei... 56 4e-08
TC92466 similar to GP|11141605|gb|AAG32022.1 LEUNIG {Arabidopsis... 55 8e-08
TC91701 similar to GP|11141605|gb|AAG32022.1 LEUNIG {Arabidopsis... 54 2e-07
TC88336 similar to PIR|T51477|T51477 glutamine-rich protein - Ar... 52 7e-07
TC81438 similar to GP|20259253|gb|AAM14362.1 unknown protein {Ar... 52 7e-07
TC84839 similar to GP|19386827|dbj|BAB86205. hypothetical protei... 51 1e-06
NP212732 NP212732|AF106929.1|AAD39890.1 putative cell wall protein 49 4e-06
TC86022 homologue to GP|19423939|gb|AAL87312.1 putative RNA heli... 47 2e-05
TC86018 similar to GP|19423939|gb|AAL87312.1 putative RNA helica... 45 8e-05
TC87088 similar to PIR|F86399|F86399 protein F17L21.22 [imported... 44 2e-04
>TC91076 similar to PIR|H96747|H96747 unknown protein T10D10.14 [imported] -
Arabidopsis thaliana, partial (9%)
Length = 834
Score = 524 bits (1350), Expect(2) = e-149
Identities = 263/263 (100%), Positives = 263/263 (100%)
Frame = +3
Query: 73 VPRFRTRMIMAEKPSDGTVALHYGDIDESDFIGAEDHLPTLPNTHFADLLADQFSSQIEH 132
VPRFRTRMIMAEKPSDGTVALHYGDIDESDFIGAEDHLPTLPNTHFADLLADQFSSQIEH
Sbjct: 3 VPRFRTRMIMAEKPSDGTVALHYGDIDESDFIGAEDHLPTLPNTHFADLLADQFSSQIEH 182
Query: 133 DGYVKEDDRIQVRPNLVNLPLGSQSSLPPNEMQQYGEPIPGQSNNEAAKLAGGSNASLNL 192
DGYVKEDDRIQVRPNLVNLPLGSQSSLPPNEMQQYGEPIPGQSNNEAAKLAGGSNASLNL
Sbjct: 183 DGYVKEDDRIQVRPNLVNLPLGSQSSLPPNEMQQYGEPIPGQSNNEAAKLAGGSNASLNL 362
Query: 193 PQSLVANARMLPPGNPQGLQMSQALLSGVSMAQRPQQLDSQQAVLQQQQQQQQLQQNQHS 252
PQSLVANARMLPPGNPQGLQMSQALLSGVSMAQRPQQLDSQQAVLQQQQQQQQLQQNQHS
Sbjct: 363 PQSLVANARMLPPGNPQGLQMSQALLSGVSMAQRPQQLDSQQAVLQQQQQQQQLQQNQHS 542
Query: 253 LLQQQNPQFQRSLLSANQLSHLNGVGQNSNMPLGNHLLNKASPLQIQMLQQQHQQQQLQQ 312
LLQQQNPQFQRSLLSANQLSHLNGVGQNSNMPLGNHLLNKASPLQIQMLQQQHQQQQLQQ
Sbjct: 543 LLQQQNPQFQRSLLSANQLSHLNGVGQNSNMPLGNHLLNKASPLQIQMLQQQHQQQQLQQ 722
Query: 313 NQQPQMQRKMMMGLGAMGMSNFR 335
NQQPQMQRKMMMGLGAMGMSNFR
Sbjct: 723 NQQPQMQRKMMMGLGAMGMSNFR 791
Score = 50.1 bits (118), Expect = 3e-06
Identities = 60/195 (30%), Positives = 79/195 (39%), Gaps = 13/195 (6%)
Frame = +3
Query: 394 QYRPGTMHSNQELLSKLRLVHNREGMSGSPQSSIASMSGARQMHPSSASL-LSQSLSNRT 452
Q RP + N L S+ L N G P ++ A+ S+ASL L QSL
Sbjct: 213 QVRPNLV--NLPLGSQSSLPPNEMQQYGEPIPGQSNNEAAKLAGGSNASLNLPQSLVANA 386
Query: 453 NMSTLQRAMGPMGPPKLMPAMSLYMNRQQQQQHQQSQQQQHQQQLQLQQQQQHIQQQLQQ 512
M G L+ +S+ R QQ QQ+ QQ QQQ QLQQ Q + Q QQ
Sbjct: 387 RMLPPGNPQGLQMSQALLSGVSM-AQRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQ--QQ 557
Query: 513 QLQQQQQQETTSQLQAVVSPPQVGSPSTMGVSS------------LSQQTHQQASPQQMS 560
Q Q+ + +QL + VG S M + + + QQ HQQ QQ
Sbjct: 558 NPQFQRSLLSANQLSHLNG---VGQNSNMPLGNHLLNKASPLQIQMLQQQHQQQQLQQNQ 728
Query: 561 QRTPMSPQQMSSGAI 575
Q M GA+
Sbjct: 729 QPQMQRKMMMGLGAM 773
Score = 22.7 bits (47), Expect(2) = e-149
Identities = 9/9 (100%), Positives = 9/9 (100%)
Frame = +2
Query: 340 GLSPMGNAM 348
GLSPMGNAM
Sbjct: 806 GLSPMGNAM 832
>BF004468 similar to PIR|H96747|H96 unknown protein T10D10.14 [imported] -
Arabidopsis thaliana, partial (4%)
Length = 618
Score = 293 bits (750), Expect(2) = 2e-82
Identities = 173/265 (65%), Positives = 182/265 (68%)
Frame = +2
Query: 136 VKEDDRIQVRPNLVNLPLGSQSSLPPNEMQQYGEPIPGQSNNEAAKLAGGSNASLNLPQS 195
+K++DRIQVRPNLVNLPLGSQSSLPPNEMQQYGEPIPGQSNNEAAKLAGGSNASLNLPQS
Sbjct: 32 LKKNDRIQVRPNLVNLPLGSQSSLPPNEMQQYGEPIPGQSNNEAAKLAGGSNASLNLPQS 211
Query: 196 LVANARMLPPGNPQGLQMSQALLSGVSMAQRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQ 255
LVANARMLPPGNPQGLQMSQALLSGVSMAQRPQQLDS QQ QQQQLQQN
Sbjct: 212 LVANARMLPPGNPQGLQMSQALLSGVSMAQRPQQLDS------QQHQQQQLQQN------ 355
Query: 256 QQNPQFQRSLLSANQLSHLNGVGQNSNMPLGNHLLNKASPLQIQMLQQQHQQQQLQQNQQ 315
Q PQ QR ++ G+G M + N
Sbjct: 356 -QQPQMQRKMMM--------GLGA---MGMSNF--------------------------- 418
Query: 316 PQMQRKMMMGLGAMGMSNFRNSLVGLSPMGNAMGIGAARGIGGTGISAPMTSITGMGNIG 375
R ++GL M GNAMGIGAARGIGGTGISAPMTSITGMGNIG
Sbjct: 419 ----RNSLVGLSPM---------------GNAMGIGAARGIGGTGISAPMTSITGMGNIG 541
Query: 376 QNPMSLGQASNISNSISQQYRPGTM 400
QNPMSLGQASNISNSISQQYRPGTM
Sbjct: 542 QNPMSLGQASNISNSISQQYRPGTM 616
Score = 40.8 bits (94), Expect = 0.002
Identities = 60/228 (26%), Positives = 88/228 (38%), Gaps = 2/228 (0%)
Frame = +2
Query: 394 QYRPGTMHSNQELLSKLRLVHNREGMSGSPQSSIASMSGARQMHPSSASL-LSQSLSNRT 452
Q RP + N L S+ L N G P ++ A+ S+ASL L QSL
Sbjct: 53 QVRPNLV--NLPLGSQSSLPPNEMQQYGEPIPGQSNNEAAKLAGGSNASLNLPQSLVANA 226
Query: 453 NMSTLQRAMGPMGPPKLMPAMSLYMNRQQQQQHQQSQQQQHQQQLQLQQQQQHIQQQLQQ 512
M + P P L + +L Q+ QQ QQHQQQ QLQQ QQ
Sbjct: 227 RM------LPPGNPQGLQMSQALLSGVSMAQRPQQLDSQQHQQQ-QLQQNQQ-------- 361
Query: 513 QLQQQQQQETTSQLQAVVSPPQVGSPSTMGVSSLSQQTHQQASPQQMSQRTPMSPQQMSS 572
PQ+ MG+ ++ + + + +PM M
Sbjct: 362 --------------------PQMQRKMMMGLGAMGMSNFRNS----LVGLSPMG-NAMGI 466
Query: 573 GAIHGMNAGNPEGPASPQLSSQTLGSVSSITNSPMDM-QGVNKSNSVN 619
GA G+ G ++P S +G++ +PM + Q N SNS++
Sbjct: 467 GAARGIGG---TGISAPMTSITGMGNIG---QNPMSLGQASNISNSIS 592
Score = 32.0 bits (71), Expect(2) = 2e-82
Identities = 13/14 (92%), Positives = 14/14 (99%)
Frame = +1
Query: 126 FSSQIEHDGYVKED 139
FSSQIEHDGYVKE+
Sbjct: 1 FSSQIEHDGYVKEE 42
>TC80581 similar to GP|19683021|gb|AAL92644.1 hypothetical protein
{Dictyostelium discoideum}, partial (5%)
Length = 850
Score = 69.7 bits (169), Expect = 3e-12
Identities = 76/271 (28%), Positives = 104/271 (38%), Gaps = 58/271 (21%)
Frame = +1
Query: 365 MTSITGMGNIGQNPMSLGQASNISNSISQQYRPGTMHSNQELLSKLRLVHNREGMSGSPQ 424
++ +T G + + L Q + +N Q G S Q + + +L+ +SGS
Sbjct: 4 LSLMTTNGALYAQQLQLSQQAPFNN----QQLVGLPQSGQPAMIQNQLLKQIPAVSGSAS 171
Query: 425 SSIASMSGARQMHPSSASLLSQSLS-NRTNMSTLQRAMGPMGPPKLMPAM---------- 473
++Q SSA L SL+ N+ + L + PMG P+L +
Sbjct: 172 LLPLQQPQSQQQLASSAQLQQSSLTLNQQQLPQLMQQPKPMGQPQLQQQLQHQQLLQQQL 351
Query: 474 ----------------------SLYMNRQQ------------QQQHQQSQQQQHQQQLQL 499
SL +N+QQ Q QQ QQQ QQQLQL
Sbjct: 352 QLQQQYQRLQQQLASSAQLQQNSLTLNQQQLPLLMQQPKSMGQPLLQQQQQQLLQQQLQL 531
Query: 500 QQQQQHIQQQLQQQLQQQQQQETTSQLQAVVSPPQVGSPSTMGVSSLSQQTH-------- 551
QQQ Q +QQQL Q QQ T +QL P + +MG L QQ
Sbjct: 532 QQQYQRLQQQLASSAQLQQNSSTLNQL-----PQLMQQQKSMGQPLLQQQLQQQQLLLQQ 696
Query: 552 -----QQASPQQMSQRTPMSPQQMSSGAIHG 577
QQ QQ QRT + QQM + I G
Sbjct: 697 QQLVLQQQQQQQQQQRTQLLQQQMQALRIPG 789
Score = 66.6 bits (161), Expect = 3e-11
Identities = 94/345 (27%), Positives = 130/345 (37%), Gaps = 3/345 (0%)
Frame = +1
Query: 209 QGLQMSQ-ALLSGVSMAQRPQQLDSQQAVLQQQQQQQ-QLQQNQHSLLQQQNPQFQRSLL 266
Q LQ+SQ A + + PQ Q A++Q Q +Q SLL Q PQ Q+ L
Sbjct: 40 QQLQLSQQAPFNNQQLVGLPQS--GQPAMIQNQLLKQIPAVSGSASLLPLQQPQSQQQLA 213
Query: 267 SANQLSHLNGVGQNSNMPLGNHLLNKASPLQIQMLQQQHQQQQLQQNQ-QPQMQRKMMMG 325
S+ QL + +P L+ + P+ LQQQ Q QQL Q Q Q Q Q + +
Sbjct: 214 SSAQLQQSSLTLNQQQLP---QLMQQPKPMGQPQLQQQLQHQQLLQQQLQLQQQYQRLQQ 384
Query: 326 LGAMGMSNFRNSLVGLSPMGNAMGIGAARGIGGTGISAPMTSITGMGNIGQNPMSLGQAS 385
A +NSL N Q P+ + Q
Sbjct: 385 QLASSAQLQQNSLTL--------------------------------NQQQLPLLMQQPK 468
Query: 386 NISNSISQQYRPGTMHSNQELLSKLRLVHNREGMSGSPQSSIASMSGARQMHPSSASLLS 445
++ + QQ + Q+LL + Q + Q +S++ L
Sbjct: 469 SMGQPLLQQQQ-------QQLLQQ--------------QLQLQQQYQRLQQQLASSAQLQ 585
Query: 446 QSLSNRTNMSTLQRAMGPMGPPKLMPAMSLYMNRQQQQQHQQSQQQQHQQQLQLQQQQQH 505
Q+ S + L + MG P L QQQ QQQL LQQQQ
Sbjct: 586 QNSSTLNQLPQLMQQQKSMGQPLL-------------------QQQLQQQQLLLQQQQLV 708
Query: 506 IQQQLQQQLQQQQQQETTSQLQAVVSPPQVGSPSTMGVSSLSQQT 550
+QQQ QQQ QQQ+ Q Q+QA+ P G S S + T
Sbjct: 709 LQQQ-QQQQQQQRTQLLQQQMQALRIPGPAGHKSLSLTGSQPEAT 840
Score = 62.8 bits (151), Expect = 4e-10
Identities = 76/259 (29%), Positives = 105/259 (40%), Gaps = 57/259 (22%)
Frame = +1
Query: 148 LVNLPLGSQSSLPPNEMQQYGEPIPG----------QSNNEAAKLAGGSNASLNLPQSLV 197
LV LP Q ++ N++ + + G QS + A A +SL L Q +
Sbjct: 85 LVGLPQSGQPAMIQNQLLKQIPAVSGSASLLPLQQPQSQQQLASSAQLQQSSLTLNQQQL 264
Query: 198 ANARMLPP--GNPQ---GLQMSQALLSGVSMAQRPQQLDSQ------------------- 233
P G PQ LQ Q L + + Q+ Q+L Q
Sbjct: 265 PQLMQQPKPMGQPQLQQQLQHQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSLTLNQQQL 444
Query: 234 -----------QAVLQQQQQQ---QQLQ-QNQHSLLQQQ---NPQFQRSLLSANQLSHLN 275
Q +LQQQQQQ QQLQ Q Q+ LQQQ + Q Q++ + NQL L
Sbjct: 445 PLLMQQPKSMGQPLLQQQQQQLLQQQLQLQQQYQRLQQQLASSAQLQQNSSTLNQLPQL- 621
Query: 276 GVGQNSNMPLGNHLLNKASPLQIQMLQQQH----QQQQLQQNQQPQMQRKMMMGLGAMGM 331
+G LL + Q +LQQQ QQQQ QQ Q+ Q+ ++ M L G
Sbjct: 622 ---MQQQKSMGQPLLQQQLQQQQLLLQQQQLVLQQQQQQQQQQRTQLLQQQMQALRIPGP 792
Query: 332 SNFRN-SLVGLSPMGNAMG 349
+ ++ SL G P A G
Sbjct: 793 AGHKSLSLTGSQPEATAFG 849
>TC79570 similar to PIR|F86427|F86427 auxin response factor 6 (ARF6)
[imported] - Arabidopsis thaliana, partial (15%)
Length = 1143
Score = 64.7 bits (156), Expect = 1e-10
Identities = 70/201 (34%), Positives = 88/201 (42%), Gaps = 22/201 (10%)
Frame = +2
Query: 428 ASMSGARQMHPSS---ASLLS----QSLSNRTNMSTLQRAMGPMGPPKLM---------P 471
A++ R + PS ASLL Q+ SN N + +Q M P P
Sbjct: 317 AALQDMRTLDPSKQHPASLLQFQQPQNFSNG-NAALMQNQMLQQSQPHQAFPKNQESQHP 493
Query: 472 AMSLYMNRQQQQ--QHQQSQQQQHQQQLQLQQQQQHIQQQLQQQLQQQQQQETTSQLQAV 529
+ S +Q QQ QHQ S Q+ Q QQQQQ QQQ QQQ QQQQQQ++ Q Q V
Sbjct: 494 SQSQAQTQQFQQLLQHQHSFTNQNYHMQQQQQQQQQQQQQQQQQHQQQQQQQSQQQ-QQV 670
Query: 530 VSPPQVGSPSTMGVSSLSQQTHQQASPQQMSQRTPMSPQQMSSGAIHGMN----AGNPEG 585
V Q+ + VS++SQ A P Q +Q P Q S H N GNP
Sbjct: 671 VDHQQMSN----AVSTMSQ--FVSAPPSQSTQ-----PMQAISSIGHQQNFSDSNGNPVS 817
Query: 586 PASPQLSSQTLGSVSSITNSP 606
P L S + S + N P
Sbjct: 818 PLHNMLGSFSNDETSHLLNFP 880
Score = 44.3 bits (103), Expect = 1e-04
Identities = 47/168 (27%), Positives = 66/168 (38%), Gaps = 3/168 (1%)
Frame = +2
Query: 161 PNEMQQYGEPIPGQSNNEAAKLAGGSNASLNLPQSLVANARMLPPGNPQGL--QMSQALL 218
P + Q+ +P SN AA + Q+ N P Q Q Q L
Sbjct: 362 PASLLQFQQP-QNFSNGNAALMQNQMLQQSQPHQAFPKNQESQHPSQSQAQTQQFQQLLQ 538
Query: 219 SGVSMAQRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQQQNPQFQRSLLSANQLSH-LNGV 277
S + + QQ QQQQQQQQ QQ QH QQQ Q Q+ ++ Q+S+ ++ +
Sbjct: 539 HQHSFTNQNYHMQQQQ---QQQQQQQQQQQQQHQQQQQQQSQQQQQVVDHQQMSNAVSTM 709
Query: 278 GQNSNMPLGNHLLNKASPLQIQMLQQQHQQQQLQQNQQPQMQRKMMMG 325
Q + P P+Q + HQQ N P M+G
Sbjct: 710 SQFVSAPPS----QSTQPMQ-AISSIGHQQNFSDSNGNPVSPLHNMLG 838
Score = 43.1 bits (100), Expect = 3e-04
Identities = 33/123 (26%), Positives = 50/123 (39%)
Frame = +2
Query: 225 QRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQQQNPQFQRSLLSANQLSHLNGVGQNSNMP 284
Q+PQ + A L Q Q QQ Q +Q Q++ +S Q L +
Sbjct: 383 QQPQNFSNGNAALMQNQMLQQSQPHQAFPKNQESQHPSQSQAQTQQFQQL--------LQ 538
Query: 285 LGNHLLNKASPLQIQMLQQQHQQQQLQQNQQPQMQRKMMMGLGAMGMSNFRNSLVGLSPM 344
+ N+ +Q Q QQQ QQQQ QQ Q Q Q++ + N++ +S
Sbjct: 539 HQHSFTNQNYHMQQQQQQQQQQQQQQQQQHQQQQQQQSQQQQQVVDHQQMSNAVSTMSQF 718
Query: 345 GNA 347
+A
Sbjct: 719 VSA 727
>TC80931 similar to GP|14190783|gb|AAF65166.2 putative phloem transcription
factor M1 {Apium graveolens}, partial (67%)
Length = 1193
Score = 58.5 bits (140), Expect = 7e-09
Identities = 54/156 (34%), Positives = 66/156 (41%), Gaps = 10/156 (6%)
Frame = +2
Query: 440 SASLLSQSLSNRTNMSTLQRAMGPMGPPKLMPAMSLYMNRQQQQQHQQSQQQQHQQQLQL 499
S + L S SNR N + + P + M +QQQQQ Q Q QQ+QQ Q+
Sbjct: 467 SHNCLVSSSSNRCNSNKCNNTSRCNRNSSIFPQLQQQMQQQQQQQ-QLPQLQQNQQLSQI 643
Query: 500 QQQQQHIQQQLQQQLQQQQQQETTSQLQAVVSPPQVGSPSTMGVSSLSQQTHQQASPQQ- 558
QQQ +QQQ QQQL Q QQQ+ + Q PQ L Q HQQ QQ
Sbjct: 644 QQQIPQLQQQ-QQQLPQLQQQQLSQLQQQQQQLPQ-----------LQQLQHQQLPQQQQ 787
Query: 559 -----MSQRTPMSP----QQMSSGAIHGMNAGNPEG 585
M Q P Q +S G + A N G
Sbjct: 788 MVGAGMGQTYVQGPGGRSQMVSQGQVSSQGATNIGG 895
Score = 46.2 bits (108), Expect = 4e-05
Identities = 36/103 (34%), Positives = 41/103 (38%)
Frame = +2
Query: 225 QRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQQQNPQFQRSLLSANQLSHLNGVGQNSNMP 284
Q+ Q QQ L Q QQ QQL Q Q + Q Q Q Q L QLS L
Sbjct: 569 QQQMQQQQQQQQLPQLQQNQQLSQIQQQIPQLQQQQQQLPQLQQQQLSQLQ--------- 721
Query: 285 LGNHLLNKASPLQIQMLQQQHQQQQLQQNQQPQMQRKMMMGLG 327
Q QQ Q QQLQ Q PQ Q+ + G+G
Sbjct: 722 --------------QQQQQLPQLQQLQHQQLPQQQQMVGAGMG 808
Score = 45.1 bits (105), Expect(2) = 1e-10
Identities = 43/110 (39%), Positives = 52/110 (47%), Gaps = 6/110 (5%)
Frame = +2
Query: 498 QLQQQQQHIQQQLQQQLQQQQQQETTSQLQAVV-----SPPQVGSPSTMGVSSLSQQTHQ 552
QLQQQ Q QQQ QQQL Q QQ + SQ+Q + Q+ +S L QQ Q
Sbjct: 563 QLQQQMQ--QQQQQQQLPQLQQNQQLSQIQQQIPQLQQQQQQLPQLQQQQLSQLQQQQQQ 736
Query: 553 QASPQQMSQRTPMSPQQMSSGAIHGMNAGNPEGPAS-PQLSSQTLGSVSS 601
QQ+ Q + QQ GA GM +GP Q+ SQ G VSS
Sbjct: 737 LPQLQQL-QHQQLPQQQQMVGA--GMGQTYVQGPGGRSQMVSQ--GQVSS 871
Score = 39.3 bits (90), Expect(2) = 1e-10
Identities = 21/48 (43%), Positives = 28/48 (57%)
Frame = +1
Query: 459 RAMGPMGPPKLMPAMSLYMNRQQQQQHQQSQQQQHQQQLQLQQQQQHI 506
R +G + P ++ +QQQQQ QQ Q QQHQ Q+Q QQQH+
Sbjct: 424 RLIGMLFPGDMVVFKPQLSGQQQQQQMQQQQMQQHQ---QMQSQQQHL 558
Score = 36.6 bits (83), Expect = 0.030
Identities = 29/107 (27%), Positives = 38/107 (35%)
Frame = +1
Query: 487 QSQQQQHQQQLQLQQQQQHIQQQLQQQLQQQQQQETTSQLQAVVSPPQVGSPSTMGVSSL 546
Q QQ QQQ+Q QQ QQH QQ QQQ T + A + S + ++
Sbjct: 472 QLSGQQQQQQMQQQQMQQH-----QQMQSQQQHLSTIAATDATTTTTATASTAATESAAF 636
Query: 547 SQQTHQQASPQQMSQRTPMSPQQMSSGAIHGMNAGNPEGPASPQLSS 593
T +P + S S A A A+ SS
Sbjct: 637 PDSTADSTTPATAAATPSTSTAAAFSAAATAATASAVTATATSTASS 777
Score = 34.3 bits (77), Expect = 0.15
Identities = 28/80 (35%), Positives = 37/80 (46%), Gaps = 1/80 (1%)
Frame = +2
Query: 205 PGNPQGLQMSQALLSGVSMAQRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQ-QQNPQFQR 263
P Q Q+SQ + Q+ QQL Q Q QQQQ Q Q LQ QQ PQ Q+
Sbjct: 608 PQLQQNQQLSQIQQQIPQLQQQQQQLPQLQQQQLSQLQQQQQQLPQLQQLQHQQLPQQQQ 787
Query: 264 SLLSANQLSHLNGVGQNSNM 283
+ + +++ G G S M
Sbjct: 788 MVGAGMGQTYVQGPGGRSQM 847
Score = 32.3 bits (72), Expect = 0.56
Identities = 19/42 (45%), Positives = 24/42 (56%)
Frame = +1
Query: 217 LLSGVSMAQRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQQQN 258
L G + +PQ QQ QQQ QQQQ+QQ+Q QQQ+
Sbjct: 439 LFPGDMVVFKPQLSGQQQ---QQQMQQQQMQQHQQMQSQQQH 555
Score = 32.0 bits (71), Expect = 0.73
Identities = 15/30 (50%), Positives = 21/30 (70%)
Frame = +1
Query: 299 QMLQQQHQQQQLQQNQQPQMQRKMMMGLGA 328
Q QQQ QQQQ+QQ+QQ Q Q++ + + A
Sbjct: 484 QQQQQQMQQQQMQQHQQMQSQQQHLSTIAA 573
Score = 28.9 bits (63), Expect = 6.2
Identities = 21/59 (35%), Positives = 28/59 (46%)
Frame = +2
Query: 266 LSANQLSHLNGVGQNSNMPLGNHLLNKASPLQIQMLQQQHQQQQLQQNQQPQMQRKMMM 324
LS N L + NSN N+ S + Q+ QQ QQQ QQ Q PQ+Q+ +
Sbjct: 464 LSHNCLVSSSSNRCNSNKCNNTSRCNRNSSIFPQLQQQMQQQQ--QQQQLPQLQQNQQL 634
>TC79752 similar to GP|11994670|dbj|BAB02898. gene_id:K7P8.17~unknown
protein {Arabidopsis thaliana}, partial (2%)
Length = 2137
Score = 62.4 bits (150), Expect = 5e-10
Identities = 87/333 (26%), Positives = 123/333 (36%), Gaps = 40/333 (12%)
Frame = +1
Query: 264 SLLSANQLSHLNGVGQNSNMPLGNHLLNKASPLQIQMLQQQH-----------QQQQLQQ 312
S+L ++ LS N S LGN+L + + P+ ++ +QQ+LQQ
Sbjct: 40 SVLPSSGLSSSNA--PPSGTSLGNNLSSPSGPMAASARDSRYGVPRGVPISADEQQRLQQ 213
Query: 313 NQQPQMQRKMMMG-LGAMGMSNFRNSLVGLSPMGNAMGIGAARGIGGTGI----SAPMTS 367
Q R M + G L G + MG GI + + G S+ M S
Sbjct: 214 YNQLISSRNMQQSSMSVPGSDRGARMLPGANGMGMMGGINRSIAMARPGFHGMTSSSMLS 393
Query: 368 ITGM---GNIGQNPMSLGQASNISN--SISQQYRPGTM--HSNQELLSKLRLVHNREGMS 420
GM +G + GQ +++ RPG H Q ++ +L + +
Sbjct: 394 SGGMLSSSMVGMPGVGAGQGNSMMRPRDTVHMMRPGHNQGHQRQMMVPELPMQVTQGNSQ 573
Query: 421 GSPQSSIASMSGARQMHPSSASLLSQSLSNRTNMSTLQ-RAMGPMGPPKLMPAMSLYMNR 479
G P S S + Q P S ++++S GP P N
Sbjct: 574 GIPAFSGMSSAFNSQTTPPSVQQYPVHAQQQSHLSNPHPHLQGPNHP-----------NN 720
Query: 480 QQQ---QQHQQSQQQQHQQQLQLQQQQ--------QHIQQQLQQQLQQQQQ-----QETT 523
QQQ + ++ Q QQ QQQ LQQQQ H+Q Q Q + QQ Q
Sbjct: 721 QQQAYIRLAKERQLQQQQQQRYLQQQQLSATNALIPHVQAQAQPPISSPQQNSSQAQPQN 900
Query: 524 SQLQAVVSPPQVGSPSTMGVSSLSQQTHQQASP 556
S Q VSP SP T S QQ H P
Sbjct: 901 SSQQVSVSPATPSSPLTPMSSQHQQQKHDLPQP 999
Score = 49.7 bits (117), Expect = 3e-06
Identities = 84/389 (21%), Positives = 137/389 (34%), Gaps = 39/389 (10%)
Frame = +1
Query: 168 GEPIPGQSNNEAAKLAGGSNASLNLPQSLVANARMLPPGNPQGLQMSQALLSGVSMAQRP 227
G P+P + + + G ++S P + P P + GV P
Sbjct: 7 GLPLPNHGSVSSVLPSSGLSSSNAPPSGTSLGNNLSSPSGPMAASARDSRY-GVPRGV-P 180
Query: 228 QQLDSQQAVLQQQQQQQQLQQNQHSLLQQQNPQFQRSLLSANQLSHLNGVGQNSNMPL-G 286
D QQ + Q Q Q S+ + + R L AN + + G+ ++ M G
Sbjct: 181 ISADEQQRLQQYNQLISSRNMQQSSMSVPGSDRGARMLPGANGMGMMGGINRSIAMARPG 360
Query: 287 NHLLNKASPLQIQMLQQQHQQQQL-----QQNQQPQMQRKMMMGLGAMGMSNFRNSLVGL 341
H + +S L + Q N + + + M + R +V
Sbjct: 361 FHGMTSSSMLSSGGMLSSSMVGMPGVGAGQGNSMMRPRDTVHMMRPGHNQGHQRQMMVPE 540
Query: 342 SPM----GNAMGIGAARGIGGTGISAPMTSITGMGNIGQNPMSLGQASNISNSISQQYRP 397
PM GN+ GI A +G+S+ S T ++ Q P+ Q S++SN +
Sbjct: 541 LPMQVTQGNSQGIPAF-----SGMSSAFNSQTTPPSVQQYPVHAQQQSHLSNP--HPHLQ 699
Query: 398 GTMHSNQELLSKLRLVHNREGMSGSPQSSIASMSGAR----------QMHPSSASLLSQS 447
G H N + + +RL R+ Q + + Q P +S S
Sbjct: 700 GPNHPNNQQQAYIRLAKERQLQQQQQQRYLQQQQLSATNALIPHVQAQAQPPISSPQQNS 879
Query: 448 LSNRTNMSTLQRAMGPMGPPKLMPAMSLYMNRQQQQQHQQSQ------------------ 489
+ S+ Q ++ P P + MS ++ QQQ+H Q
Sbjct: 880 SQAQPQNSSQQVSVSPATPSSPLTPMS---SQHQQQKHDLPQPGFSRNPGSSVTSQAVKQ 1050
Query: 490 -QQQHQQQLQLQQQQQHIQQQLQQQLQQQ 517
Q+Q QQ+ Q +QH Q Q QQQ
Sbjct: 1051RQRQAQQRQYQQSARQHPNQPQHAQAQQQ 1137
Score = 38.1 bits (87), Expect = 0.010
Identities = 23/67 (34%), Positives = 40/67 (59%), Gaps = 3/67 (4%)
Frame = +3
Query: 444 LSQSLSNRTNMSTLQRAMGPMGPPKLMPAMSLYMNRQ--QQQQHQQSQQQQH-QQQLQLQ 500
LS +L++ ++ S QR P+ P K + +++Q Q+ +HQQ +Q+QH + + LQ
Sbjct: 1926 LSANLASHSHNSGAQRQHQPL-PLKQQSTLKPNLSQQSCQEPEHQQQEQEQHFPEDVALQ 2102
Query: 501 QQQQHIQ 507
Q QH+Q
Sbjct: 2103 HQPQHVQ 2123
Score = 37.4 bits (85), Expect = 0.017
Identities = 39/163 (23%), Positives = 66/163 (39%)
Frame = +1
Query: 153 LGSQSSLPPNEMQQYGEPIPGQSNNEAAKLAGGSNASLNLPQSLVANARMLPPGNPQGLQ 212
L + ++L P+ Q PI N + A N+S + S + L P + Q Q
Sbjct: 802 LSATNALIPHVQAQAQPPISSPQQNSSQ--AQPQNSSQQVSVSPATPSSPLTPMSSQHQQ 975
Query: 213 MSQALLSGVSMAQRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQQQNPQFQRSLLSANQLS 272
L ++ P + QAV +Q+Q+Q QQ Q+ +Q+P + + Q
Sbjct: 976 QKHDLPQP-GFSRNPGSSVTSQAV---KQRQRQAQQRQYQQSARQHPNQPQHAQAQQQAK 1143
Query: 273 HLNGVGQNSNMPLGNHLLNKASPLQIQMLQQQHQQQQLQQNQQ 315
L G+G+ + + H N P I L Q +++ Q
Sbjct: 1144ILKGIGRGNTL---IHQNNSVDPSHINGLSVAPGTQPVEKGDQ 1263
Score = 29.6 bits (65), Expect = 3.6
Identities = 26/94 (27%), Positives = 42/94 (44%), Gaps = 14/94 (14%)
Frame = +3
Query: 441 ASLLSQSLSNRTNMSTLQRAMGPMGPPKLMPAMSLYM-NRQQQQQHQQ------------ 487
+SL S ++ N +L G +GP +L ++ + N Q+QHQ
Sbjct: 1845 SSLESTAVENSAATESLTVNQG-LGPRQLSANLASHSHNSGAQRQHQPLPLKQQSTLKPN 2021
Query: 488 -SQQQQHQQQLQLQQQQQHIQQQLQQQLQQQQQQ 520
SQQ + + Q Q+Q+QH + + Q Q Q Q
Sbjct: 2022 LSQQSCQEPEHQQQEQEQHFPEDVALQHQPQHVQ 2123
>TC89480 similar to GP|20260684|gb|AAM13240.1 unknown protein {Arabidopsis
thaliana}, partial (43%)
Length = 1068
Score = 58.9 bits (141), Expect = 6e-09
Identities = 33/56 (58%), Positives = 38/56 (66%), Gaps = 6/56 (10%)
Frame = +1
Query: 476 YMNRQQQQQHQQSQ------QQQHQQQLQLQQQQQHIQQQLQQQLQQQQQQETTSQ 525
++++QQQQ QQSQ QQQ QQQL QQQQQ I QQ QQQLQQ QQQ+ Q
Sbjct: 181 FLHQQQQQPFQQSQTTQSQFQQQQQQQLYQQQQQQRILQQQQQQLQQPQQQQNLHQ 348
Score = 45.4 bits (106), Expect = 6e-05
Identities = 30/77 (38%), Positives = 34/77 (43%), Gaps = 5/77 (6%)
Frame = +1
Query: 463 PMGPPKLMPAMSLYMNRQQQQQHQQSQQQQHQQQLQLQQQQQHIQQQLQQQ-----LQQQ 517
P P++ N+ QQ QQ HQQQ Q QQ Q Q Q QQQ QQQ
Sbjct: 97 PNSNPQIQSQSQSQSNQDPNLYLQQQQQFLHQQQQQPFQQSQTTQSQFQQQQQQQLYQQQ 276
Query: 518 QQQETTSQLQAVVSPPQ 534
QQQ Q Q + PQ
Sbjct: 277 QQQRILQQQQQQLQQPQ 327
Score = 42.7 bits (99), Expect = 4e-04
Identities = 33/100 (33%), Positives = 46/100 (46%), Gaps = 10/100 (10%)
Frame = +1
Query: 445 SQSLSNRTNMSTLQRAMGPMGPPKLMP------AMSLYMNRQQQQQHQQSQQ----QQHQ 494
SQS SN+ LQ+ + + P S + +QQQQ +QQ QQ QQ Q
Sbjct: 127 SQSQSNQDPNLYLQQQQQFLHQQQQQPFQQSQTTQSQFQQQQQQQLYQQQQQQRILQQQQ 306
Query: 495 QQLQLQQQQQHIQQQLQQQLQQQQQQETTSQLQAVVSPPQ 534
QQLQ QQQQ++ Q L E +++ +P Q
Sbjct: 307 QQLQQPQQQQNLHQSLASHFHLLNLVENLAEVVEHGNPDQ 426
Score = 41.2 bits (95), Expect = 0.001
Identities = 31/100 (31%), Positives = 40/100 (40%)
Frame = +1
Query: 229 QLDSQQAVLQQQQQQQQLQQNQHSLLQQQNPQFQRSLLSANQLSHLNGVGQNSNMPLGNH 288
Q+ SQ Q LQQ Q L QQQ FQ+S + +Q
Sbjct: 112 QIQSQSQSQSNQDPNLYLQQQQQFLHQQQQQPFQQSQTTQSQFQQ--------------- 246
Query: 289 LLNKASPLQIQMLQQQHQQQQLQQNQQPQMQRKMMMGLGA 328
+ L Q QQ+ QQQ QQ QQPQ Q+ + L +
Sbjct: 247 --QQQQQLYQQQQQQRILQQQQQQLQQPQQQQNLHQSLAS 360
Score = 40.4 bits (93), Expect = 0.002
Identities = 51/184 (27%), Positives = 77/184 (41%), Gaps = 26/184 (14%)
Frame = +1
Query: 164 MQQYGEP------IPGQSNNEAAKLAGGSNASLNLPQSLVANARMLPPGNPQGLQMSQAL 217
M+ Y P IP ++N + S ++ + L + L Q Q SQ
Sbjct: 49 MEHYSSPGGTWTMIPTPNSNPQIQSQSQSQSNQDPNLYLQQQQQFLHQQQQQPFQQSQTT 228
Query: 218 LSGVSMAQRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQQQNPQFQRSLLS----ANQLSH 273
S Q+ Q QQ QQQQQQLQQ Q QQQN +SL S N + +
Sbjct: 229 QSQFQQQQQQQLYQQQQQQRILQQQQQQLQQPQ----QQQN--LHQSLASHFHLLNLVEN 390
Query: 274 LNGVGQNSN---------MPLGNH------LLNKAS-PLQIQMLQQQHQQQQLQQNQQPQ 317
L V ++ N L NH LLN S + + + + Q+++L++++Q
Sbjct: 391 LAEVVEHGNPDQQSDALITELSNHFEKCQQLLNSISASISTKAMTVEGQKKKLEESEQLL 570
Query: 318 MQRK 321
QR+
Sbjct: 571 NQRR 582
Score = 36.6 bits (83), Expect = 0.030
Identities = 49/192 (25%), Positives = 67/192 (34%), Gaps = 25/192 (13%)
Frame = +1
Query: 448 LSNRTNMSTLQRAMGPMGPPKLMPAMSLYMNRQQQQQHQQSQQQQHQQQLQLQQQQQHIQ 507
L R ++ P G ++P N Q Q Q Q L LQQQQQ +
Sbjct: 22 LKRRCKDRAMEHYSSPGGTWTMIPTP----NSNPQIQSQSQSQSNQDPNLYLQQQQQFLH 189
Query: 508 QQLQQQLQQQQQQETTSQLQAVVSPPQVGSPSTMGVSSLSQQTHQQASPQQMSQRTPMSP 567
QQQQ+ Q Q S Q L QQ QQ QQ Q+
Sbjct: 190 ---------QQQQQPFQQSQTTQSQFQ-----QQQQQQLYQQQQQQRILQQQQQQLQQPQ 327
Query: 568 QQMS-----SGAIHGMNA----------GNPEGPASPQLS---------SQTLGSVS-SI 602
QQ + + H +N GNP+ + ++ Q L S+S SI
Sbjct: 328 QQQNLHQSLASHFHLLNLVENLAEVVEHGNPDQQSDALITELSNHFEKCQQLLNSISASI 507
Query: 603 TNSPMDMQGVNK 614
+ M ++G K
Sbjct: 508 STKAMTVEGQKK 543
>BG644345 weakly similar to GP|19697333|gb putative protein potential
transcriptional repressor Not4hp - Mus musculus, partial
(7%)
Length = 635
Score = 58.5 bits (140), Expect = 7e-09
Identities = 40/97 (41%), Positives = 46/97 (47%)
Frame = +1
Query: 424 QSSIASMSGARQMHPSSASLLSQSLSNRTNMSTLQRAMGPMGPPKLMPAMSLYMNRQQQQ 483
Q S+ G +Q P QS +T G + P + QQQQ
Sbjct: 148 QPSLFQTPGQQQASPFQTPGQQQSSPFQTPGQQQASPFQTPGQQQPSPFSQITPFSQQQQ 327
Query: 484 QHQQSQQQQHQQQLQLQQQQQHIQQQLQQQLQQQQQQ 520
Q Q QQQQ QQQ QQQQQ +QQQ Q QLQQQQQQ
Sbjct: 328 QLQFQQQQQQQQQQLQQQQQQQLQQQQQLQLQQQQQQ 438
Score = 54.3 bits (129), Expect = 1e-07
Identities = 30/46 (65%), Positives = 30/46 (65%)
Frame = +1
Query: 481 QQQQHQQSQQQQHQQQLQLQQQQQHIQQQLQQQLQQQQQQETTSQL 526
QQQQ QQ QQ Q QQQ QLQQQQQ QQ QQQL QQ Q QL
Sbjct: 340 QQQQQQQQQQLQQQQQQQLQQQQQLQLQQQQQQLLQQHQSSQQQQL 477
Score = 52.0 bits (123), Expect = 7e-07
Identities = 29/53 (54%), Positives = 32/53 (59%)
Frame = +1
Query: 479 RQQQQQHQQSQQQQHQQQLQLQQQQQHIQQQLQQQLQQQQQQETTSQLQAVVS 531
+QQQ Q QQ QQ Q QQQLQLQQQQQ + QQ Q QQQ T + A S
Sbjct: 358 QQQQLQQQQQQQLQQQQQLQLQQQQQQLLQQHQSSQQQQLYLFTNDKTPATYS 516
Score = 47.0 bits (110), Expect = 2e-05
Identities = 52/171 (30%), Positives = 59/171 (34%), Gaps = 2/171 (1%)
Frame = +1
Query: 152 PLGSQSSLPPNEMQQYGEPI--PGQSNNEAAKLAGGSNASLNLPQSLVANARMLPPGNPQ 209
P Q SL QQ P PGQ + + G AS PG Q
Sbjct: 136 PQAQQPSLFQTPGQQQASPFQTPGQQQSSPFQTPGQQQAS-----------PFQTPGQQQ 282
Query: 210 GLQMSQALLSGVSMAQRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQQQNPQFQRSLLSAN 269
SQ +Q+ QQL QQ QQQQQQQQLQQ Q LQQQ
Sbjct: 283 PSPFSQI----TPFSQQQQQLQFQQ---QQQQQQQQLQQQQQQQLQQQQ----------- 408
Query: 270 QLSHLNGVGQNSNMPLGNHLLNKASPLQIQMLQQQHQQQQLQQNQQPQMQR 320
Q+ QQ QQQ LQQ+Q Q Q+
Sbjct: 409 -----------------------------QLQLQQQQQQLLQQHQSSQQQQ 474
Score = 43.5 bits (101), Expect = 2e-04
Identities = 41/115 (35%), Positives = 51/115 (43%), Gaps = 1/115 (0%)
Frame = +1
Query: 213 MSQALLSGVSMAQRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQQQNPQFQRSLLSANQLS 272
MS+AL ++ PQ +QQ L Q QQQ Q QQ +P FQ Q S
Sbjct: 94 MSEALQ*PMAFTFTPQ---AQQPSLFQTPGQQQASPFQTPGQQQSSP-FQTP--GQQQAS 255
Query: 273 HLNGVGQNSNMPLGNHLLNKASPLQIQMLQQQHQQ-QQLQQNQQPQMQRKMMMGL 326
GQ P Q+Q QQQ QQ QQLQQ QQ Q+Q++ + L
Sbjct: 256 PFQTPGQQQPSPFSQITPFSQQQQQLQFQQQQQQQQQQLQQQQQQQLQQQQQLQL 420
Score = 41.2 bits (95), Expect = 0.001
Identities = 38/104 (36%), Positives = 44/104 (41%), Gaps = 2/104 (1%)
Frame = +1
Query: 458 QRAMGPMGPPKLMPAMSLYMNRQQQQQ--HQQSQQQQHQQQLQLQQQQQHIQQQLQQQLQ 515
Q+ P P A QQQ Q + Q QQQLQ QQQQQ QQQLQ
Sbjct: 208 QQQSSPFQTPGQQQASPFQTPGQQQPSPFSQITPFSQQQQQLQFQQQQQQQQQQLQ---- 375
Query: 516 QQQQQETTSQLQAVVSPPQVGSPSTMGVSSLSQQTHQQASPQQM 559
QQQQ+ Q Q + Q L QQ HQ + QQ+
Sbjct: 376 -QQQQQQLQQQQQLQLQQQ--------QQQLLQQ-HQSSQQQQL 477
Score = 33.9 bits (76), Expect = 0.19
Identities = 28/89 (31%), Positives = 33/89 (36%), Gaps = 7/89 (7%)
Frame = +1
Query: 480 QQQQQHQQSQQQQHQQQLQLQQQQQHIQ-------QQLQQQLQQQQQQETTSQLQAVVSP 532
QQQ Q+ QQ Q QQQ Q QQQLQ QQQQ+ Q
Sbjct: 208 QQQSSPFQTPGQQQASPFQTPGQQQPSPFSQITPFSQQQQQLQFQQQQQQQQQQLQQQQQ 387
Query: 533 PQVGSPSTMGVSSLSQQTHQQASPQQMSQ 561
Q+ + + QQ QQ Q Q
Sbjct: 388 QQLQQQQQLQLQQQQQQLLQQHQSSQQQQ 474
Score = 30.0 bits (66), Expect = 2.8
Identities = 31/107 (28%), Positives = 39/107 (35%)
Frame = +1
Query: 463 PMGPPKLMPAMSLYMNRQQQQQHQQSQQQQHQQQLQLQQQQQHIQQQLQQQLQQQQQQET 522
PM P + + QQ Q+ QQ Q QQQ Q Q QQ +T
Sbjct: 91 PMSEALQ*PMAFTFTPQAQQPSLFQTPGQQQASPFQTPGQQQSSPFQTPGQ-QQASPFQT 267
Query: 523 TSQLQAVVSPPQVGSPSTMGVSSLSQQTHQQASPQQMSQRTPMSPQQ 569
Q Q SP +P + L Q QQ QQ+ Q+ QQ
Sbjct: 268 PGQQQP--SPFSQITPFSQQQQQLQFQQQQQQQQQQLQQQQQQQLQQ 402
>AW586908 weakly similar to GP|2104814|emb| pleiotropic effects on cellular
differentiation and slug behaviour {Dictyostelium
discoideum}, partial (8%)
Length = 668
Score = 58.5 bits (140), Expect = 7e-09
Identities = 83/292 (28%), Positives = 113/292 (38%), Gaps = 5/292 (1%)
Frame = +2
Query: 238 QQQQQQQQLQQNQHSLLQQQNPQFQRSLLSANQLSHLNGVGQNSNMPLGNHLLNKASPLQ 297
QQQQQQQ++Q SLL Q Q+ QL+ G+ L + Q
Sbjct: 26 QQQQQQQKMQ----SLLPMPLDQLQQQQQRQQQLA-------------GSQNLPQPQQQQ 154
Query: 298 IQMLQQQHQQ-QQLQQNQQPQMQRKMMMGLGAMGMSNFRNSLVGLSPMGNAMGIGAARGI 356
Q+ QQ QQ QQ QQ+QQP + M G +G + N V
Sbjct: 155 PQLTQQSPQQPQQQQQHQQP--CQNTTMNNGTIGSNQIPNQCV----------------- 277
Query: 357 GGTGISAPMTSITGMGNIGQNPMSLGQASNISNSISQQYRPGTMHSNQELLSKLRLVHNR 416
Q P++ Q + QQ G+M S Q L S + N
Sbjct: 278 -------------------QQPVTYSQ-------LQQQLLSGSMQSQQNLQSAGK---NG 370
Query: 417 EGMSGSPQSSIASMSGARQMHPSSASLL----SQSLSNRTNMSTLQRAMGPMGPPKLMPA 472
M+ PQ S +Q+ A LL Q+ ++++ LQ++M GP
Sbjct: 371 LMMTSLPQDS----QFQQQIDQQQAGLLQRQQQQNQLQQSSLQLLQQSMLQRGP------ 520
Query: 473 MSLYMNRQQQQQHQQSQQQQHQQQLQLQQQQQHIQQQLQQQLQQQQQQETTS 524
QQ Q SQ Q + QQQQ + Q+LQQQ QQQQQQ+ S
Sbjct: 521 ----------QQPQMSQTV--PQNISDQQQQLQLLQKLQQQQQQQQQQQPLS 640
Score = 57.4 bits (137), Expect = 2e-08
Identities = 58/173 (33%), Positives = 77/173 (43%), Gaps = 9/173 (5%)
Frame = +2
Query: 153 LGSQSSLPPNEMQQYGEPIPGQS-NNEAAKLAGGSNASLNLPQSLVANARMLPPGNPQGL 211
L QS P + QQ+ +P + NN N + P + + L G+ Q
Sbjct: 161 LTQQSPQQPQQQQQHQQPCQNTTMNNGTIGSNQIPNQCVQQPVTYSQLQQQLLSGSMQSQ 340
Query: 212 Q-MSQALLSGVSMAQRPQ------QLDSQQA-VLQQQQQQQQLQQNQHSLLQQQNPQFQR 263
Q + A +G+ M PQ Q+D QQA +LQ+QQQQ QLQQ+ LLQQ QR
Sbjct: 341 QNLQSAGKNGLMMTSLPQDSQFQQQIDQQQAGLLQRQQQQNQLQQSSLQLLQQS--MLQR 514
Query: 264 SLLSANQLSHLNGVGQNSNMPLGNHLLNKASPLQIQMLQQQHQQQQLQQNQQP 316
Q V QN + Q+Q+LQ+ QQQQ QQ QQP
Sbjct: 515 ---GPQQPQMSQTVPQNIS----------DQQQQLQLLQKLQQQQQQQQQQQP 634
Score = 56.2 bits (134), Expect = 4e-08
Identities = 82/285 (28%), Positives = 104/285 (35%), Gaps = 2/285 (0%)
Frame = +2
Query: 242 QQQQLQQNQHSLLQQQNPQFQRSLLSANQLSHLNGVGQNSNMPLGNHLLNKASPLQIQML 301
QQQQ QQ SLL Q Q+ QL+ G+ L + Q Q+
Sbjct: 26 QQQQQQQKMQSLLPMPLDQLQQQQQRQQQLA-------------GSQNLPQPQQQQPQLT 166
Query: 302 QQQHQQ-QQLQQNQQPQMQRKMMMGLGAMGMSNFRNSLVGLSPMGNAMGIGAARGIGGTG 360
QQ QQ QQ QQ+QQP + M G +G + N V
Sbjct: 167 QQSPQQPQQQQQHQQPC--QNTTMNNGTIGSNQIPNQCV--------------------- 277
Query: 361 ISAPMTSITGMGNIGQNPMSLGQASNISNSISQQYRPGTMHSNQELLSKLRLVHNREGMS 420
Q P++ Q + QQ G+M S Q L S + N M+
Sbjct: 278 ---------------QQPVTYSQ-------LQQQLLSGSMQSQQNLQSAGK---NGLMMT 382
Query: 421 GSPQSSIASMSGARQMHPSSASLLS-QSLSNRTNMSTLQRAMGPMGPPKLMPAMSLYMNR 479
PQ S +Q+ A LL Q N+ S+LQ L+ L
Sbjct: 383 SLPQDS----QFQQQIDQQQAGLLQRQQQQNQLQQSSLQ----------LLQQSMLQRGP 520
Query: 480 QQQQQHQQSQQQQHQQQLQLQQQQQHIQQQLQQQLQQQQQQETTS 524
QQ Q Q Q QQ QLQ Q+ QQ QQQ QQQQ T+S
Sbjct: 521 QQPQMSQTVPQNISDQQQQLQLLQK--LQQQQQQQQQQQPLSTSS 649
Score = 46.6 bits (109), Expect = 3e-05
Identities = 40/120 (33%), Positives = 53/120 (43%), Gaps = 17/120 (14%)
Frame = +2
Query: 476 YMNRQQQQQHQQS----------QQQQHQQQL-------QLQQQQQHIQQQLQQQLQQQQ 518
+ +QQQQQ QS QQQQ QQQL Q QQQQ + QQ QQ QQQQ
Sbjct: 20 WSQQQQQQQKMQSLLPMPLDQLQQQQQRQQQLAGSQNLPQPQQQQPQLTQQSPQQPQQQQ 199
Query: 519 QQETTSQLQAVVSPPQVGSPSTMGVSSLSQQTHQQASPQQMSQRTPMSPQQMSSGAIHGM 578
Q + Q ++ +GS T+ Q QQ+ + S Q + S +G+
Sbjct: 200 QHQQPCQ-NTTMNNGTIGSNQIPNQCVQQPVTYSQLQ-QQLLSGSMQSQQNLQSAGKNGL 373
Score = 44.7 bits (104), Expect = 1e-04
Identities = 32/86 (37%), Positives = 38/86 (43%)
Frame = +2
Query: 481 QQQQHQQSQQQQHQQQLQLQQQQQHIQQQLQQQLQQQQQQETTSQLQAVVSPPQVGSPST 540
Q Q QQ QQ LQ QQQQ +QQ Q LQQ Q Q Q + PQ S
Sbjct: 392 QDSQFQQQIDQQQAGLLQRQQQQNQLQQSSLQLLQQSMLQRGPQQPQMSQTVPQNISDQQ 571
Query: 541 MGVSSLSQQTHQQASPQQMSQRTPMS 566
+ L + QQ QQ Q+ P+S
Sbjct: 572 QQLQLLQKLQQQQ---QQQQQQQPLS 640
Score = 44.3 bits (103), Expect = 1e-04
Identities = 41/147 (27%), Positives = 61/147 (40%), Gaps = 26/147 (17%)
Frame = +2
Query: 203 LPPGNPQGLQMSQALLSGVSMAQRPQQLDSQQAVLQQQQQQQQLQQNQ------------ 250
+P Q Q Q L+G +PQQ Q QQ QQQ Q Q
Sbjct: 68 MPLDQLQQQQQRQQQLAGSQNLPQPQQQQPQLTQQSPQQPQQQQQHQQPCQNTTMNNGTI 247
Query: 251 ------HSLLQQ--QNPQFQRSLLSANQLS--HLNGVGQN----SNMPLGNHLLNKASPL 296
+ +QQ Q Q+ LLS + S +L G+N +++P + +
Sbjct: 248 GSNQIPNQCVQQPVTYSQLQQQLLSGSMQSQQNLQSAGKNGLMMTSLPQDSQFQQQIDQQ 427
Query: 297 QIQMLQQQHQQQQLQQNQQPQMQRKMM 323
Q +LQ+Q QQ QLQQ+ +Q+ M+
Sbjct: 428 QAGLLQRQQQQNQLQQSSLQLLQQSML 508
Score = 42.4 bits (98), Expect = 5e-04
Identities = 34/99 (34%), Positives = 43/99 (43%), Gaps = 2/99 (2%)
Frame = +2
Query: 470 MPAMSLYMNRQQQQQHQQSQQ--QQHQQQLQLQQQQQHIQQQLQQQLQQQQQQETTSQLQ 527
MP L +Q+QQQ SQ Q QQQ QL QQ QQ QQQ Q QQ + T+
Sbjct: 68 MPLDQLQQQQQRQQQLAGSQNLPQPQQQQPQLTQQS---PQQPQQQQQHQQPCQNTTMNN 238
Query: 528 AVVSPPQVGSPSTMGVSSLSQQTHQQASPQQMSQRTPMS 566
+ Q+ + + SQ Q S SQ+ S
Sbjct: 239 GTIGSNQIPNQCVQQPVTYSQLQQQLLSGSMQSQQNLQS 355
Score = 42.0 bits (97), Expect = 7e-04
Identities = 41/123 (33%), Positives = 48/123 (38%), Gaps = 1/123 (0%)
Frame = +2
Query: 483 QQHQQSQQQQHQQQLQLQQQQQHIQQQLQQQLQQQQQQETTSQLQAVVSPPQVGSPSTMG 542
QQ S QQ QQQ ++Q QLQQQ Q+QQQ + L PQ
Sbjct: 2 QQPPASWSQQQQQQQKMQSLLPMPLDQLQQQQQRQQQLAGSQNL------PQPQQQQPQL 163
Query: 543 VSSLSQQTHQQASPQQMSQRTPMSPQQMSSGAIHGMNAGNPEGPASPQLSSQTL-GSVSS 601
QQ QQ QQ Q T M+ + S I P QL Q L GS+ S
Sbjct: 164 TQQSPQQPQQQQQHQQPCQNTTMNNGTIGSNQIPNQCVQQP--VTYSQLQQQLLSGSMQS 337
Query: 602 ITN 604
N
Sbjct: 338 QQN 346
Score = 40.0 bits (92), Expect = 0.003
Identities = 51/185 (27%), Positives = 62/185 (32%), Gaps = 40/185 (21%)
Frame = +2
Query: 424 QSSIASMSGARQMHPSSASLLSQSLSNRTNMSTLQRAMGPMGPPKLMPAMSLYMNRQQQQ 483
Q AS S +Q SLL L Q+ +L + +L +QQQ
Sbjct: 2 QQPPASWSQQQQQQQKMQSLLPMPLDQLQQQQQRQQ--------QLAGSQNLPQPQQQQP 157
Query: 484 QHQQSQQQQHQQQLQLQQ----------------------QQQHIQQQLQQQL-----QQ 516
Q Q QQ QQQ Q QQ QQ QLQQQL Q
Sbjct: 158 QLTQQSPQQPQQQQQHQQPCQNTTMNNGTIGSNQIPNQCVQQPVTYSQLQQQLLSGSMQS 337
Query: 517 QQQQETT-------------SQLQAVVSPPQVGSPSTMGVSSLSQQTHQQASPQQMSQRT 563
QQ ++ SQ Q + Q G + QQ+ Q Q M QR
Sbjct: 338 QQNLQSAGKNGLMMTSLPQDSQFQQQIDQQQAGLLQRQQQQNQLQQSSLQLLQQSMLQRG 517
Query: 564 PMSPQ 568
P PQ
Sbjct: 518 PQQPQ 532
Score = 34.3 bits (77), Expect = 0.15
Identities = 28/67 (41%), Positives = 33/67 (48%), Gaps = 2/67 (2%)
Frame = +2
Query: 211 LQMSQALLSGVSMAQR-PQQLDSQQAVLQQ-QQQQQQLQQNQHSLLQQQNPQFQRSLLSA 268
LQ S L SM QR PQQ Q V Q QQQQLQ Q QQQ Q Q+ L ++
Sbjct: 467 LQQSSLQLLQQSMLQRGPQQPQMSQTVPQNISDQQQQLQLLQKLQQQQQQQQQQQPLSTS 646
Query: 269 NQLSHLN 275
+ L+
Sbjct: 647 SHFCSLS 667
Score = 32.3 bits (72), Expect = 0.56
Identities = 20/45 (44%), Positives = 24/45 (52%), Gaps = 1/45 (2%)
Frame = +2
Query: 208 PQGLQMSQALLSGVS-MAQRPQQLDSQQAVLQQQQQQQQLQQNQH 251
PQ QMSQ + +S Q+ Q L Q QQQQQQQ L + H
Sbjct: 518 PQQPQMSQTVPQNISDQQQQLQLLQKLQQQQQQQQQQQPLSTSSH 652
>TC85969 similar to GP|21553908|gb|AAM62991.1 unknown {Arabidopsis thaliana},
partial (31%)
Length = 2756
Score = 57.4 bits (137), Expect = 2e-08
Identities = 84/351 (23%), Positives = 137/351 (38%), Gaps = 49/351 (13%)
Frame = +2
Query: 306 QQQQLQQNQQPQMQ-------RKMMMGLGAMGMSNFRNSLVGLSPMGNAMGIGAARGIGG 358
QQQQLQQ QQ Q +M++G+G +G R G + M + +
Sbjct: 998 QQQQLQQQQQDQKVLGIDEQFAQMVVGVGGIGQ---RLQDEGFAVMSSPPHPPVPTTLAA 1168
Query: 359 TGISAPMTSITGMGNIGQNPMSLGQASNISNSISQQYRPGTM--------HSNQELLSKL 410
G+ P+ S +G+ S + S+ + + P + + ++
Sbjct: 1169 VGV--PIGSAVVVGDYQNRVFSDDERSDHGVPVGYRKPPTPQPQVVQVQPQAQNQAQTQS 1342
Query: 411 RLVHNREGMSGSPQSSIASMSGARQMHP----SSASLLSQSLSNRTNMSTLQRAMGPMGP 466
+ + S +PQ S G + SS S LS ++S + + ++ G
Sbjct: 1343 QAQSQAQTQSQAPQFQQKSSGGGSDLPSPDSVSSDSSLSNAMSRQKPVVYQEQIQIQPGT 1522
Query: 467 PKLMPAMSLYMN------RQQQQQH---------QQSQQQQHQQQLQLQQQQQHIQQQ-- 509
++ + +N R Q QQH QQ + QQ QQQ +LQQQQQ Q Q
Sbjct: 1523 TRVSNPVDPKLNLSDPHSRIQVQQHVQDPGYLLQQQFELQQQQQQFELQQQQQQPQHQPQ 1702
Query: 510 ---------LQQQLQQQQQQETTSQLQAVVSPPQVGSPSTMGVSSLSQQTHQQ---ASPQ 557
QQQLQ QQQ+ + P + P+ V QQ H Q +
Sbjct: 1703 QSQPQHQPQQQQQLQHHQQQQFIHGGHYIHHNPAI--PTYYPVYPSQQQPHHQVYYVPAR 1876
Query: 558 QMSQRTPMSPQQMSSG-AIHGMNAGNPEGPASPQLSSQTLGSVSSITNSPM 607
Q Q +S QQ + G + + + P+ P +P Q + + I ++P+
Sbjct: 1877 QPQQGYNISVQQANMGESATTIPSSRPQNPPNPTTLVQQNAAYNPIRSAPL 2029
Score = 36.2 bits (82), Expect = 0.039
Identities = 34/126 (26%), Positives = 51/126 (39%), Gaps = 4/126 (3%)
Frame = +2
Query: 211 LQMSQALLSGVSMAQRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQQQNPQFQRSLLSANQ 270
+Q+ Q + + Q+ +L QQ + QQQQQQ Q Q PQ Q+ L Q
Sbjct: 1580 IQVQQHVQDPGYLLQQQFELQQQQQQFELQQQQQQPQHQPQQSQPQHQPQQQQQLQHHQQ 1759
Query: 271 LSHLNGVGQNSNMPLGNHL-LNKASPLQIQML---QQQHQQQQLQQNQQPQMQRKMMMGL 326
++G G+++ N A P + QQ H Q +QPQ + +
Sbjct: 1760 QQFIHG---------GHYIHHNPAIPTYYPVYPSQQQPHHQVYYVPARQPQQGYNISVQQ 1912
Query: 327 GAMGMS 332
MG S
Sbjct: 1913 ANMGES 1930
Score = 33.5 bits (75), Expect = 0.25
Identities = 30/96 (31%), Positives = 34/96 (35%)
Frame = +2
Query: 222 SMAQRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQQQNPQFQRSLLSANQLSHLNGVGQNS 281
S Q Q + +LQQQ + QQ QQ QQQ PQ Q
Sbjct: 1574 SRIQVQQHVQDPGYLLQQQFELQQQQQQFELQQQQQQPQHQ------------------- 1696
Query: 282 NMPLGNHLLNKASPLQIQMLQQQHQQQQLQQNQQPQ 317
P Q Q Q QQQQLQ +QQ Q
Sbjct: 1697 -------------PQQSQPQHQPQQQQQLQHHQQQQ 1765
>TC93379 similar to GP|19569994|gb|AAL92295.1 hypothetical protein
{Dictyostelium discoideum}, partial (1%)
Length = 535
Score = 56.2 bits (134), Expect = 4e-08
Identities = 28/42 (66%), Positives = 32/42 (75%)
Frame = +2
Query: 480 QQQQQHQQSQQQQHQQQLQLQQQQQHIQQQLQQQLQQQQQQE 521
QQQQQHQQ QQQQ + +QQQQQ QQ+QQQLQQQQ Q+
Sbjct: 110 QQQQQHQQQQQQQQYMAMMMQQQQQ---QQMQQQLQQQQMQQ 226
Score = 48.1 bits (113), Expect = 1e-05
Identities = 25/42 (59%), Positives = 29/42 (68%)
Frame = +2
Query: 476 YMNRQQQQQHQQSQQQQHQQQLQLQQQQQHIQQQLQQQLQQQ 517
Y +QQQ Q QQ QQQQ+ + QQQQQ +QQQLQQQ QQ
Sbjct: 104 YHQQQQQHQQQQ-QQQQYMAMMMQQQQQQQMQQQLQQQQMQQ 226
Score = 46.6 bits (109), Expect = 3e-05
Identities = 26/51 (50%), Positives = 31/51 (59%), Gaps = 1/51 (1%)
Frame = +2
Query: 476 YMNRQQQQQHQ-QSQQQQHQQQLQLQQQQQHIQQQLQQQLQQQQQQETTSQ 525
Y QQ Q + QQQQHQQQ QQQQQ++ +QQQ QQQ QQ+ Q
Sbjct: 71 YQGMQQMQPNPYHQQQQQHQQQ---QQQQQYMAMMMQQQQQQQMQQQLQQQ 214
Score = 45.1 bits (105), Expect = 8e-05
Identities = 25/56 (44%), Positives = 30/56 (52%)
Frame = +2
Query: 472 AMSLYMNRQQQQQHQQSQQQQHQQQLQLQQQQQHIQQQLQQQLQQQQQQETTSQLQ 527
A + MN Q QQ Q + QQ Q QQQQ QQ + +QQQQQQ+ QLQ
Sbjct: 41 AAAAAMNGGYYQGMQQMQPNPYHQQQQQHQQQQQQQQYMAMMMQQQQQQQMQQQLQ 208
Score = 35.4 bits (80), Expect = 0.066
Identities = 25/64 (39%), Positives = 30/64 (46%), Gaps = 3/64 (4%)
Frame = +2
Query: 198 ANARMLPPGNPQGLQMSQALLSGVSMAQRPQQLDSQQ---AVLQQQQQQQQLQQNQHSLL 254
A A + G QG+Q Q Q QQ QQ ++QQQQQQQ QQ Q +
Sbjct: 41 AAAAAMNGGYYQGMQQMQPNPYHQQQQQHQQQQQQQQYMAMMMQQQQQQQMQQQLQQQQM 220
Query: 255 QQQN 258
QQ N
Sbjct: 221 QQGN 232
Score = 32.7 bits (73), Expect = 0.43
Identities = 23/71 (32%), Positives = 29/71 (40%)
Frame = +2
Query: 205 PGNPQGLQMSQALLSGVSMAQRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQQQNPQFQRS 264
P M+ G+ Q P QQ QQ QQQQQ QQ ++QQQ Q +
Sbjct: 32 PAGAAAAAMNGGYYQGMQQMQ-PNPYHQQQ---QQHQQQQQQQQYMAMMMQQQQQQQMQQ 199
Query: 265 LLSANQLSHLN 275
L Q+ N
Sbjct: 200 QLQQQQMQQGN 232
Score = 32.7 bits (73), Expect = 0.43
Identities = 28/100 (28%), Positives = 35/100 (35%)
Frame = +2
Query: 225 QRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQQQNPQFQRSLLSANQLSHLNGVGQNSNMP 284
Q QQ+ QQQQ QQQ QQ Q+ + Q Q Q+
Sbjct: 74 QGMQQMQPNPYHQQQQQHQQQQQQQQYMAMMMQQQQQQQ--------------------- 190
Query: 285 LGNHLLNKASPLQIQMLQQQHQQQQLQQNQQPQMQRKMMM 324
+QQQ QQQQ+QQ M + MM
Sbjct: 191 ----------------MQQQLQQQQMQQGNMNNMYPQHMM 262
Score = 30.8 bits (68), Expect = 1.6
Identities = 31/91 (34%), Positives = 35/91 (38%), Gaps = 2/91 (2%)
Frame = +2
Query: 498 QLQQQQQHIQQQLQQQLQQQQQQETTSQLQAVVSPPQVGSPSTMGVSSLSQQTHQQASPQ 557
Q QQ Q QQQ QQQQQ+ Q A++ Q QQ QQ Q
Sbjct: 74 QGMQQMQPNPYHQQQQQHQQQQQQ--QQYMAMMMQQQ-----------QQQQMQQQLQQQ 214
Query: 558 QMSQ--RTPMSPQQMSSGAIHGMNAGNPEGP 586
QM Q M PQ M G + N GP
Sbjct: 215 QMQQGNMNNMYPQHMMYGGGRPHPSMNYMGP 307
Score = 30.4 bits (67), Expect = 2.1
Identities = 17/39 (43%), Positives = 20/39 (50%)
Frame = +2
Query: 476 YMNRQQQQQHQQSQQQQHQQQLQLQQQQQHIQQQLQQQL 514
YM QQQ QQ QQQ QQQ Q QQ ++ Q +
Sbjct: 152 YMAMMMQQQQQQQMQQQLQQQ---QMQQGNMNNMYPQHM 259
Score = 30.0 bits (66), Expect = 2.8
Identities = 17/35 (48%), Positives = 21/35 (59%), Gaps = 1/35 (2%)
Frame = +2
Query: 473 MSLYMNRQQQQQ-HQQSQQQQHQQQLQLQQQQQHI 506
M++ M +QQQQQ QQ QQQQ QQ QH+
Sbjct: 155 MAMMMQQQQQQQMQQQLQQQQMQQGNMNNMYPQHM 259
Score = 30.0 bits (66), Expect = 2.8
Identities = 18/53 (33%), Positives = 24/53 (44%), Gaps = 9/53 (16%)
Frame = +2
Query: 290 LNKASPLQIQMLQQQHQQQQ---------LQQNQQPQMQRKMMMGLGAMGMSN 333
+ + P QQQHQQQQ +QQ QQ QMQ+++ G N
Sbjct: 80 MQQMQPNPYHQQQQQHQQQQQQQQYMAMMMQQQQQQQMQQQLQQQQMQQGNMN 238
Score = 28.9 bits (63), Expect = 6.2
Identities = 20/57 (35%), Positives = 27/57 (47%)
Frame = +2
Query: 205 PGNPQGLQMSQALLSGVSMAQRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQQQNPQF 261
P + Q Q Q MA QQ QQ +QQQ QQQQ+QQ + + Q+ +
Sbjct: 101 PYHQQQQQHQQQQQQQQYMAMMMQQQQQQQ--MQQQLQQQQMQQGNMNNMYPQHMMY 265
>TC92466 similar to GP|11141605|gb|AAG32022.1 LEUNIG {Arabidopsis thaliana},
partial (14%)
Length = 632
Score = 55.1 bits (131), Expect = 8e-08
Identities = 39/105 (37%), Positives = 48/105 (45%)
Frame = +1
Query: 479 RQQQQQHQQSQQQQHQQQLQLQQQQQHIQQQLQQQLQQQQQQETTSQLQAVVSPPQVGSP 538
R+QQQQ QQ QQQ QQ Q QQQQ Q+QQ L Q+ Q+ Q Q P
Sbjct: 367 REQQQQQQQQQQQPQPQQSQHAQQQQQQHMQMQQLLMQRHAQQQQQQQQHQQQP------ 528
Query: 539 STMGVSSLSQQTHQQASPQQMSQRTPMSPQQMSSGAIHGMNAGNP 583
Q QQ PQQ R + +G+ +G+ AGNP
Sbjct: 529 ---------QSQPQQPQPQQQQNR---DRTHLLNGSANGL-AGNP 624
Score = 47.0 bits (110), Expect = 2e-05
Identities = 31/66 (46%), Positives = 37/66 (55%), Gaps = 4/66 (6%)
Frame = +1
Query: 479 RQQQQQHQQSQQ---QQHQQQLQLQQQ-QQHIQQQLQQQLQQQQQQETTSQLQAVVSPPQ 534
+QQQQQH Q QQ Q+H QQ Q QQQ QQ Q Q QQ QQQQ + L +
Sbjct: 433 QQQQQQHMQMQQLLMQRHAQQQQQQQQHQQQPQSQPQQPQPQQQQNRDRTHLLNGSANGL 612
Query: 535 VGSPST 540
G+P+T
Sbjct: 613 AGNPAT 630
Score = 45.8 bits (107), Expect = 5e-05
Identities = 33/89 (37%), Positives = 38/89 (42%)
Frame = +1
Query: 233 QQAVLQQQQQQQQLQQNQHSLLQQQNPQFQRSLLSANQLSHLNGVGQNSNMPLGNHLLNK 292
QQ QQQQQQ Q QQ+QH+ QQQ + LL
Sbjct: 373 QQQQQQQQQQQPQPQQSQHAQQQQQQHMQMQQLLMQRH---------------------- 486
Query: 293 ASPLQIQMLQQQHQQQQLQQNQQPQMQRK 321
Q Q QQQHQQQ Q QQPQ Q++
Sbjct: 487 ---AQQQQQQQQHQQQPQSQPQQPQPQQQ 564
Score = 36.2 bits (82), Expect = 0.039
Identities = 24/60 (40%), Positives = 31/60 (51%), Gaps = 4/60 (6%)
Frame = +1
Query: 209 QGLQMSQALLSGVSMAQRPQQLDSQQAVLQ----QQQQQQQLQQNQHSLLQQQNPQFQRS 264
Q Q Q S + Q+ Q + QQ ++Q QQQQQQQ QQ S QQ PQ Q++
Sbjct: 391 QQQQQPQPQQSQHAQQQQQQHMQMQQLLMQRHAQQQQQQQQHQQQPQSQPQQPQPQQQQN 570
Score = 35.0 bits (79), Expect = 0.086
Identities = 22/48 (45%), Positives = 26/48 (53%), Gaps = 2/48 (4%)
Frame = +1
Query: 225 QRPQQLDSQQAVLQQQQ--QQQQLQQNQHSLLQQQNPQFQRSLLSANQ 270
Q+PQ SQ A QQQQ Q QQL +H+ QQQ Q Q+ S Q
Sbjct: 400 QQPQPQQSQHAQQQQQQHMQMQQLLMQRHAQQQQQQQQHQQQPQSQPQ 543
Score = 30.0 bits (66), Expect = 2.8
Identities = 25/70 (35%), Positives = 30/70 (42%), Gaps = 2/70 (2%)
Frame = +1
Query: 205 PGNPQGLQMSQALLSGVSMAQRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQQQNPQFQRS 264
P Q Q + M Q Q +QQ QQQ QQQ Q Q QQQ + +
Sbjct: 406 PQPQQSQHAQQQQQQHMQMQQLLMQRHAQQQQQQQQHQQQPQSQPQQPQPQQQQNRDRTH 585
Query: 265 LL--SANQLS 272
LL SAN L+
Sbjct: 586 LLNGSANGLA 615
>TC91701 similar to GP|11141605|gb|AAG32022.1 LEUNIG {Arabidopsis thaliana},
partial (18%)
Length = 1174
Score = 53.9 bits (128), Expect = 2e-07
Identities = 44/133 (33%), Positives = 57/133 (42%), Gaps = 2/133 (1%)
Frame = +1
Query: 465 GPPKLMPAMSLYMNRQQQQQHQQSQQQQHQQQLQLQQQQQHIQQQLQQQLQQQQQQETTS 524
GPP + M + Q QQ QQQQHQQQ + QQQ +QQ Q Q +
Sbjct: 40 GPPFPRGDTDMLMKLKLAQLQQQQQQQQHQQQSSINAQQQQLQQHALSNQQSQTSNHSMH 219
Query: 525 QLQAVVSPPQVGSPSTMGVSSLSQQTHQQASPQQM--SQRTPMSPQQMSSGAIHGMNAGN 582
Q V GS + G S S + + Q S QM ++ P+S SSG + N
Sbjct: 220 QQDKVGGGG--GSVTMDGSMSNSYRGNDQVSKNQMGRKRKQPVS----SSGPANSSGTAN 381
Query: 583 PEGPASPQLSSQT 595
GP SP + T
Sbjct: 382 TAGP-SPSSAPST 417
Score = 33.5 bits (75), Expect = 0.25
Identities = 33/109 (30%), Positives = 47/109 (42%)
Frame = +1
Query: 235 AVLQQQQQQQQLQQNQHSLLQQQNPQFQRSLLSANQLSHLNGVGQNSNMPLGNHLLNKAS 294
A LQQQQQQQQ QQ QQQ Q Q+ LS NQ S + + +G +
Sbjct: 91 AQLQQQQQQQQHQQQSSINAQQQ--QLQQHALS-NQQSQTSNHSMHQQDKVGGG--GGSV 255
Query: 295 PLQIQMLQQQHQQQQLQQNQQPQMQRKMMMGLGAMGMSNFRNSLVGLSP 343
+ M Q+ +NQ + +++ + G S N+ G SP
Sbjct: 256 TMDGSMSNSYRGNDQVSKNQMGRKRKQPVSSSGPANSSGTANT-AGPSP 399
Score = 30.4 bits (67), Expect = 2.1
Identities = 29/93 (31%), Positives = 41/93 (43%), Gaps = 6/93 (6%)
Frame = +1
Query: 206 GNPQGLQMSQALLSGVSMAQRPQQLDSQQAVLQQQ--QQQQQLQQNQHSLLQQQNPQFQR 263
G P + +L + +AQ QQ QQ Q QQQQLQ QH+L QQ+
Sbjct: 37 GGPPFPRGDTDMLMKLKLAQLQQQQQQQQHQQQSSINAQQQQLQ--QHALSNQQSQTSNH 210
Query: 264 SLLSANQLSHLNGV----GQNSNMPLGNHLLNK 292
S+ +++ G G SN GN ++K
Sbjct: 211 SMHQQDKVGGGGGSVTMDGSMSNSYRGNDQVSK 309
Score = 30.0 bits (66), Expect = 2.8
Identities = 15/29 (51%), Positives = 18/29 (61%)
Frame = +1
Query: 292 KASPLQIQMLQQQHQQQQLQQNQQPQMQR 320
K + LQ Q QQQHQQQ QQ Q+Q+
Sbjct: 85 KLAQLQQQQQQQQHQQQSSINAQQQQLQQ 171
>TC88336 similar to PIR|T51477|T51477 glutamine-rich protein - Arabidopsis
thaliana, partial (67%)
Length = 1297
Score = 52.0 bits (123), Expect = 7e-07
Identities = 38/101 (37%), Positives = 52/101 (50%), Gaps = 13/101 (12%)
Frame = +2
Query: 480 QQQQQHQQSQQQQHQQQLQLQQQQQH--IQQQLQQQLQQQQQQETTSQLQA-------VV 530
QQQQQ QQ QQQ Q+ Q QQQQQH + QQLQ+Q QQQQQ S+ + +
Sbjct: 197 QQQQQQQQQQQQLLIQEQQRQQQQQHFMLLQQLQKQQQQQQQAAAISRFPSNIDAHLRPI 376
Query: 531 SPP----QVGSPSTMGVSSLSQQTHQQASPQQMSQRTPMSP 567
PP Q +P+ L+ + ++ QQ Q+ + P
Sbjct: 377 RPPLNLQQNPNPNPNSNPILNLHQNPNSNHQQQQQQKMIRP 499
Score = 47.4 bits (111), Expect = 2e-05
Identities = 26/48 (54%), Positives = 32/48 (66%), Gaps = 3/48 (6%)
Frame = +2
Query: 477 MNRQQQQQHQQSQQQQHQQQLQLQQQQQHIQQQ---LQQQLQQQQQQE 521
M + QQ QQQQ QQQL +Q+QQ+ QQQ L QQLQ+QQQQ+
Sbjct: 173 MEEAKALHQQQQQQQQQQQQLLIQEQQRQQQQQHFMLLQQLQKQQQQQ 316
Score = 37.7 bits (86), Expect = 0.013
Identities = 36/107 (33%), Positives = 46/107 (42%), Gaps = 7/107 (6%)
Frame = +2
Query: 225 QRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQQQNPQFQRSLLSANQLSHLN------GVG 278
Q+ QQL Q+ QQQQQ L Q QQQ S +N +HL +
Sbjct: 218 QQQQQLLIQEQQRQQQQQHFMLLQQLQKQQQQQQQAAAISRFPSNIDAHLRPIRPPLNLQ 397
Query: 279 QNSN-MPLGNHLLNKASPLQIQMLQQQHQQQQLQQNQQPQMQRKMMM 324
QN N P N +LN Q HQQQQ Q+ +P Q ++ M
Sbjct: 398 QNPNPNPNSNPILNLH-----QNPNSNHQQQQQQKMIRPGNQMELQM 523
Score = 36.6 bits (83), Expect = 0.030
Identities = 23/61 (37%), Positives = 33/61 (53%), Gaps = 7/61 (11%)
Frame = +2
Query: 478 NRQQQQQHQQSQQQQHQQQLQLQQQQQHIQQQLQQQ-------LQQQQQQETTSQLQAVV 530
+R +++ + Q QQQ Q QQQQ IQ+Q +QQ LQQ Q+Q+ Q A +
Sbjct: 155 SRFRKKMEEAKALHQQQQQQQQQQQQLLIQEQQRQQQQQHFMLLQQLQKQQQQQQQAAAI 334
Query: 531 S 531
S
Sbjct: 335 S 337
>TC81438 similar to GP|20259253|gb|AAM14362.1 unknown protein {Arabidopsis
thaliana}, partial (21%)
Length = 1334
Score = 52.0 bits (123), Expect = 7e-07
Identities = 45/139 (32%), Positives = 55/139 (39%)
Frame = +2
Query: 468 KLMPAMSLYMNRQQQQQHQQSQQQQHQQQLQLQQQQQHIQQQLQQQLQQQQQQETTSQLQ 527
K MPAMS + + QQHQ+ QQQQ QLQQ + QQ Q QQQ+ QL
Sbjct: 35 KQMPAMSGPASPLRLQQHQR-QQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLH 211
Query: 528 AVVSPPQVGSPSTMGVSSLSQQTHQQASPQQMSQRTPMSPQQMSSGAIHGMNAGNPEGPA 587
PQ QQ Q PQQ SQ QQ +H P G
Sbjct: 212 QQQPSPQ------------QQQQQQLLQPQQQSQLQASVHQQQH---LHSPRVAGPTGQK 346
Query: 588 SPQLSSQTLGSVSSITNSP 606
S L+ + +S +P
Sbjct: 347 SISLTGSQPDATASGATTP 403
Score = 49.3 bits (116), Expect = 4e-06
Identities = 45/122 (36%), Positives = 53/122 (42%), Gaps = 1/122 (0%)
Frame = +2
Query: 477 MNRQQQQQHQQSQQQQHQQQLQLQQQQQHIQQQLQQQLQQQQQQETTSQLQAVVSPPQ-V 535
M QQQ Q QQQ+ Q QL QQQ QQQ QQQL Q QQQ SQLQA V Q +
Sbjct: 140 MTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQQQQQLLQPQQQ---SQLQASVHQQQHL 310
Query: 536 GSPSTMGVSSLSQQTHQQASPQQMSQRTPMSPQQMSSGAIHGMNAGNPEGPASPQLSSQT 595
SP G + Q+ T P +SGA G + ++Q
Sbjct: 311 HSPRVAGPTG-----------QKSISLTGSQPDATASGA---TTPGGSSSQGTEAATNQV 448
Query: 596 LG 597
LG
Sbjct: 449 LG 454
Score = 44.3 bits (103), Expect = 1e-04
Identities = 49/165 (29%), Positives = 60/165 (35%), Gaps = 8/165 (4%)
Frame = +2
Query: 218 LSGVSMAQRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQQQNPQFQRSLLSANQLSHLNGV 277
+SG + R QQ QQ QQ QLQQN +L QQQ QF + S
Sbjct: 50 MSGPASPLRLQQHQRQQ---QQLASSGQLQQNSMTLNQQQLSQFMQQQKS---------- 190
Query: 278 GQNSNMPLGNHLLNKASPLQIQMLQQQHQQQQLQQNQQPQMQRKMMMG--------LGAM 329
+G L++ P QQQ QQQ LQ QQ Q+Q + G
Sbjct: 191 -------MGQPQLHQQQPSP----QQQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAGPT 337
Query: 330 GMSNFRNSLVGLSPMGNAMGIGAARGIGGTGISAPMTSITGMGNI 374
G + SL G P A G G G A + G I
Sbjct: 338 GQKSI--SLTGSQPDATASGATTPGGSSSQGTEAATNQVLGKRKI 466
Score = 39.7 bits (91), Expect = 0.003
Identities = 39/122 (31%), Positives = 53/122 (42%), Gaps = 3/122 (2%)
Frame = +2
Query: 400 MHSNQELLSKLRLVHNREGMSGSPQSSIASMSGARQMHP--SSASLLSQSLS-NRTNMST 456
MH++ L++ + + MSG P S + RQ SS L S++ N+ +S
Sbjct: 2 MHNS---LTQSQWLKQMPAMSG-PASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQ 169
Query: 457 LQRAMGPMGPPKLMPAMSLYMNRQQQQQHQQSQQQQHQQQLQLQQQQQHIQQQLQQQLQQ 516
+ MG P+L HQQ Q QQQ QL Q QQ Q QLQ + Q
Sbjct: 170 FMQQQKSMGQPQL---------------HQQQPSPQQQQQQQLLQPQQ--QSQLQASVHQ 298
Query: 517 QQ 518
QQ
Sbjct: 299 QQ 304
Score = 35.4 bits (80), Expect = 0.066
Identities = 25/51 (49%), Positives = 27/51 (52%)
Frame = +2
Query: 212 QMSQALLSGVSMAQRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQQQNPQFQ 262
Q+SQ + SM Q QL QQ QQQQQQQ LQ Q S LQ Q Q
Sbjct: 158 QLSQFMQQQKSMGQ--PQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQQQ 304
Score = 30.4 bits (67), Expect = 2.1
Identities = 24/66 (36%), Positives = 31/66 (46%), Gaps = 1/66 (1%)
Frame = +2
Query: 211 LQMSQALLSGVSMAQRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQ-QQNPQFQRSLLSAN 269
LQ + L+ ++Q QQ S QQQ QQ Q LLQ QQ Q Q S+ +
Sbjct: 125 LQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQQQQQLLQPQQQSQLQASV---H 295
Query: 270 QLSHLN 275
Q HL+
Sbjct: 296 QQQHLH 313
>TC84839 similar to GP|19386827|dbj|BAB86205. hypothetical protein~similar
to Arabidopsis thaliana chromosome 5 T6I14_10, partial
(20%)
Length = 819
Score = 51.2 bits (121), Expect = 1e-06
Identities = 44/137 (32%), Positives = 52/137 (37%), Gaps = 2/137 (1%)
Frame = +1
Query: 455 STLQRAMGPMGPPKLMPAMSLYMNRQQQQQHQQSQQQQHQQQLQLQQQQQHIQQQLQQQL 514
+ LQ + + P L+ M + Q QQHQQ QQQQ QQQ QQ QQH QQQ
Sbjct: 97 TALQSQILILPTP*LVTMMYNDPHNPQHQQHQQHQQQQQQQQ---QQYQQHQQQQQPPHF 267
Query: 515 QQQQQQETTSQLQAVVSPPQVGSPSTMGVSSLSQQTHQQASPQQMSQRTPMSPQQMSSGA 574
QQQQ+ S H+ P Q PM Q +S
Sbjct: 268 HHQQQQQQ---------------------SGPGPDFHRGPPPPMPQQPPPMMRQPSASST 384
Query: 575 IHGMN--AGNPEGPASP 589
G G P GP P
Sbjct: 385 NLGSEFLPGGPPGPPGP 435
Score = 33.5 bits (75), Expect = 0.25
Identities = 19/39 (48%), Positives = 19/39 (48%)
Frame = +1
Query: 223 MAQRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQQQNPQF 261
M P QQ QQQQQQQ QQ Q QQQ P F
Sbjct: 151 MYNDPHNPQHQQHQQHQQQQQQQQQQYQQHQQQQQPPHF 267
Score = 32.0 bits (71), Expect = 0.73
Identities = 24/62 (38%), Positives = 25/62 (39%), Gaps = 3/62 (4%)
Frame = +1
Query: 205 PGNPQGLQMSQALLSGVSMAQRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQQQN---PQF 261
P NPQ Q Q QQ QQ QQ QQQQQ H QQQ+ P F
Sbjct: 163 PHNPQHQQHQQ----------HQQQQQQQQQQYQQHQQQQQPPHFHHQQQQQQSGPGPDF 312
Query: 262 QR 263
R
Sbjct: 313 HR 318
>NP212732 NP212732|AF106929.1|AAD39890.1 putative cell wall protein
Length = 576
Score = 49.3 bits (116), Expect = 4e-06
Identities = 27/82 (32%), Positives = 45/82 (53%), Gaps = 5/82 (6%)
Frame = +2
Query: 475 LYMNRQQQQQHQQSQQQQHQQQLQLQQQQQHIQQQLQQ-----QLQQQQQQETTSQLQAV 529
L+ ++Q Q+QHQ+ Q+QHQ Q Q++QH Q+ ++ Q+Q+Q++Q + Q
Sbjct: 302 LHQHQQHQEQHQEQHQEQHQHQEDHHQKEQHHHQRKEEQPHLLQVQEQERQLLQQERQEA 481
Query: 530 VSPPQVGSPSTMGVSSLSQQTH 551
P Q P T V +L + H
Sbjct: 482 QLPKQQQEPET--VLNLKSEFH 541
Score = 36.2 bits (82), Expect = 0.039
Identities = 23/96 (23%), Positives = 43/96 (43%), Gaps = 4/96 (4%)
Frame = +2
Query: 478 NRQQQQQHQQSQQQQ----HQQQLQLQQQQQHIQQQLQQQLQQQQQQETTSQLQAVVSPP 533
++ Q+ HQQ+Q Q+ HQ++ L+++ ++ QL++++Q Q + + Q Q +
Sbjct: 89 HQHQEHHHQQNQHQEHHHHHQKERMLKERIHQMEMQLKERMQHQMELKEKMQHQMELKER 268
Query: 534 QVGSPSTMGVSSLSQQTHQQASPQQMSQRTPMSPQQ 569
P + L HQQ Q Q Q
Sbjct: 269 M---PLKKELREL*LHQHQQHQEQHQEQHQEQHQHQ 367
>TC86022 homologue to GP|19423939|gb|AAL87312.1 putative RNA helicase
{Arabidopsis thaliana}, partial (89%)
Length = 1982
Score = 47.0 bits (110), Expect = 2e-05
Identities = 32/83 (38%), Positives = 42/83 (50%), Gaps = 1/83 (1%)
Frame = +2
Query: 480 QQQQQHQQSQQQQHQQQLQLQQQ-QQHIQQQLQQQLQQQQQQETTSQLQAVVSPPQVGSP 538
QQ+ HQ QQQQ+ Q+ +Q Q QQH Q Q Q Q Q QQQQ Q Q + Q+G
Sbjct: 182 QQRPHHQYQQQQQYVQRHMMQNQHQQHYQNQQQNQQQNQQQQ----QQQQWLRRNQLGGG 349
Query: 539 STMGVSSLSQQTHQQASPQQMSQ 561
+ V ++T Q + SQ
Sbjct: 350 TDTNVVEEVEKTVQSEANDSSSQ 418
Score = 38.1 bits (87), Expect = 0.010
Identities = 17/33 (51%), Positives = 23/33 (69%)
Frame = +2
Query: 476 YMNRQQQQQHQQSQQQQHQQQLQLQQQQQHIQQ 508
+M + Q QQH Q+QQQ QQ Q QQQQQ +++
Sbjct: 233 HMMQNQHQQHYQNQQQNQQQNQQQQQQQQWLRR 331
Score = 31.6 bits (70), Expect = 0.95
Identities = 23/81 (28%), Positives = 28/81 (34%)
Frame = +2
Query: 247 QQNQHSLLQQQNPQFQRSLLSANQLSHLNGVGQNSNMPLGNHLLNKASPLQIQMLQQQHQ 306
QQ H QQQ QR ++ H QQQ+Q
Sbjct: 182 QQRPHHQYQQQQQYVQRHMMQNQHQQHYQN-------------------------QQQNQ 286
Query: 307 QQQLQQNQQPQMQRKMMMGLG 327
QQ QQ QQ Q R+ +G G
Sbjct: 287 QQNQQQQQQQQWLRRNQLGGG 349
Score = 31.2 bits (69), Expect = 1.2
Identities = 18/62 (29%), Positives = 33/62 (53%), Gaps = 4/62 (6%)
Frame = +2
Query: 476 YMNRQQQQQHQQSQQQQHQQQLQLQQ----QQQHIQQQLQQQLQQQQQQETTSQLQAVVS 531
Y N+QQ QQ Q QQQQ QQ L+ Q ++ +++++ +Q + ++ +A +
Sbjct: 263 YQNQQQNQQ-QNQQQQQQQQWLRRNQLGGGTDTNVVEEVEKTVQSEANDSSSQDWKARLK 439
Query: 532 PP 533
P
Sbjct: 440 LP 445
Score = 29.3 bits (64), Expect = 4.7
Identities = 27/73 (36%), Positives = 34/73 (45%), Gaps = 8/73 (10%)
Frame = +2
Query: 219 SGVSMAQRP------QQLDSQQAVLQQQQQQ--QQLQQNQHSLLQQQNPQFQRSLLSANQ 270
S QRP QQ Q+ ++Q Q QQ Q QQNQ Q Q Q Q+ L NQ
Sbjct: 167 SNTGFQQRPHHQYQQQQQYVQRHMMQNQHQQHYQNQQQNQQ---QNQQQQQQQQWLRRNQ 337
Query: 271 LSHLNGVGQNSNM 283
L G G ++N+
Sbjct: 338 L----GGGTDTNV 364
>TC86018 similar to GP|19423939|gb|AAL87312.1 putative RNA helicase
{Arabidopsis thaliana}, partial (12%)
Length = 885
Score = 45.1 bits (105), Expect = 8e-05
Identities = 24/46 (52%), Positives = 27/46 (58%)
Frame = +1
Query: 476 YMNRQQQQQHQQSQQQQHQQQLQLQQQQQHIQQQLQQQLQQQQQQE 521
Y +Q Q+H Q QHQQ Q QQ Q QQ QQQ QQQQQQ+
Sbjct: 571 YQQQQYVQRHLMQNQNQHQQHYQHHQQNQ---QQYQQQNQQQQQQQ 699
Score = 42.4 bits (98), Expect = 5e-04
Identities = 30/99 (30%), Positives = 45/99 (45%), Gaps = 12/99 (12%)
Frame = +1
Query: 476 YMNRQ-QQQQHQQSQQQQHQQQLQLQQQQQHIQQQLQQQLQQQQ-----------QQETT 523
Y+ R Q Q+Q Q QH QQ Q Q QQQ+ QQQ QQ L++ Q + E T
Sbjct: 586 YVQRHLMQNQNQHQQHYQHHQQNQQQYQQQNQQQQQQQWLRRNQLGGGTDTNVVEEVEKT 765
Query: 524 SQLQAVVSPPQVGSPSTMGVSSLSQQTHQQASPQQMSQR 562
Q + + ++G + + +T + + MS R
Sbjct: 766 VQSETMTLVHKIGRKTKASTADTRYRTEDVTATKGMSLR 882
Score = 39.7 bits (91), Expect = 0.003
Identities = 26/49 (53%), Positives = 29/49 (59%), Gaps = 4/49 (8%)
Frame = +1
Query: 481 QQQQHQQSQQ--QQHQQQLQLQQQQQHIQ--QQLQQQLQQQQQQETTSQ 525
QQ+ H Q QQ Q+H Q Q Q QQH Q QQ QQQ QQQ QQ+ Q
Sbjct: 556 QQRPHYQQQQYVQRHLMQNQ-NQHQQHYQHHQQNQQQYQQQNQQQQQQQ 699
Score = 37.4 bits (85), Expect = 0.017
Identities = 22/46 (47%), Positives = 26/46 (55%), Gaps = 1/46 (2%)
Frame = +1
Query: 483 QQHQQSQQQQHQQQLQLQQQQQHIQQ-QLQQQLQQQQQQETTSQLQ 527
QQ QQQQ+ Q+ +Q Q QH Q Q QQ QQQ QQ+ Q Q
Sbjct: 556 QQRPHYQQQQYVQRHLMQNQNQHQQHYQHHQQNQQQYQQQNQQQQQ 693
Score = 32.3 bits (72), Expect = 0.56
Identities = 26/66 (39%), Positives = 33/66 (49%), Gaps = 7/66 (10%)
Frame = +1
Query: 225 QRP---QQLDSQQAVLQQQQQQQQ----LQQNQHSLLQQQNPQFQRSLLSANQLSHLNGV 277
QRP QQ Q+ ++Q Q Q QQ QQNQ QQ Q Q+ L NQL G
Sbjct: 559 QRPHYQQQQYVQRHLMQNQNQHQQHYQHHQQNQQQYQQQNQQQQQQQWLRRNQL----GG 726
Query: 278 GQNSNM 283
G ++N+
Sbjct: 727 GTDTNV 744
Score = 29.6 bits (65), Expect = 3.6
Identities = 27/95 (28%), Positives = 35/95 (36%)
Frame = +1
Query: 231 DSQQAVLQQQQQQQQLQQNQHSLLQQQNPQFQRSLLSANQLSHLNGVGQNSNMPLGNHLL 290
++ A QQ+ QQ Q Q L+Q QN Q H QN
Sbjct: 535 NNANAGFQQRPHYQQQQYVQRHLMQNQNQHQQ----------HYQHHQQN---------- 654
Query: 291 NKASPLQIQMLQQQHQQQQLQQNQQPQMQRKMMMG 325
QQQ+QQQ QQ QQ ++R + G
Sbjct: 655 -----------QQQYQQQNQQQQQQQWLRRNQLGG 726
>TC87088 similar to PIR|F86399|F86399 protein F17L21.22 [imported] -
Arabidopsis thaliana, partial (4%)
Length = 2997
Score = 43.5 bits (101), Expect = 2e-04
Identities = 35/126 (27%), Positives = 63/126 (49%), Gaps = 12/126 (9%)
Frame = +3
Query: 213 MSQALLSGVSMAQRPQQLDSQQAVLQQQQQQQQLQQNQHSLLQQQNPQ--FQRSLLSANQ 270
M Q L Q+ ++L QQA QQQQ ++LQQ Q LQQQ + Q++ +S++
Sbjct: 345 MLQKLQKERLQQQQAERLQQQQAERLQQQQAERLQQQQAERLQQQQAERLQQQTNISSHF 524
Query: 271 LSHLNG----------VGQNSNMPLGNHLLNKASPLQIQMLQQQHQQQQLQQNQQPQMQR 320
++LNG Q++ + + N + + ++ + Q QQ+QL+ QQ +
Sbjct: 525 PAYLNGSDLDRFPGFSPSQSNKSGIQQMMQNPGTDFE-RLFELQAQQRQLEIQQQDMHHQ 701
Query: 321 KMMMGL 326
+++ L
Sbjct: 702 QLLQQL 719
Score = 40.0 bits (92), Expect = 0.003
Identities = 38/140 (27%), Positives = 52/140 (37%), Gaps = 14/140 (10%)
Frame = +3
Query: 401 HSNQELLSKLRLVHNREGMSGSPQSSIASMSGARQMHPSSASLLSQSLSNRTNMSTLQRA 460
H + L K RL + Q+ A ++ A L Q + R T +
Sbjct: 339 HLMLQKLQKERLQQQQAERLQQQQAERLQQQQAERLQQQQAERLQQQQAERLQQQTNISS 518
Query: 461 MGPMGP--------PKLMPAMSLYMNRQQQQQHQQS------QQQQHQQQLQLQQQQQHI 506
P P P+ S QQ Q+ + + Q Q+QL++QQQ H
Sbjct: 519 HFPAYLNGSDLDRFPGFSPSQSNKSGIQQMMQNPGTDFERLFELQAQQRQLEIQQQDMHH 698
Query: 507 QQQLQQQLQQQQQQETTSQL 526
QQ LQQ Q QQQ QL
Sbjct: 699 QQLLQQLKLQPQQQSQVQQL 758
Score = 40.0 bits (92), Expect = 0.003
Identities = 24/51 (47%), Positives = 33/51 (64%)
Frame = +3
Query: 475 LYMNRQQQQQHQQSQQQQHQQQLQLQQQQQHIQQQLQQQLQQQQQQETTSQ 525
L R QQQQ ++ QQQQ ++LQ QQQ + +QQQ ++LQQQQ + Q
Sbjct: 357 LQKERLQQQQAERLQQQQ-AERLQ-QQQAERLQQQQAERLQQQQAERLQQQ 503
Score = 37.0 bits (84), Expect = 0.023
Identities = 21/44 (47%), Positives = 26/44 (58%), Gaps = 1/44 (2%)
Frame = +3
Query: 479 RQQQQQHQQSQQQQHQQQLQLQ-QQQQHIQQQLQQQLQQQQQQE 521
RQ + Q Q QQ QQL+LQ QQQ +QQ L +QL QQ +
Sbjct: 663 RQLEIQQQDMHHQQLLQQLKLQPQQQSQVQQLLLEQLMHQQMSD 794
Score = 36.2 bits (82), Expect = 0.039
Identities = 21/45 (46%), Positives = 25/45 (54%)
Frame = +3
Query: 484 QHQQSQQQQHQQQLQLQQQQQHIQQQLQQQLQQQQQQETTSQLQA 528
Q Q ++ Q QQ +LQQQQ QQ Q + QQQQ E Q QA
Sbjct: 351 QKLQKERLQQQQAERLQQQQAERLQQQQAERLQQQQAERLQQQQA 485
Score = 35.8 bits (81), Expect = 0.050
Identities = 21/50 (42%), Positives = 32/50 (64%), Gaps = 4/50 (8%)
Frame = +3
Query: 482 QQQHQQSQQQQHQQQLQLQQQQQHIQQQLQQQLQQQQ----QQETTSQLQ 527
Q+ ++ QQQ ++LQ QQQ + +QQQ ++LQQQQ QQ+ +LQ
Sbjct: 351 QKLQKERLQQQQAERLQ-QQQAERLQQQQAERLQQQQAERLQQQQAERLQ 497
Score = 34.3 bits (77), Expect = 0.15
Identities = 83/366 (22%), Positives = 137/366 (36%), Gaps = 38/366 (10%)
Frame = +3
Query: 231 DSQQAVLQQQQQQQQLQQNQHSLLQQQNPQFQRSLLSANQLSHLNGVGQNSNMPLGNHLL 290
D ++ Q+ Q+++LQQ Q LQQQ Q L Q L
Sbjct: 327 DVADHLMLQKLQKERLQQQQAERLQQQ----QAERLQQQQAERL---------------- 446
Query: 291 NKASPLQIQMLQQQHQQQQLQQNQQPQMQRKMMMGLGAMGMSNFRNSLVGLSPMGNAMGI 350
QQ Q ++LQQ Q ++Q++ + S+F L G S +
Sbjct: 447 ------------QQQQAERLQQQQAERLQQQTNIS------SHFPAYLNG-SDLDRF--- 560
Query: 351 GAARGIGGTGISAPMTSITGMGNIGQNP------MSLGQASNISNSISQQYRPGTMHSNQ 404
G S ++ +G+ + QNP + QA I QQ MH +Q
Sbjct: 561 --------PGFSPSQSNKSGIQQMMQNPGTDFERLFELQAQQRQLEIQQQ----DMH-HQ 701
Query: 405 ELLSKLRLVHNREG------MSGSPQSSIASMSGARQMHPSSASLLSQSLSNRTNMSTLQ 458
+LL +L+L ++ + ++ + + H S L + R + LQ
Sbjct: 702 QLLQQLKLQPQQQSQVQQLLLEQLMHQQMSDPNFGQSKHDPSRDNLLDQVQLRRYLHDLQ 881
Query: 459 RAMGPMGPPKLMPAMSLY------MNRQQQQQHQQSQ---QQQH------QQQLQLQQQQ 503
+ +G L P+M + +N Q +Q S+ Q +H QQL+ QQ Q
Sbjct: 882 QNPHSLG--HLDPSMEQFIQANIGLNAAQGRQADLSELLLQARHGNILPSDQQLRFQQDQ 1055
Query: 504 ---QHIQQQLQQQLQQQQQQ--------ETTSQLQAVVSPPQVGSPSTMGVSSLSQQTHQ 552
Q + L+QQL ++ T QL S Q+G + VS + +Q
Sbjct: 1056LQAQQLSMALKQQLGLDGERHFGRSRPINETGQLVRNPSNHQLGHSAGFNVSEIHKQQQM 1235
Query: 553 QASPQQ 558
A+ ++
Sbjct: 1236LAAQEE 1253
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.310 0.124 0.338
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,559,472
Number of Sequences: 36976
Number of extensions: 213753
Number of successful extensions: 5972
Number of sequences better than 10.0: 223
Number of HSP's better than 10.0 without gapping: 1941
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3465
length of query: 623
length of database: 9,014,727
effective HSP length: 102
effective length of query: 521
effective length of database: 5,243,175
effective search space: 2731694175
effective search space used: 2731694175
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146708.10


